Comparative Transcriptome Analysis of Purple and Green Non-Heading Chinese Cabbage and Function Analyses of BcTT8 Gene
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials
2.2. Determination of Total Anthocyanin Content
2.3. Transcriptome Analysis of Green and Purple NHCC
2.4. Gene Function Annotation and Differential Expressed Gene Identification
2.5. Expression of Anthocyanin-Related Pathway Genes in Green and Purple NHCC
2.6. Sequence Analysis of BcTT8
2.7. Subcellular Localization Assays and Analysis of Phylogenetic Tree
2.8. Silencing of BcTT8 through VIGS System
2.9. Overexpression of BcTT8 in Arabidopsis
2.10. Statistical Analysis
3. Results
3.1. Samples Expression Pattern and Differentially Expressed Genes Clustering
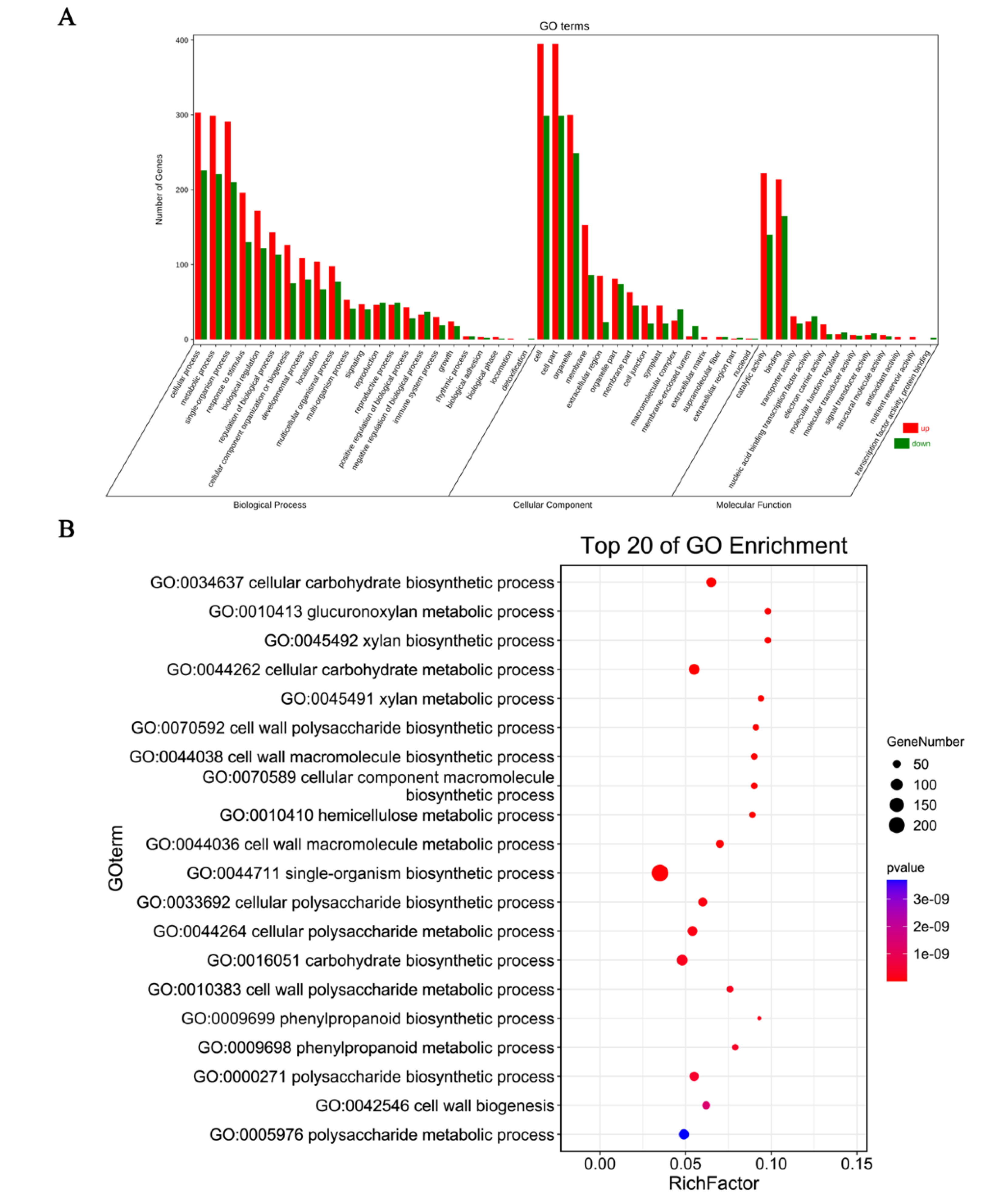
3.2. Differentially Expressed Genes GO Enrichment
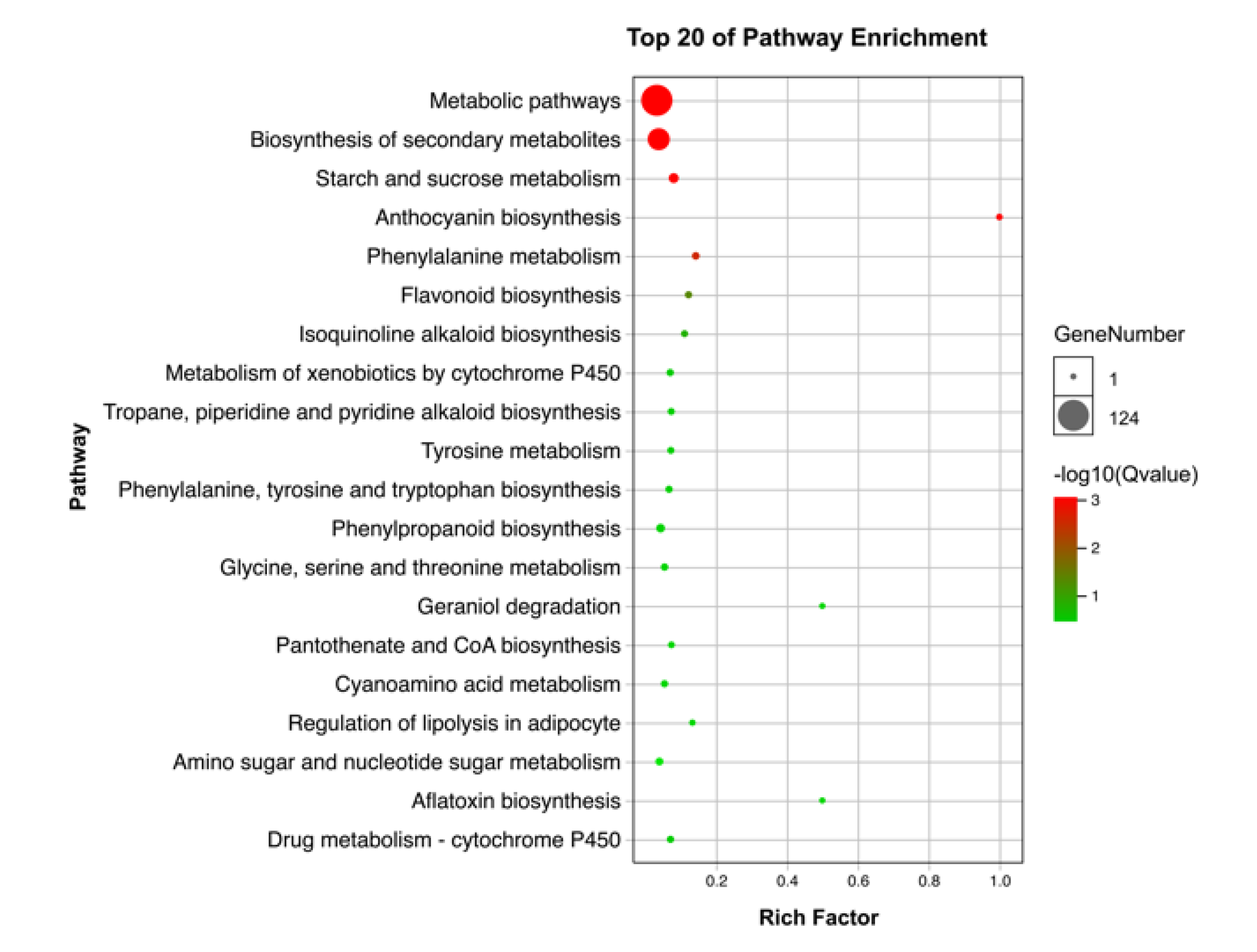
3.3. Differentially Expressed Genes KEGG Enrichment
3.4. Verification of Transcriptome Result by RT-qPCR
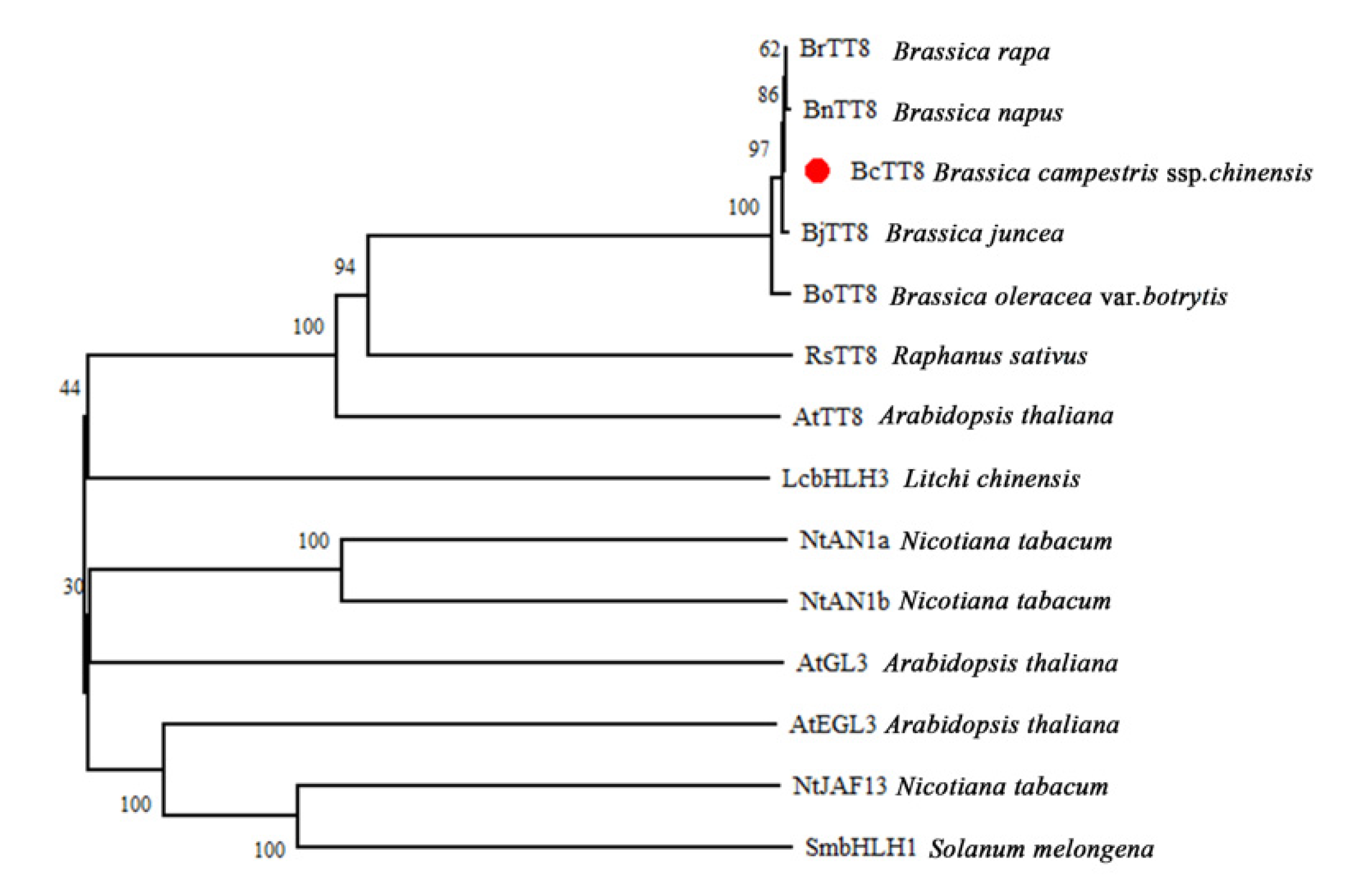
3.5. Characterization and Phylogenetic Analysis of BcTT8
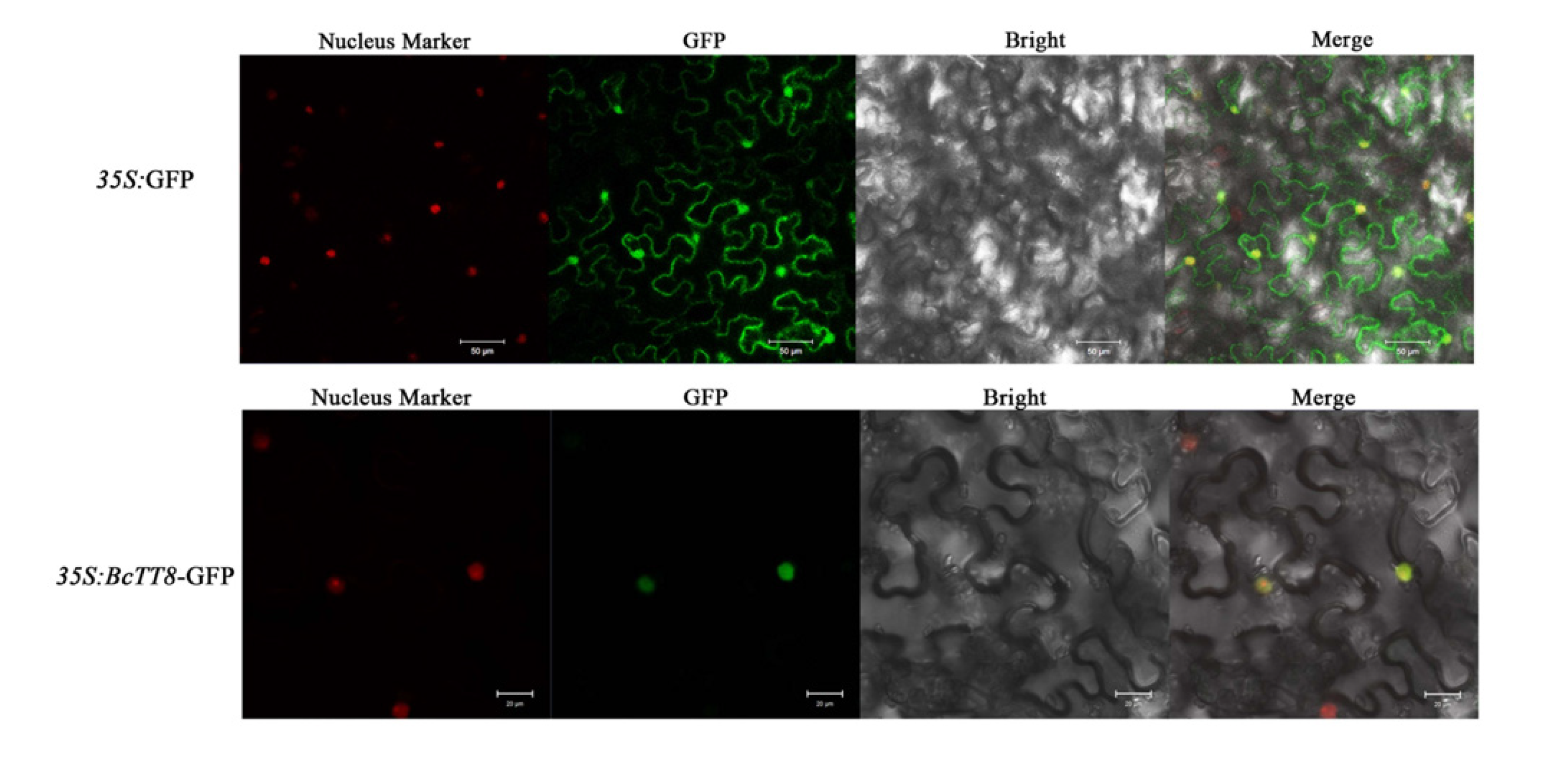
3.6. Subcellular Localization of BcTT8
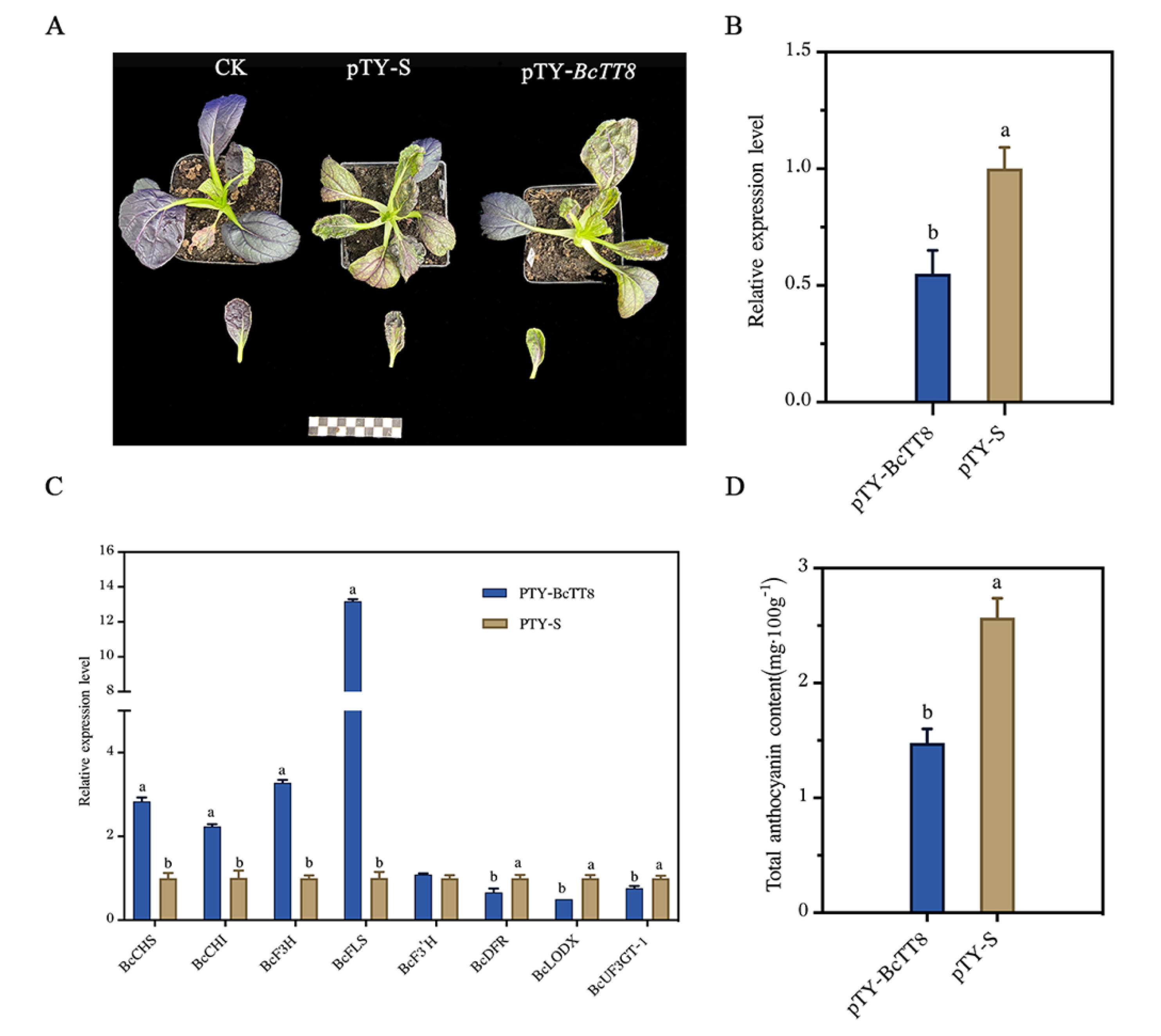
3.7. Expression Analysis of Structural Genes after Silencing of BcTT8
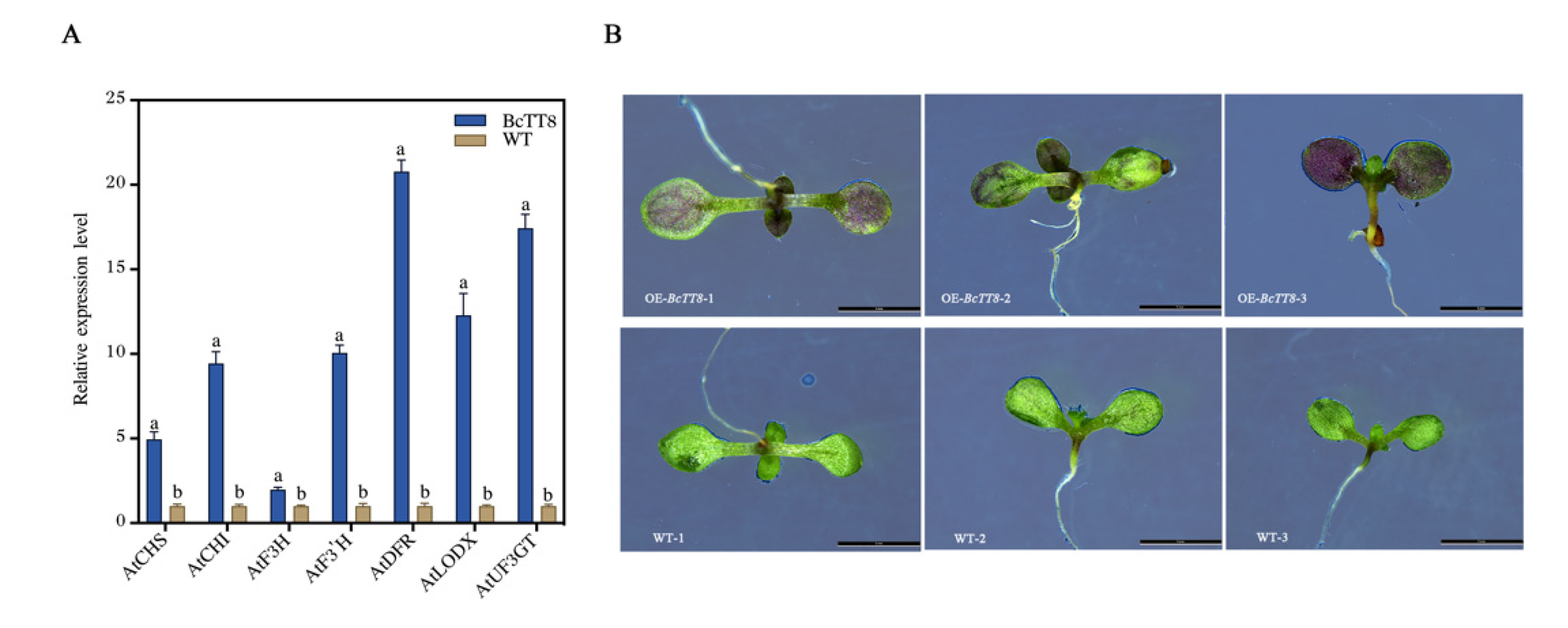
3.8. Heterologous Expression Analysis of BcTT8 in Arabidopsis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Davies, K.M.; Albert, N.W.; Schwinn, K.E. From landing lights to mimicry: The molecular regulation of flower colouration and mechanisms for pigmentation patterning. Funct. Plant Biol. 2012, 39, 619–638. [Google Scholar] [CrossRef] [PubMed]
- Mattioli, R.; Francioso, A.; Mosca, L.; Silva, P. Anthocyanins: A comprehensive review of their chemical properties and health effects on cardiovascular and neurodegenerative diseases. Molecules 2020, 25, 17. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Ren, Y.; Zhao, W.; Li, R.; Zhang, L. Low temperature promotes anthocyanin biosynthesis and related gene expression in the seedlings of purple head Chinese cabbage (Brassica rapa L.). Genes 2020, 11, 81. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, Y.; Teng, S.; Yin, H.; Zhang, S.; Tuo, X.; Tran, L.S.P. Transcriptome analysis reveals roles of anthocyanin-and jasmonic acid-biosynthetic pathways in rapeseed in response to high light stress. Int. J. Mol. Sci. 2021, 22, 13027. [Google Scholar] [CrossRef]
