The Value of the Stemness Index in Ovarian Cancer Prognosis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Obtained
2.2. Calculation of the Stemness Index and Immune Score
2.3. Correlation Analysis of the Stemness Index and Immune Infiltration
2.4. Differentially Expressed and Functional Enrichment Analyses
2.5. Data-Obtained Identification of OC’s Stemness-Related Molecular Subtypes
2.6. Evaluation of the Relationship between Stemness Subtypes and Immune Infiltration
2.7. Construction and Validation of the Prognostic Signature
2.8. Identification of Prognostic Factors and Nomogram Construction
2.9. Statistical Analysis
3. Results
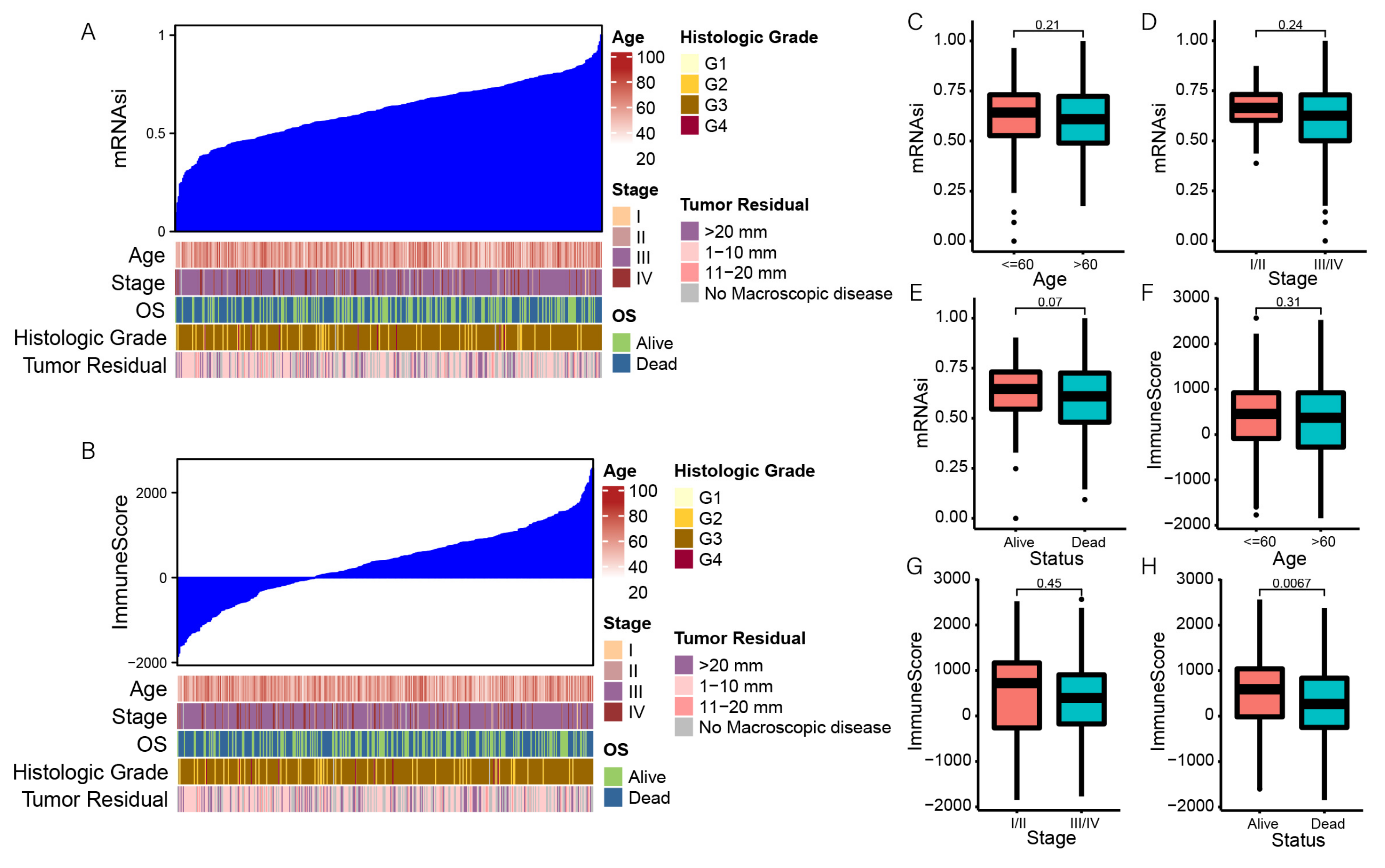
3.1. Correlation between the Stemness Index and Clinical Characteristics
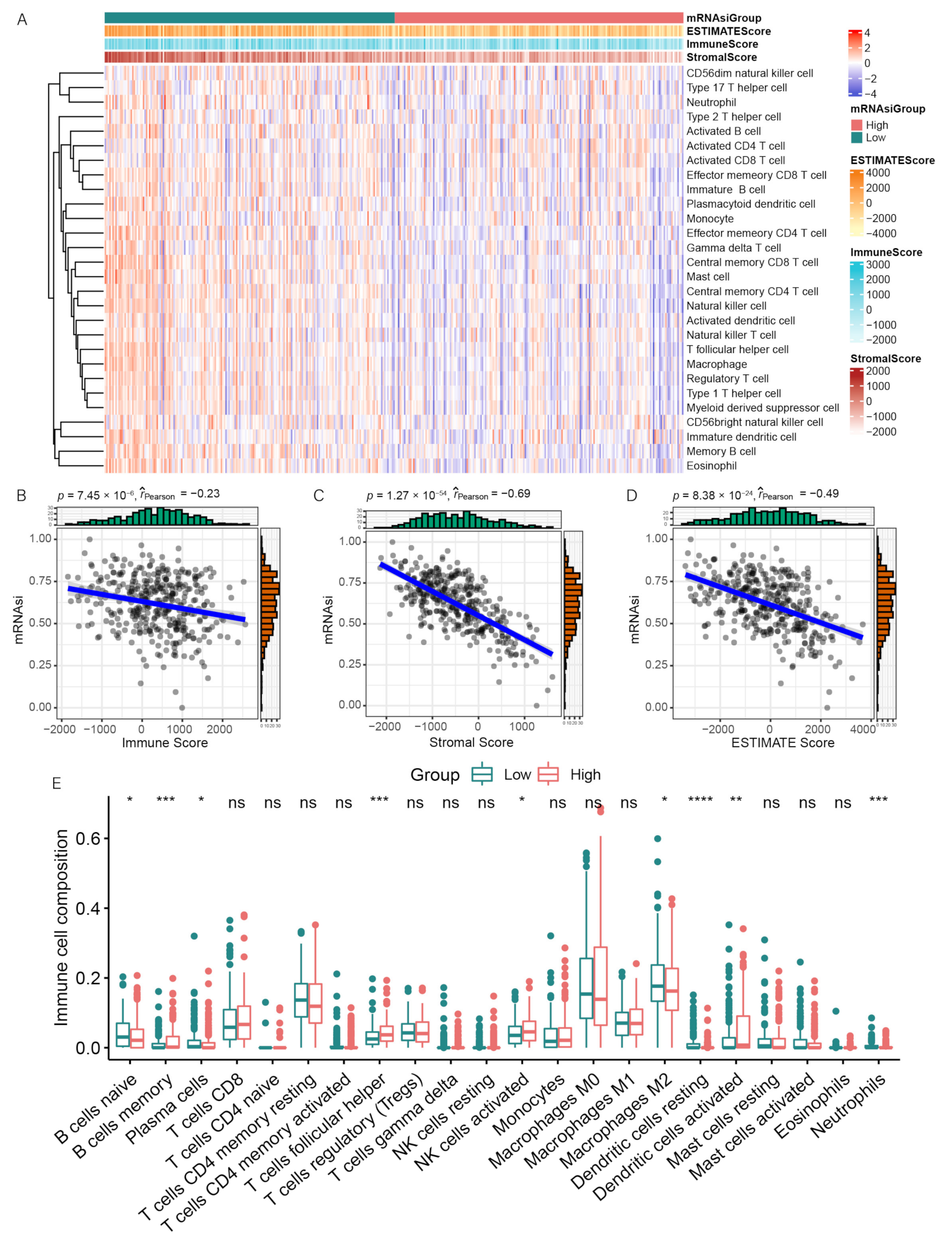
3.2. Correlation between mRNAsi and Immune Infiltration
3.3. Differentially Expressed and Functional Enrichment Analyses
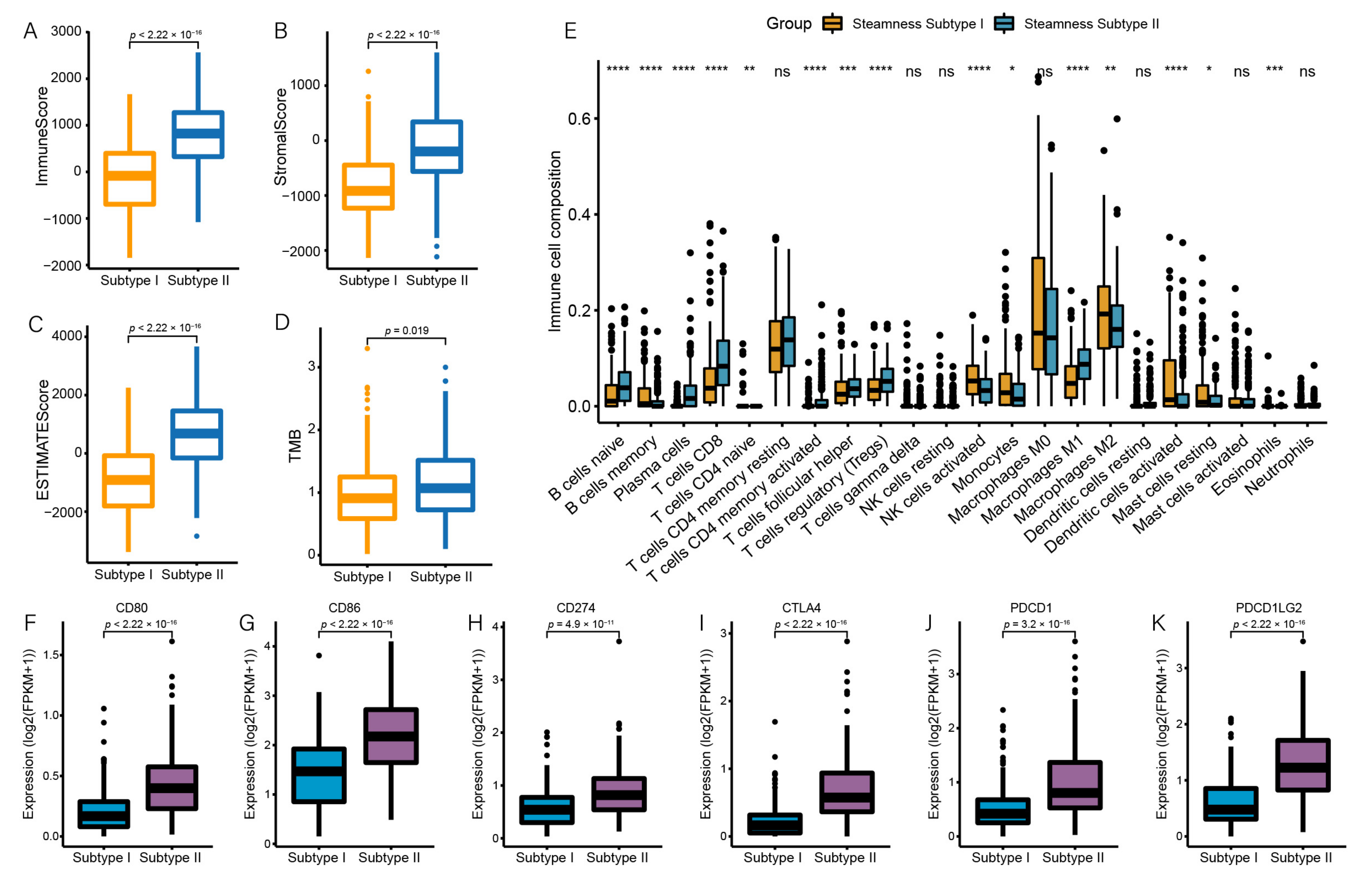
3.4. Identification of Two Stemness Subtypes with Distinct Characteristics
3.5. Stemness Subtype Differences in Immune Infiltration
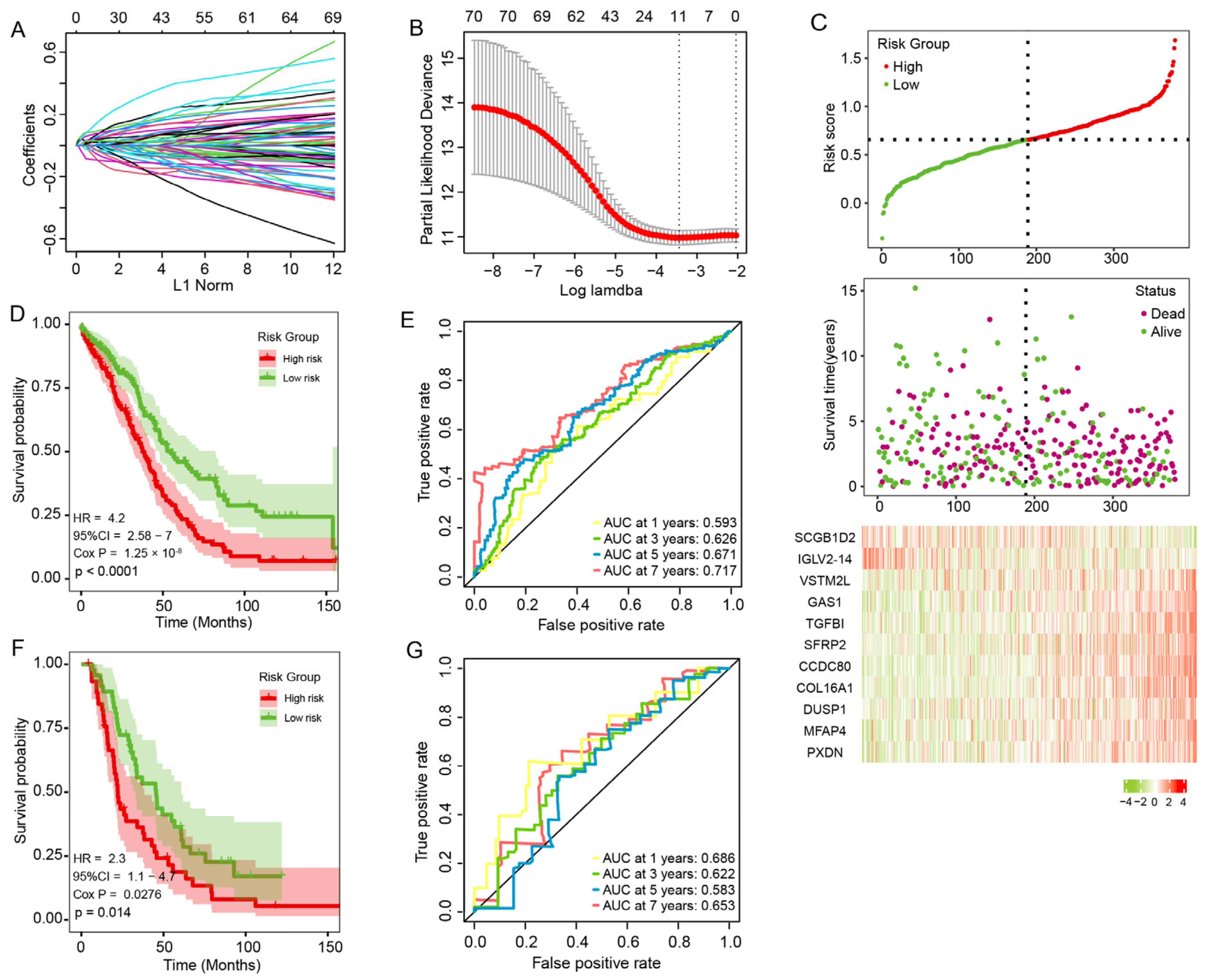
3.6. Construction and Validation of the Prognosis Risk Signature
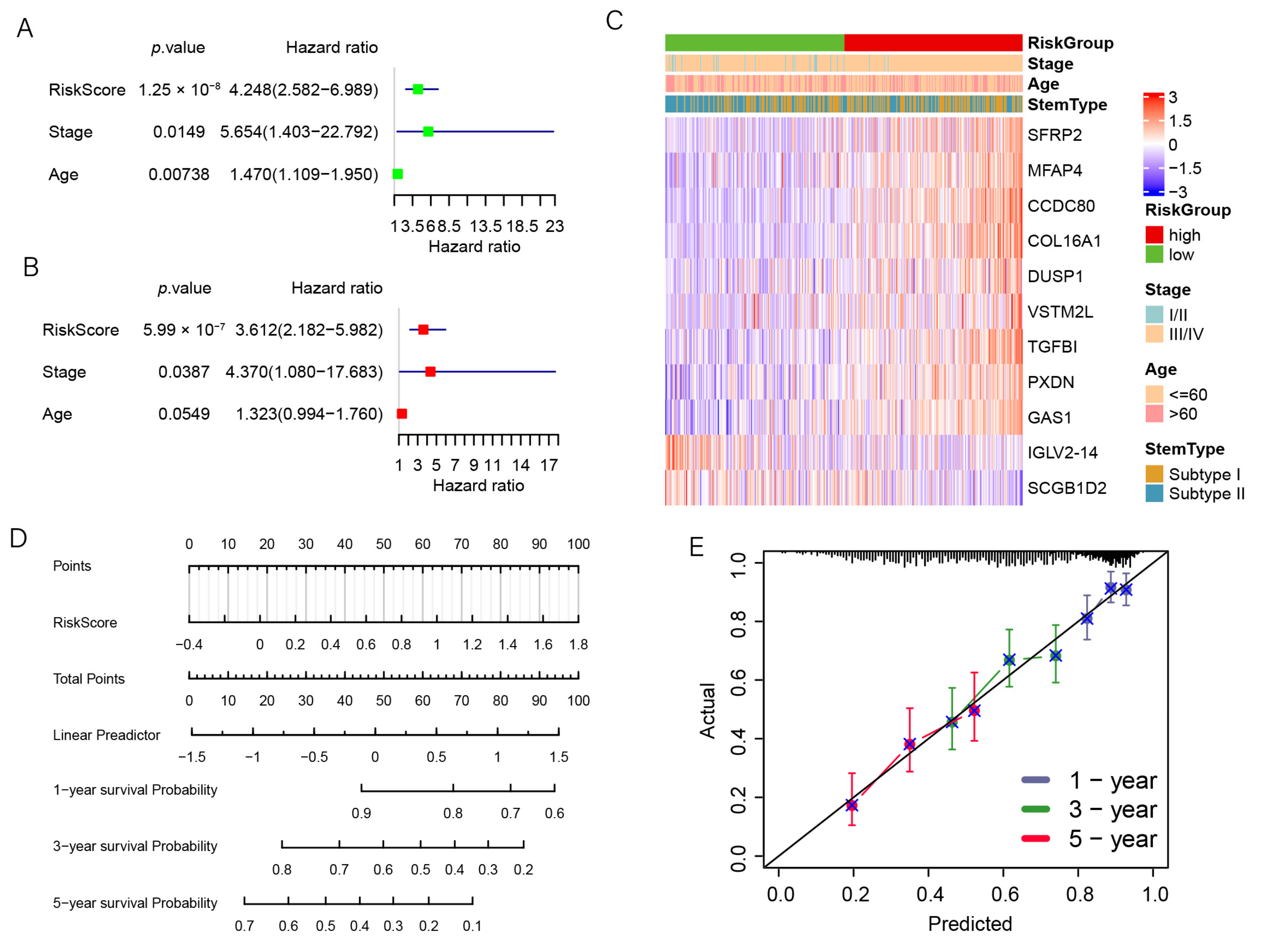
3.7. The Prognostic Signature Is an Independent Prognostic Factor for OC
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Lheureux, S.; Gourley, C.; Vergote, I.; Oza, A.M. Epithelial ovarian cancer. Lancet 2019, 393, 1240–1253. [Google Scholar] [CrossRef] [Green Version]
- Gupta, S.; Nag, S.; Aggarwal, S.; Rauthan, A.; Warrier, N. Maintenance therapy for recurrent epithelial ovarian cancer: Current therapies and future perspectives—a review. J. Ovarian Res. 2019, 12, 103. [Google Scholar] [CrossRef] [PubMed]
