Susceptibility Loci for Type 2 Diabetes in the Ethnically Endogamous Indian Sindhi Population: A Pooled Blood Genome-Wide Association Study
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Definitions of Metabolic Conditions
2.3. Pool Definitions for GWAS
2.4. Whole Blood Pool Construction and Genotyping
2.5. Statistical Analyses
2.6. Functional Relevance of Strongly Associated Variants and Genes
3. Results
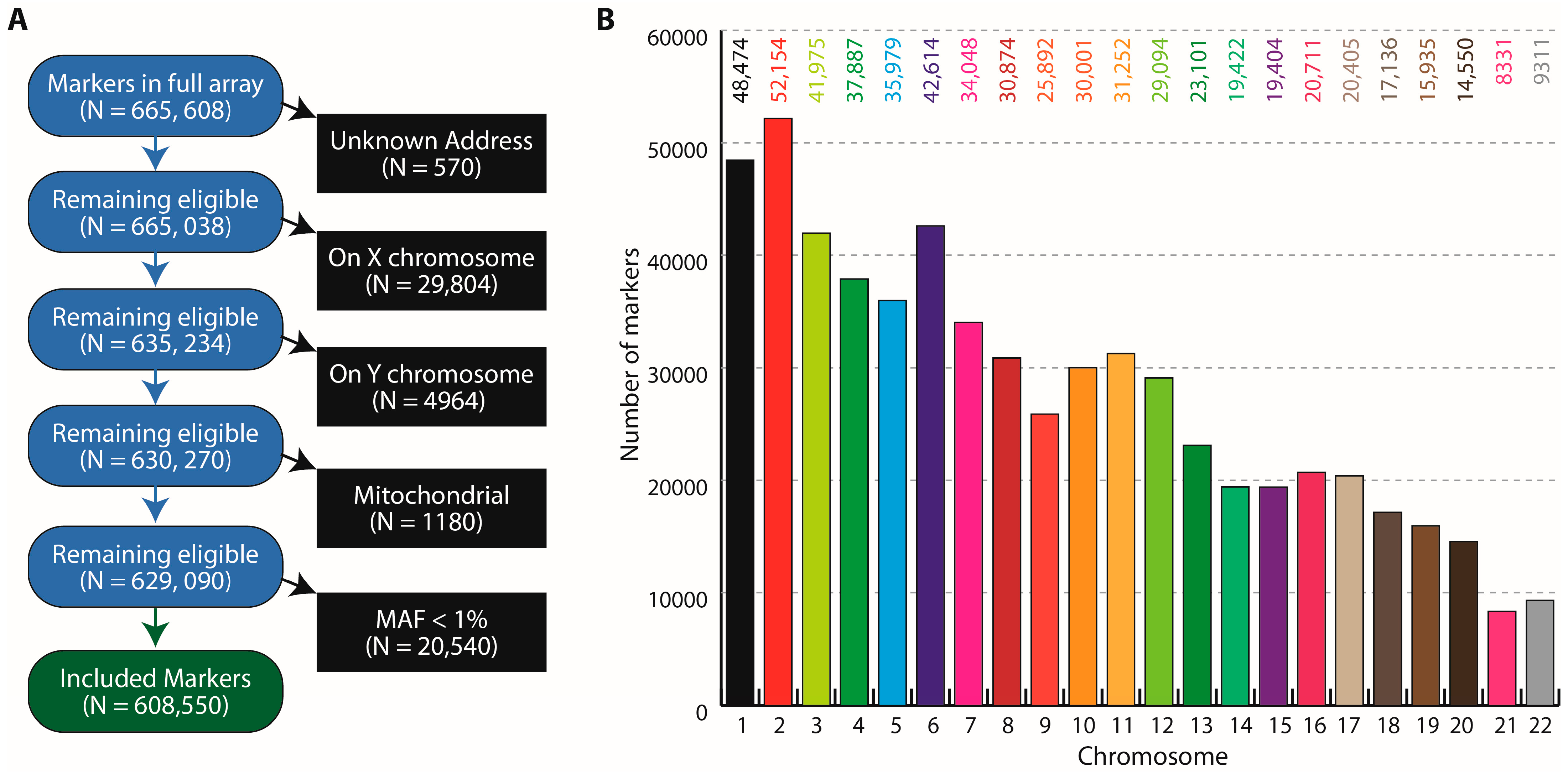
3.1. Study Participants, Pools and the GSA Variants
3.2. Pooled Blood and Genotyping Quality Control
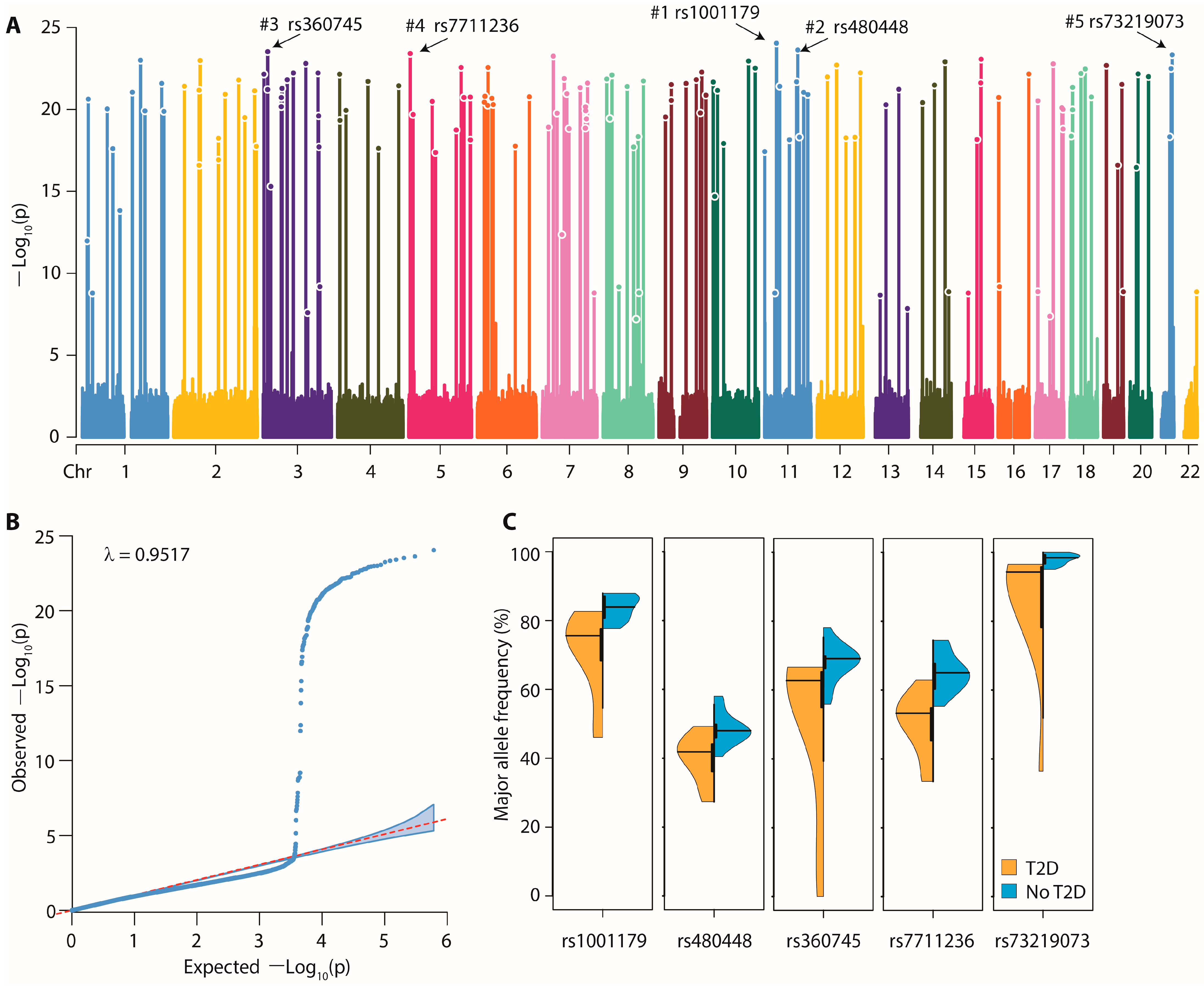
3.3. Pooled GWAS Results—Variants
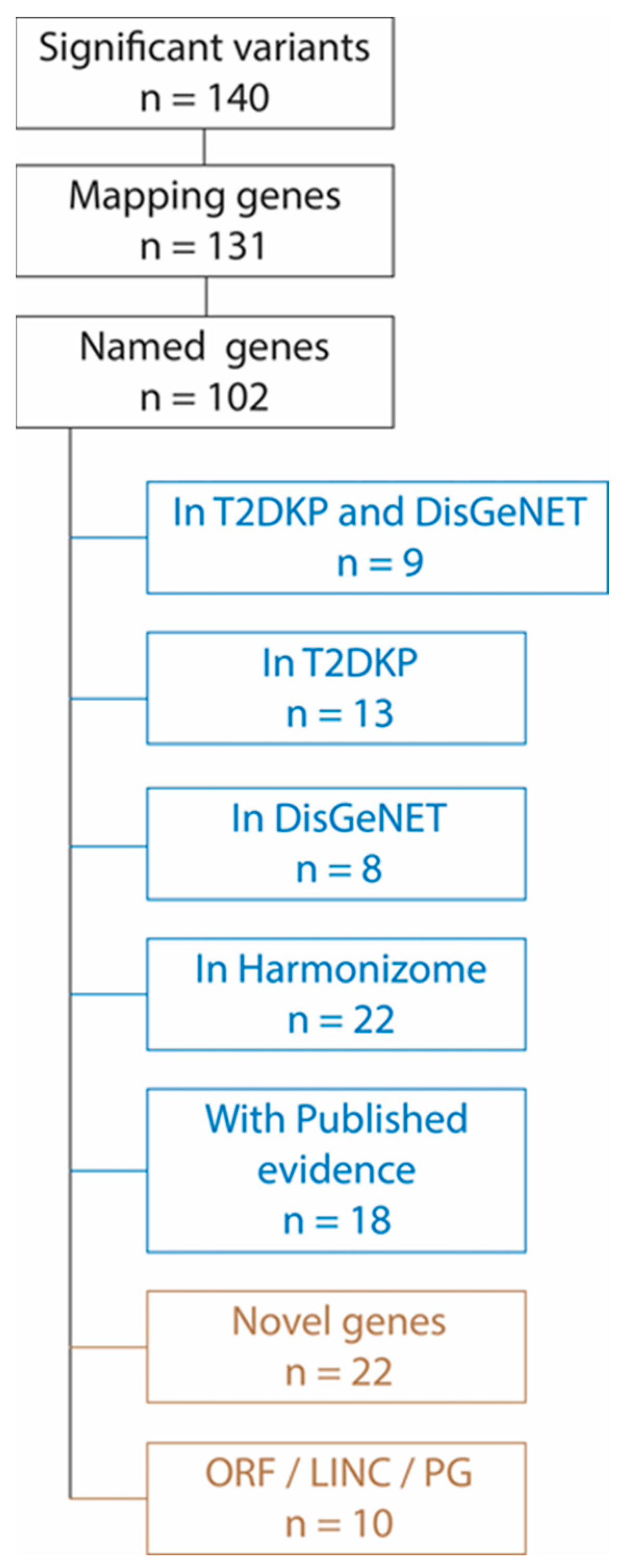
3.4. Pooled GWAS Results—Genes
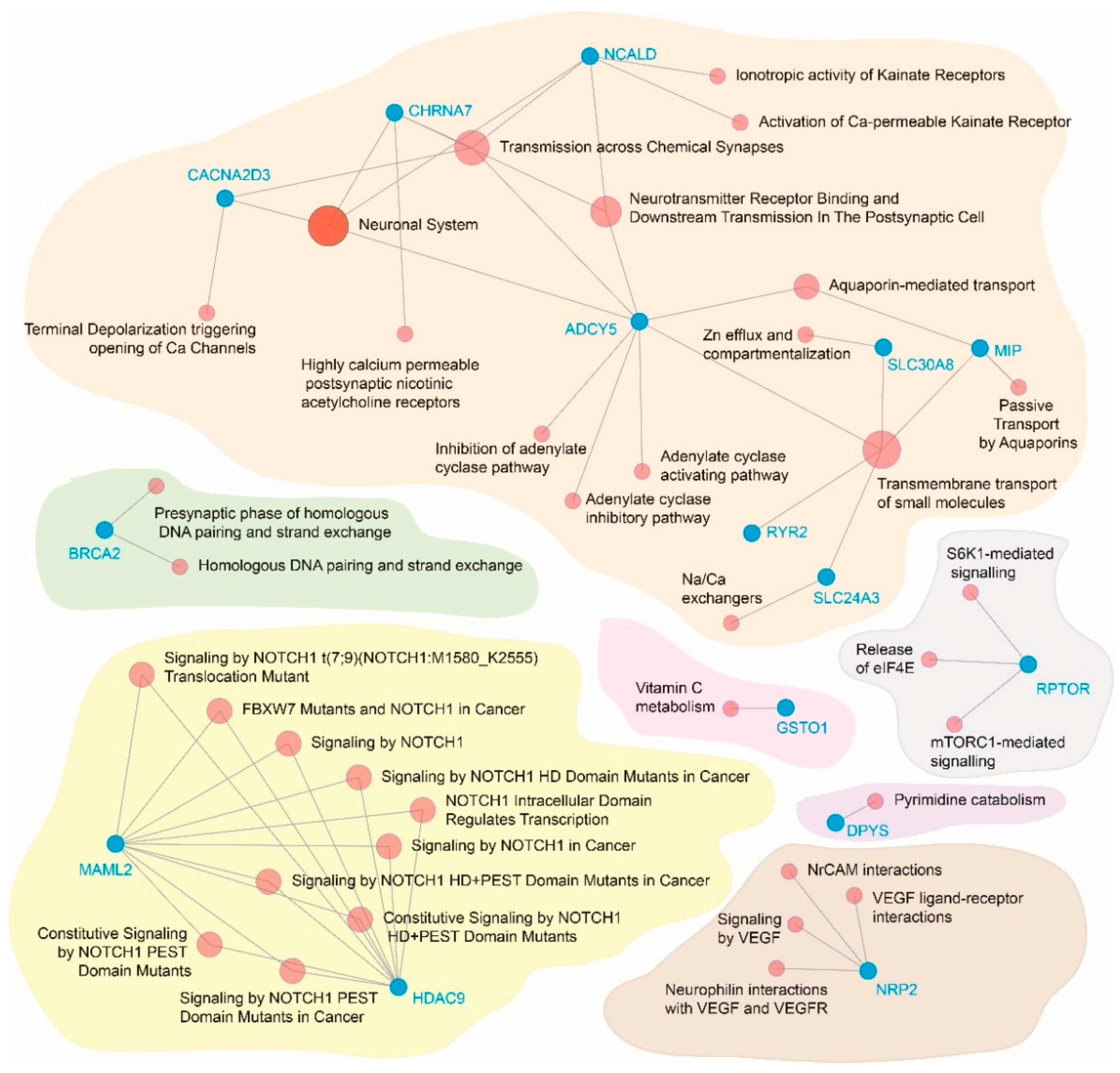
3.5. Functional Relevance of the Significantly Associated Genes