- Guo, R.; Yuan, G.; Wang, Q. Sucrose enhances the accumulation of anthocyanins and glucosinolates in broccoli sprouts. Food Chem. 2011, 129, 1080–1087. [Google Scholar] [CrossRef] [PubMed]
- Dębski, H.; Szwed, M.; Wiczkowski, W.; Szawara-Nowak, D.; Bączek, N.; Horbowicz, M. UV-B radiation increases anthocyanin levels in cotyledons and inhibits the growth of common buckwheat seedlings. Acta Biol. Hung. 2016, 67, 403–411. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakabayashi, R.; Yonekura-Sakakibara, K.; Urano, K.; Suzuki, M.; Yamada, Y.; Nishizawa, T.; Matsuda, F.; Kojima, M.; Sakakibara, H.; Shinozaki, K.; et al. Enhancement of oxidative and drought tolerance in Arabidopsis by overaccumulation of antioxidant flavonoids. Plant J. 2014, 77, 367–379. [Google Scholar] [CrossRef]
- Martens, S.; Preuß, A.; Matern, U. Multifunctional flavonoid dioxygenases: Flavonol and anthocyanin biosynthesis in Arabidopsis thaliana L. Phytochemistry 2010, 71, 1040–1049. [Google Scholar] [CrossRef]
- An, J.P.; Zhang, X.W.; Liu, Y.J.; Wang, X.F.; You, C.X.; Hao, Y.J. ABI5 regulates ABA-induced anthocyanin biosynthesis by modulating the MYB1-bHLH3 complex in apple. J. Exp. Bot. 2021, 72, 1460–1472. [Google Scholar] [CrossRef]
- Sun, C.; Deng, L.; Du, M.; Zhao, J.; Chen, Q.; Huang, T.; Jiang, H.; Li, C.B.; Li, C. A transcriptional network promotes anthocyanin biosynthesis in tomato flesh. Mol. Plant. 2020, 13, 42–58. [Google Scholar] [CrossRef]
- Biswas, T.; Mathur, A.; Gupta, V.; Luqman, S.; Mathur, A.K. Elicitation and phenylalanine precursor feeding based modulation of in vitro anthocyanin production, enzyme activity and gene expression in an Indian ginseng congener-panax sikkimensis ban. Ind. Crops Prod. 2020, 145, 111986. [Google Scholar] [CrossRef]
- Ramsay, N.A.; Glover, B.J. MYB–bHLH–WD40 protein complex and the evolution of cellular diversity. Trends Plant Sci. 2005, 10, 63–70. [Google Scholar] [CrossRef] [PubMed]