- Akter, S.; Rahman, M.A.; Hasan, M.N.; Akhter, H.; Noor, P.; Islam, R.; Shin, Y.; Rahman, M.D.H.; Gazi, M.S.; Huda, M.N.; et al. Recent Advances in Ovarian Cancer: Therapeutic Strategies, Potential Biomarkers, and Technological Improvements. Cells 2022, 11, 650. [Google Scholar] [CrossRef] [PubMed]
- Krishnan, V.; Berek, J.S.; Dorigo, O. Immunotherapy in ovarian cancer. Curr. Probl. Cancer 2017, 41, 48–63. [Google Scholar] [CrossRef] [PubMed]
- Vlashi, E.; Pajonk, F. Cancer stem cells, cancer cell plasticity and radiation therapy. Semin. Cancer Biol. 2015, 31, 28–35. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scheel, C.; Weinberg, R.A. Cancer stem cells and epithelial-mesenchymal transition: Concepts and molecular links. Semin. Cancer Biol. 2012, 22, 396–403. [Google Scholar] [CrossRef]
- Heng, W.S.; Gosens, R.; Kruyt, F.A.E. Lung cancer stem cells: Origin, features, maintenance mechanisms and therapeutic targeting. Biochem. Pharmacol. 2019, 160, 121–133. [Google Scholar] [CrossRef]
- Motohara, T.; Yoshida, G.J.; Katabuchi, H. The hallmarks of ovarian cancer stem cells and niches: Exploring their harmonious interplay in therapy resistance. Semin. Cancer Biol. 2021, 77, 182–193. [Google Scholar] [CrossRef]
- Baccelli, I.; Schneeweiss, A.; Riethdorf, S.; Stenzinger, A.; Schillert, A.; Vogel, V.; Klein, C.; Saini, M.; Bauerle, T.; Wallwiener, M.; et al. Identification of a population of blood circulating tumor cells from breast cancer patients that initiates metastasis in a xenograft assay. Nat. Biotechnol. 2013, 31, 539–544. [Google Scholar] [CrossRef]
- Nassar, D.; Blanpain, C. Cancer Stem Cells: Basic Concepts and Therapeutic Implications. Annu. Rev. Pathol. 2016, 11, 47–76. [Google Scholar] [CrossRef] [PubMed]