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rana, S.; Ali, S.; Wani, H.A.; Mushtaq, Q.D.; Sharma, S.; Rehman, M.U. Metabolic syndrome and underlying genetic determinants-A systematic review. J. Diabetes Metab. Disord. 2022, 21, 1095–1104. [Google Scholar] [CrossRef] [PubMed]
- Mamtani, M.; Jaisinghani, M.T.; Jaiswal, S.G.; Pipal, K.V.; Patel, A.A.; Kulkarni, H. Genetic association of anthropometric traits with type 2 diabetes in ethnically endogamous Sindhi families. PLoS ONE 2021, 16, e0257390. [Google Scholar] [CrossRef] [PubMed]
- Asamoah, E.A.; Obirikorang, C.; Acheampong, E.; Annani-Akollor, M.E.; Laing, E.F.; Owiredu, E.W.; Anto, E.O. Heritability and Genetics of Type 2 Diabetes Mellitus in Sub-Saharan Africa: A Systematic Review and Meta-Analysis. J. Diabetes Res. 2020, 2020, 3198671. [Google Scholar] [CrossRef]
- Cuschieri, S. The genetic side of type 2 diabetes—A review. Diabetes Metab. Syndr. 2019, 13, 2503–2506. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.F. Genetic and Epigenetic Studies in Diabetic Kidney Disease. Front. Genet. 2019, 10, 507. [Google Scholar] [CrossRef] [PubMed]