- Lipsick, J.S. One billion years of Myb. Oncogene 1996, 13, 223–235. [Google Scholar] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Wang, X.C.; Wu, J.; Guan, M.L.; Zhao, C.H.; Geng, P.; Zhao, Q. Arabidopsis MYB4 plays dual roles in flavonoid biosynthesis. Plant J. 2020, 101, 637–652. [Google Scholar] [CrossRef]
- Jia, N.; Wang, J.J.; Liu, J.; Jiang, J.; Sun, J.; Yan, P.; Sun, Y.; Wan, P.; Ye, W.; Fan, B. DcTT8, a bHLH transcription factor, regulates anthocyanin biosynthesis in Dendrobium candidum. Plant Physiol. Biochem. 2021, 162, 603–612. [Google Scholar] [CrossRef]
- Qi, Y.; Zhou, L.; Han, L.; Zou, H.; Miao, K.; Wang, Y. PsbHLH1, a novel transcription factor involved in regulating anthocyanin biosynthesis in tree peony (Paeonia suffruticosa). Plant Physiol. Biochem. 2021, 154, 396–408. [Google Scholar] [CrossRef]
- Deng, J.; Li, J.J.; Su, M.Y.; Lin, Z.Y.; Chen, L.; Yang, P.F. A bHLH gene NnTT8 of Nelumbo nucifera regulates anthocyanin biosynthesis. Plant Physiol. Biochem. 2021, 158, 518–523. [Google Scholar] [CrossRef]
- Xu, W.J.; Grain, D.; Bobet, S.; le Gourrierec, J.; Thevenin, J.; Kelemen, Z.; Lepiniec, L.; Dubos, C. Complexity and robustness of the flavonoid transcriptional regulatory network revealed by comprehensive analyses of MYB-bHLH-WDR complexes and their targets in Arabidopsis seed. New Phytol. 2014, 202, 132–144. [Google Scholar] [CrossRef]
- Li, P.; Chen, B.; Zhang, G.; Chen, L.; Dong, Q.; Wen, J.; Mysore, K.S.; Zhao, J. Regulation of anthocyanin and proanthocyanidin biosynthesis by Medicago truncatula bHLH transcription factor MtTT8. New Phytol. 2016, 210, 905–921. [Google Scholar] [CrossRef]
- Lim, S.H.; Kim, D.H.; Kim, J.K.; Lee, J.Y.; Ha, S.H. A radish basic helix-loop-helix transcription factor, RsTT8 acts a positive regulator for anthocyanin biosynthesis. Front. Plant Sci. 2017, 8, 1917. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Zhang, J.; Xiang, Y.; Xiang, L.; Liu, Y.; He, X.; Zhou, X.; Liu, X.; Huang, Z. Extracts of Tsai Tai (Brassica chinensis): Enhanced antioxidant activity and anti-aging effects both in vitro and in Caenorhabditis elegans. Food Funct. 