- Vlashi, E.; Lagadec, C.; Vergnes, L.; Matsutani, T.; Masui, K.; Poulou, M.; Popescu, R.; Della Donna, L.; Evers, P.; Dekmezian, C.; et al. Metabolic state of glioma stem cells and nontumorigenic cells. Proc. Natl. Acad. Sci. USA 2011, 108, 16062–16067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Marzagalli, M.; Raimondi, M.; Fontana, F.; Montagnani Marelli, M.; Moretti, R.M.; Limonta, P. Cellular and molecular biology of cancer stem cells in melanoma: Possible therapeutic implications. Semin. Cancer Biol. 2019, 59, 221–235. [Google Scholar] [CrossRef] [PubMed]
- Binju, M.; Amaya-Padilla, M.A.; Wan, G.; Gunosewoyo, H.; Suryo Rahmanto, Y.; Yu, Y. Therapeutic Inducers of Apoptosis in Ovarian Cancer. Cancers 2019, 11, 1786. [Google Scholar] [CrossRef] [Green Version]
- McMullen, M.; Madariaga, A.; Lheureux, S. New approaches for targeting platinum-resistant ovarian cancer. Semin. Cancer Biol. 2021, 77, 167–181. [Google Scholar] [CrossRef]
- Mihanfar, A.; Aghazadeh Attari, J.; Mohebbi, I.; Majidinia, M.; Kaviani, M.; Yousefi, M.; Yousefi, B. Ovarian cancer stem cell: A potential therapeutic target for overcoming multidrug resistance. J. Cell Physiol. 2019, 234, 3238–3253. [Google Scholar] [CrossRef]
- Keyvani, V.; Farshchian, M.; Esmaeili, S.A.; Yari, H.; Moghbeli, M.; Nezhad, S.K.; Abbaszadegan, M.R. Ovarian cancer stem cells and targeted therapy. J. Ovarian Res. 2019, 12, 120. [Google Scholar] [CrossRef] [Green Version]
- Malta, T.M.; Sokolov, A.; Gentles, A.J.; Burzykowski, T.; Poisson, L.; Weinstein, J.N.; Kaminska, B.; Huelsken, J.; Omberg, L.; Gevaert, O.; et al. Machine Learning Identifies Stemness Features Associated with Oncogenic Dedifferentiation. Cell 2018, 173, 338–354.e15. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.M.; Liu, C.L.; Green, M.R.; Gentles, A.J.; Feng, W.; Xu, Y.; Hoang, C.D.; Diehn, M.; Alizadeh, A.A. Robust enumeration of cell subsets from tissue expression profiles. Nat. Methods 2015, 12, 453–457. [Google Scholar] [CrossRef] [Green Version]
- Ye, Y.; Dai, Q.; Qi, H. A novel defined pyroptosis-related gene signature for predicting the prognosis of ovarian cancer. Cell Death Discov. 2021, 7, 71. [Google Scholar] [CrossRef]
- Munoz-Galvan, S.; Carnero, A. Targeting Cancer Stem Cells to Overcome Therapy Resistance in Ovarian Cancer. Cells 2020, 9, 1402. [Google Scholar] [CrossRef] [PubMed]
- Motohara, T.; Katabuchi, H. Ovarian Cancer Stemness: Biological and Clinical Implications for Metastasis and Chemotherapy Resistance. Cancers 2019, 11, 907. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pan, S.; Zhan, Y.; Chen, X.; Wu, B.; Liu, B. Identification of Biomarkers for Controlling Cancer Stem Cell Characteristics in Bladder Cancer by Network Analysis of Transcriptome Data Stemness Indices. Front. Oncol. 2019, 9, 613. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pei, J.; Wang, Y.; Li, Y. Identification of key genes controlling breast cancer stem cell characteristics via stemness indices analysis. J. Transl. Med. 2020, 18, 74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Tseng, J.T.; Lien, I.C.; Li, F.; Wu, W.; Li, H. mRNAsi Index: Machine Learning in Mining Lung Adenocarcinoma Stem Cell Biomarkers. Genes 2020, 11, 257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, Y.; Xiao, H.; Cheng, M.; Fan, X. Bioinformatics Analysis Reveals Biomarkers With Cancer Stem Cell Characteristics in Lung Squamous Cell Carcinoma. Front. Genet. 2020, 11, 427. [Google Scholar] [CrossRef]