2016, 7, 943–952. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Ding, Q.; Hou, X.L.; You, X. Analysis of flavonoid metabolites in watercress (Nasturtium officinale R. Br.) and the non-heading Chinese cabbage (Brassica rapa ssp. chinensis cv. Aijiaohuang) using UHPLC-ESI-MS/MS. Molecules 2021, 26, 5825. [Google Scholar] [CrossRef] [PubMed]
- Abbaoui, B.; Lucas, C.R.; Riedl, K.M.; Clinton, S.K.; Mortazavi, A. Cruciferous vegetables, isothiocyanates, and bladder cancer prevention. Mol. Nutr. Food Res. 2018, 62, 1800079. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chen, G.; Dong, T.; Pan, Y.; Zhao, Z.; Tian, S.; Hu, Z. Anthocyanin accumulation and transcriptional regulation of anthocyanin biosynthesis in purple bok choy (Brassica rapa var. chinensis). J. Agric. Food Chem. 2014, 62, 12366–12376. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, B.; Wu, T.; Yang, Y.; Fan, L.; Wen, M.; Sui, J. Transcriptomic profiling of two Pak Choi varieties with contrasting anthocyanin contents provides an insight into structural and regulatory genes in anthocyanin biosynthetic pathway. BMC Genom. 2017, 18, 288. [Google Scholar] [CrossRef] [Green Version]
- Guo, N.; Cheng, F.; Wu, J.; Liu, B.; Zheng, S.; Liang, J.; Wang, X. Anthocyanin biosynthetic genes in Brassica rapa. BMC Genom. 2014, 15, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.; Durst, R.W.; Wrolstad, R.E. Determination of total monomeric anthocyanin pigment content of fruit juices, beverages, natural colorants, and wines by the pH differential method: Collaborative study. J. AOAC Int. 2005, 88, 1269–1278. [Google Scholar] [CrossRef] [Green Version]
- Li, Y.; Liu, G.F.; Ma, L.M.; Liu, T.K.; Zhang, C.W.; Xiao, D.; Zheng, H.K.; Chen, F.; Hou, X.L. A chromosome-level reference genome of non-heading Chinese cabbage [Brassica campestris (syn. Brassica rapa) ssp. chinensis]. Hortic. Res. 2020, 7, 212. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Yang, X.D.; Wang, Q.; Gao, L.W.; Yang, Y.; Xiao, D.; Liu, T.K.; Li, Y.; Hou, X.L.; Zhang, C.W. Efficient virus-induced gene silencing in Brassica rapa using a turnip yellow mosaic virus vector. Biol. Plant. 2018, 62, 826–834. [Google Scholar] [CrossRef]