- Zhang, Q.; Wang, J.; Liu, M.; Zhu, Q.; Li, Q.; Xie, C.; Han, C.; Wang, Y.; Gao, M.; Liu, J. Weighted correlation gene network analysis reveals a new stemness index-related survival model for prognostic prediction in hepatocellular carcinoma. Aging 2020, 12, 13502–13517. [Google Scholar] [CrossRef]
- Suo, H.D.; Tao, Z.; Zhang, L.; Jin, Z.N.; Li, X.Y.; Ma, W.; Wang, Z.; Qiu, Y.; Jin, F.; Chen, B.; et al. Coexpression Network Analysis of Genes Related to the Characteristics of Tumor Stemness in Triple-Negative Breast Cancer. Biomed. Res. Int. 2020, 2020, 7575862. [Google Scholar] [CrossRef]
- Chen, X.; Zhang, D.; Jiang, F.; Shen, Y.; Li, X.; Hu, X.; Wei, P.; Shen, X. Prognostic Prediction Using a Stemness Index-Related Signature in a Cohort of Gastric Cancer. Front. Mol. Biosci. 2020, 7, 570702. [Google Scholar] [CrossRef]
- Wang, Z.; Wang, Y.; Yang, T.; Xing, H.; Wang, Y.; Gao, L.; Guo, X.; Xing, B.; Wang, Y.; Ma, W. Machine learning revealed stemness features and a novel stemness-based classification with appealing implications in discriminating the prognosis, immunotherapy and temozolomide responses of 906 glioblastoma patients. Brief. Bioinform. 2021, 22, bbab032. [Google Scholar] [CrossRef]
- Tan, J.; Zhu, H.; Tang, G.; Liu, H.; Wanggou, S.; Cao, Y.; Xin, Z.; Zhou, Q.; Zhan, C.; Wu, Z.; et al. Molecular Subtypes Based on the Stemness Index Predict Prognosis in Glioma Patients. Front. Genet. 2021, 12, 616507. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Liu, Q.; Huang, J.; Diao, G.; Liang, Z. Transcriptome-based stemness indices analysis reveals platinum-based chemo-theraputic response indicators in advanced-stage serous ovarian cancer. Bioengineered 2021, 12, 3753–3771. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Wu, D.; Xia, Y.; Yang, B.; Xu, T. Identification of hub genes and compounds controlling ovarian cancer stem cell characteristics via stemness indices analysis. Ann. Transl. Med. 2021, 9, 379. [Google Scholar] [CrossRef]