- Zhang, X.; Henriques, R.; Lin, S.S.; Niu, Q.W.; Chua, N.H. Agrobacterium-mediated transformation of Arabidopsis thaliana using the floral dip method. Nat. Protoc. 2006, 1, 641–646. [Google Scholar] [CrossRef]
- Stein, O.; Granot, D. An overview of sucrose synthases in plants. Front. Plant Sci. 2019, 10, 95. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rameneni, J.J.; Choi, S.R.; Chhapekar, S.S.; Kim, M.S.; Singh, S.; Yi, S.Y.; Oh, S.H.; Kim, H.; Lee, C.Y.; Oh, M.H.; et al. Red Chinese cabbage transcriptome analysis reveals structural genes and multiple transcription factors regulating reddish purple color. Int. J. Mol. Sci. 2020, 21, 2901. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Butelli, E.; Martin, C. Engineering anthocyanin biosynthesis in plants. Curr. Opin. Plant Biol. 2014, 19, 81–90. [Google Scholar] [CrossRef]
- Huang, G.; Zeng, Y.; Wei, L.; Yao, Y.; Dai, J.; Liu, G.; Gui, Z. Comparative transcriptome analysis of mulberry reveals anthocyanin biosynthesis mechanisms in black (Morus atropurpurea Roxb.) and white (Morus alba L.) fruit genotypes. BMC Plant Biol. 2020, 20, 1–12. [Google Scholar] [CrossRef]
- Li, J.; An, Y.; Wang, L. Transcriptomic analysis of Ficus carica peels with a focus on the key genes for anthocyanin biosynthesis. Int. J. Mol. Sci. 2020, 21, 1245. [Google Scholar] [CrossRef] [Green Version]
- Lim, S.H.; Kim, D.H.; Jung, J.A.; Lee, J.Y. Alternative splicing of the basic helix-loop-helix transcription factor gene CmbHLH2 affects anthocyanin biosynthesis in ray florets of chrysanthemum (Chrysanthemum morifolium). Front. Plant Sci. 2021, 12, 1116. [Google Scholar] [CrossRef]
- Li, Y.; Shan, X.; Gao, R.; Yang, S.; Wang, S.; Gao, X.; Wang, L. Two IIIf clade-bHLHs from Freesia hybrida play divergent roles in flavonoid biosynthesis and trichome formation when ectopically expressed in Arabidopsis. Sci. Rep. 2016, 6, 30514. [Google Scholar] [CrossRef] [Green Version]
- Xu, W.; Grain, D.; le Gourrierec, J.; Harscoët, E.; Berger, A.; Jauvion, V.; Scagnelli, A.; Berger, N.; Bidzinski, P.; Kelemen, Z.; et al. Regulation of flavonoid biosynthesis involves an unexpected complex transcriptional regulation of TT8 expression in Arabidopsis. New Phytol. 2013, 198, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Ratcliff, F.; Martin-Hernandez, A.M.; Baulcombe, D.C. Technical advance: Tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 2001, 25, 237–245. [Google Scholar] [CrossRef] [PubMed]
- Fischer, T.C.; Mirbeth, B.; Rentsch, J.; Sutter, C.; Ring, L.; Flachowsky, H.; Habegger, R.; Hoffmann, T.; Hanke, M.V.; Schwab, W. Premature and ectopic anthocyanin formation by silencing of anthocyanidin reductase in strawberry (Fragaria × ananassa). New Phytol. 2014, 201, 440–451. [Google Scholar] [CrossRef] [PubMed]
- Jin, X.H.; Huang, H.; Wang, L.; Sun, Y.; Dai, S.L. Transcriptomics and metabolite analysis reveals the molecular mechanism of anthocyanin biosynthesis branch pathway in different Seneclo cruentus cultivars. Front. Plant Sci. 2016, 7, 1307. [Google Scholar] [CrossRef] [Green Version]
- Sakuta, M.; Tanaka, A.; Iwase, K.; Miyasaka, M.; Ichiki, S.; Hatai, M.; Inoue, Y.T.; Yamagami, A.; Nakano, T.; Yoshida, K.; et al. Anthocyanin synthesis potential in betalain-producing Caryophyllales plants. J. Plant Res. 2021, 134, 1335–1349. [Google Scholar] [CrossRef]
- Soundararajan, P.; Kim, J.S. Anti-carcinogenic glucosinolates in cruciferous vegetables and their antagonistic effects on prevention of cancers. Molecules 2018, 23, 2983. [Google Scholar] [CrossRef] [Green Version]
- Melim, C.; Lauro, M.R.; Pires, I.M.; Oliveira, P.J.; Cabral, C. The role of glucosinolates from cruciferous vegetables (Brassicaceae) in gastrointestinal cancers: From prevention to therapeutics. Pharmaceutics 2022, 14, 190. [Google Scholar] [CrossRef]
- He, J.; Giusti, M.M. Anthocyanins: Natural colorants with health-promoting properties. Annu. Rev. Food Sci. Technol. 2010, 1, 163–187. [Google Scholar] [CrossRef]
- Pojer, E.; Mattivi, F.; Johnson, D.; Stockley, C.S. The case for anthocyanin consumption to promote human health: A review. Compr. Rev. Food Sci. Food Saf. 2013, 12, 483–508. [Google Scholar] [CrossRef]