- Jain, S.; Annett, S.L.; Morgan, M.P.; Robson, T. The Cancer Stem Cell Niche in Ovarian Cancer and Its Impact on Immune Surveillance. Int. J. Mol. Sci. 2021, 22, 4091. [Google Scholar] [CrossRef] [PubMed]
- Miranda, A.; Hamilton, P.T.; Zhang, A.W.; Pattnaik, S.; Becht, E.; Mezheyeuski, A.; Bruun, J.; Micke, P.; de Reynies, A.; Nelson, B.H. Cancer stemness, intratumoral heterogeneity, and immune response across cancers. Proc. Natl. Acad. Sci. USA 2019, 116, 9020–9029. [Google Scholar] [CrossRef] [Green Version]
- Khosravi, N.; Mokhtarzadeh, A.; Baghbanzadeh, A.; Hajiasgharzadeh, K.; Shahgoli, V.K.; Hemmat, N.; Safarzadeh, E.; Baradaran, B. Immune checkpoints in tumor microenvironment and their relevance to the development of cancer stem cells. Life Sci. 2020, 256, 118005. [Google Scholar] [CrossRef]
- Jia, L.; Wang, Y.; Wang, C.Y. circFAT1 Promotes Cancer Stemness and Immune Evasion by Promoting STAT3 Activation. Adv. Sci. 2021, 8, 2003376. [Google Scholar] [CrossRef]
- Jardim, D.L.; Goodman, A.; de Melo Gagliato, D.; Kurzrock, R. The Challenges of Tumor Mutational Burden as an Immunotherapy Biomarker. Cancer Cell 2021, 39, 154–173. [Google Scholar] [CrossRef]
- Chan, T.A.; Yarchoan, M.; Jaffee, E.; Swanton, C.; Quezada, S.A.; Stenzinger, A.; Peters, S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann. Oncol. 2019, 30, 44–56. [Google Scholar] [CrossRef]
- Kline, J.; Godfrey, J.; Ansell, S.M. The immune landscape and response to immune checkpoint blockade therapy in lymphoma. Blood 2020, 135, 523–533. [Google Scholar] [CrossRef]
- Kaur, A.; Webster, M.R.; Marchbank, K.; Behera, R.; Ndoye, A.; Kugel, C.H., 3rd; Dang, V.M.; Appleton, J.; O’Connell, M.P.; Cheng, P.; et al. sFRP2 in the aged microenvironment drives melanoma metastasis and therapy resistance. Nature 2016, 532, 250–254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montagner, M.; Bhome, R.; Hooper, S.; Chakravarty, P.; Qin, X.; Sufi, J.; Bhargava, A.; Ratcliffe, C.D.H.; Naito, Y.; Pocaterra, A.; et al. Crosstalk with lung epithelial cells regulates Sfrp2-mediated latency in breast cancer dissemination. Nat. Cell Biol. 2020, 22, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Wu, Q.; Yin, X.; Zhao, W.; Xu, W.; Chen, L. Downregulation of SFRP2 facilitates cancer stemness and radioresistance of glioma cells via activating Wnt/β-catenin signaling. PLoS ONE 2021, 16, e0260864. [Google Scholar] [CrossRef]
- Li, P.; Zhao, S.; Hu, Y. SFRP2 modulates nonsmall cell lung cancer A549 cell apoptosis and metastasis by regulating mitochondrial fission via Wnt pathways. Mol. Med. Rep. 2019, 20, 1925–1932. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Sun, Q.; Li, L.; Zhou, J.; Zhang, C.; Hu, T.; Zhou, X.; Zhang, L.; Wang, B.; Li, B.; et al. High Expression Levels of AGGF1 and MFAP4 Predict Primary Platinum-Based Chemoresistance and are Associated with Adverse Prognosis in Patients with Serous Ovarian Cancer. J. Cancer 2019, 10, 397–407. [Google Scholar] [CrossRef]
- Wang, W.D.; Wu, G.Y.; Bai, K.H.; Shu, L.L.; Chi, P.D.; He, S.Y.; Huang, X.; Zhang, Q.Y.; Li, L.; Wang, D.W.; et al. A prognostic stemness biomarker CCDC80 reveals acquired drug resistance and immune infiltration in colorectal cancer. Clin. Transl. Med. 2020, 10, e225. [Google Scholar] [CrossRef]