- Khoo, H.E.; Azlan, A.; Tang, S.T.; Lim, S.M. Anthocyanidins and anthocyanins: Colored pigments as food, pharmaceutical ingredients, and the potential health benefits. Food Nutr. Res. 2017, 61, 1361779. [Google Scholar] [CrossRef] [Green Version]


| Gene Name | Gene ID | Mean FPKM (Purple) | Mean FPKM (Green) | Log2FC |
|---|---|---|---|---|
| BcTT8 | BraC09g027820 | 20.971186 | 0.128603 | 7.1793481 |
| BcEGL1 | BraC09g014020 | 0.164681 | 0.77609633 | −2.7202394 |
| BcGL3 | BraC04g016160 | 0.374962 | 1.52723467 | −1.6471003 |
| BcPRE1 | BraC07g019880 | 1.330022 | 6.18879667 | −2.371988 |
| Gene Name | Gene ID | Mean FPKM (Purple) | Mean FPKM (Green) | Log2FC | Up or Down Regulated |
|---|---|---|---|---|---|
| BcCHI-1 | BraC09g053560 | 71.52371067 | 30.162151 | 1.4134447 | up |
| BcCHI-2 | BraC09g053860 | 49.64995433 | 19.18305767 | 1.54414961 | up |
| BcDFR | BraC09g018850 | 332.763204 | 0.084531 | 12.0172491 | up |
| BcLODX | BraC03g052160 | 35.953383 | 0.135272333 | 8.22759096 | up |
| BcNCED4 | BraC08g01423 | 0 | 0.688357333 | −8.7651817 | down |
| BcFLS | BraC10g030090 | 107.904424 | 259.9946647 | −1.1015793 | down |
| BcUF3GT-1 | BraC06g022480 | 205.7734173 | 0.050154333 | 12.021179 | up |
| BcUF3GT-2 | BraC10g012540 | 57.12839133 | 0 | 15.1169748 | up |
| BcUF75C1 | BraC08g010530 | 322.273295 | 0.611051 | 9.20124194 | up |
| BcTT19-1 | BraC02g007050 | 82.251634 | 10.887416 | 3.10472421 | up |
| BcTT19-2 | BraC10g024210 | 90.53810367 | 0.047466333 | 11.1466138 | up |
| Bc5MAT | BraC09g003150 | 83.30002233 | 0.080537667 | 10.1547666 | up |
| BcTT8 | BraC09g027820 | 16.56795833 | 0.128603 | 7.1793481 | up |
| BcMYBL2-1 | BraC07g035800 | 42.067511 | 1.699416333 | 4.83412855 | up |
| BcMYBL2-2 | BraC02g021000 | 14.61018833 | 3.159260333 | 2.38862003 | up |
| Pathway | Gene Name | Gene ID | Mean FPKM (Purple) | Mean FPKM (Green) | Log2FC | Up or Downregulated |
|---|---|---|---|---|---|---|
| Flavonoid Biosynthesis | BcCHI-1 | BraC09g053560 | 71.52371067 | 30.162151 | 1.4134447 | up |
| BcCHI-2 | BraC09g053860 | 49.64995433 | 19.18305767 | 1.54414961 | up | |
| BcDFR | BraC09g018850 | 332.763204 | 0.084531 | 12.0172491 | up | |