- Wang, W.; Xu, C.; Ren, Y.; Wang, S.; Liao, C.; Fu, X.; Hu, H. A Novel Cancer Stemness-Related Signature for Predicting Prognosis in Patients with Colon Adenocarcinoma. Stem Cells Int. 2021, 2021, 7036059. [Google Scholar] [CrossRef]
- Jiang, W.; Zhu, D.; Wang, C.; Zhu, Y. An immune relevant signature for predicting prognoses and immunotherapeutic responses in patients with muscle-invasive bladder cancer (MIBC). Cancer Med. 2020, 9, 2774–2790. [Google Scholar] [CrossRef] [Green Version]
- Teng, F.; Xu, Z.; Chen, J.; Zheng, G.; Zheng, G.; Lv, H.; Wang, Y.; Wang, L.; Cheng, X. DUSP1 induces apatinib resistance by activating the MAPK pathway in gastric cancer. Oncol. Rep. 2018, 40, 1203–1222. [Google Scholar] [CrossRef]
- Fang, J.; Ye, Z.; Gu, F.; Yan, M.; Lin, Q.; Lin, J.; Wang, Z.; Xu, Y.; Wang, Y. DUSP1 enhances the chemoresistance of gallbladder cancer via the modulation of the p38 pathway and DNA damage/repair system. Oncol. Lett. 2018, 16, 1869–1875. [Google Scholar] [CrossRef] [Green Version]
- Fico, F.; Santamaria-Martinez, A. TGFBI modulates tumour hypoxia and promotes breast cancer metastasis. Mol. Oncol. 2020, 14, 3198–3210. [Google Scholar] [CrossRef] [PubMed]


Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yuan, H.; Yu, Q.; Pang, J.; Chen, Y.; Sheng, M.; Tang, W. The Value of the Stemness Index in Ovarian Cancer Prognosis. Genes 2022, 13, 993. https://doi.org/10.3390/genes13060993
Yuan H, Yu Q, Pang J, Chen Y, Sheng M, Tang W. The Value of the Stemness Index in Ovarian Cancer Prognosis. Genes. 2022; 13(6):993. https://doi.org/10.3390/genes13060993
Chicago/Turabian StyleYuan, Hongjun, Qian Yu, Jianyu Pang, Yongzhi Chen, Miaomiao Sheng, and Wenru Tang. 2022. "The Value of the Stemness Index in Ovarian Cancer Prognosis" Genes 13, no. 6: 993. https://doi.org/10.3390/genes13060993