| BcLODX | BraC03g052160 | 35.953383 | 0.135272333 | 8.22759096 | up | |
| BcC4H | BraC03g016590 | 156.2112757 | 18.594747 | 3.233418596 | up | |
| BcFLS | BraC10g030090 | 107.904424 | 259.9946647 | −1.1015793 | down | |
| Anthocyanin biosynthesis | BcUF3GT-1 | BraC06g022480 | 205.7734173 | 0.050154333 | 12.021179 | up |
| BcUF3GT-2 | BraC10g012540 | 57.12839133 | 0 | 15.1169748 | up | |
| BcUF75C1 | BraC08g010530 | 322.273295 | 0.611051 | 9.20124194 | up |
| Gene Name | Gene ID | Mean FPKM (Purple) | Mean FPKM (Green) | Log2FC | Up or Down Regulated |
|---|---|---|---|---|---|
| BcCHS2 | BraC10g026540 | 471.590159 | 471.7846887 | 0.000594985 | |
| BcCHI-1 | BraC09g053560 | 71.52371067 | 30.162151 | 1.4134447 | up |
| BcF3H | BraC02g029180 | 4.807456333 | 7.09875 | −0.525317861 | |
| BcF3′H | BraC08g015770 | 24.735745 | 30.335388 | −0.13065833 | |
| BcDFR | BraC09g018850 | 332.763204 | 0.084531 | 12.0172491 | up |
| BcLODX | BraC03g052160 | 35.953383 | 0.135272333 | 8.22759096 | up |
| BcFLS | BraC10g030090 | 107.904424 | 259.9946647 | −1.1015793 | down |
| BcUF3GT-1 | BraC06g022480 | 205.7734173 | 0.050154333 | 12.021179 | up |
| BcTT8 | BraC09g027820 | 16.56795833 | 0.128603 | 7.1793481 | up |
| BcMYBL2-1 | BraC07g035800 | 42.067511 | 1.699416333 | 4.83412855 | up |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tang, L.; Xiao, D.; Yin, Y.; Wang, H.; Wang, J.; Liu, T.; Hou, X.; Li, Y. Comparative Transcriptome Analysis of Purple and Green Non-Heading Chinese Cabbage and Function Analyses of BcTT8 Gene. Genes 2022, 13, 988. https://doi.org/10.3390/genes13060988
Tang L, Xiao D, Yin Y, Wang H, Wang J, Liu T, Hou X, Li Y. Comparative Transcriptome Analysis of Purple and Green Non-Heading Chinese Cabbage and Function Analyses of BcTT8 Gene. Genes. 2022; 13(6):988. https://doi.org/10.3390/genes13060988
Chicago/Turabian StyleTang, Liping, Dong Xiao, Yuqin Yin, Haibin Wang, Jianjun Wang, Tongkun Liu, Xilin Hou, and Ying Li. 2022. "Comparative Transcriptome Analysis of Purple and Green Non-Heading Chinese Cabbage and Function Analyses of BcTT8 Gene" Genes 13, no. 6: 988. https://doi.org/10.3390/genes13060988