- DeForest, N.; Majithia, A.R. Genetics of Type 2 Diabetes: Implications from Large-Scale Studies. Curr. Diabetes Rep. 2022, 22, 227–235. [Google Scholar] [CrossRef] [PubMed]
- Xia, W.; Chen, N.; Peng, W.; Jia, X.; Yu, Y.; Wu, X.; Gao, H. Systematic Meta-analysis Revealed an Association of PGC-1alpha rs8192678 Polymorphism in Type 2 Diabetes Mellitus. Dis. Markers 2019, 2019, 2970401. [Google Scholar] [CrossRef]
- Chowdhury, R.; Narayan, K.M.; Zabetian, A.; Raj, S.; Tabassum, R. Genetic studies of type 2 diabetes in South Asians: A systematic overview. Curr. Diabetes Rev. 2014, 10, 258–274. [Google Scholar] [CrossRef]
- Fu, J.; Festen, E.A.; Wijmenga, C. Multi-ethnic studies in complex traits. Hum. Mol. Genet. 2011, 20, R206–R213. [Google Scholar] [CrossRef]
- Ukkola, O. Genetic variants of ghrelin in metabolic disorders. Peptides 2011, 32, 2319–2322. [Google Scholar] [CrossRef]
- Carter, M. Indians and the Colonial Diaspora; Institute of Southeast Asian Studies: Singapore, 2008. [Google Scholar]
- Florez, J.C.; Pearson, E.R. A roadmap to achieve pharmacological precision medicine in diabetes. Diabetologia 2022. [Google Scholar] [CrossRef] [PubMed]
- Kumar, R.; Nandhini, L.P.; Kamalanathan, S.; Sahoo, J.; Vivekanadan, M. Evidence for current diagnostic criteria of diabetes mellitus. World J. Diabetes 2016, 7, 396–405. [Google Scholar] [CrossRef] [PubMed]
- Whelton, P.K.; Carey, R.M.; Aronow, W.S.; Casey, D.E., Jr.; Collins, K.J.; Dennison Himmelfarb, C.; DePalma, S.M.; Gidding, S.; Jamerson, K.A.; Jones, D.W.; et al. 2017 ACC/AHA/AAPA/ABC/ACPM/AGS/APhA/ASH/ASPC/NMA/PCNA Guideline for the Prevention, Detection, Evaluation, and Management of High Blood Pressure in Adults: A Report of the American College of Cardiology/American Heart Association Task Force on Clinical Practice Guidelines. Hypertension 2018, 71, e116–e135. [Google Scholar] [CrossRef]
- Snehalatha, C.; Viswanathan, V.; Ramachandran, A. Cutoff values for normal anthropometric variables in asian Indian adults. Diabetes Care 2003, 26, 1380–1384. [Google Scholar] [CrossRef] [PubMed]
- Sharma, Y.K.; Prakash, N.; Gupta, A. Prevalence of metabolic syndrome as per the NCEP and IDF definitions vis-a-vis severity and duration of psoriasis in a semi-urban Maharashtrian population: A case control study. Diabetes Metab. Syndr. 2016, 10, S72–S76. [Google Scholar] [CrossRef]
- Craig, J.E.; Hewitt, A.W.; McMellon, A.E.; Henders, A.K.; Ma, L.; Wallace, L.; Sharma, S.; Burdon, K.P.; Visscher, P.M.; Montgomery, G.W.; et al. Rapid inexpensive genome-wide association using pooled whole blood. Genome Res. 2009, 19, 2075–2080. [Google Scholar] [CrossRef]
- Turner, S. qqman: An R package for visualizing GWAS results using Q-Q and manhattan plots. J. Open Source Softw. 2018, 3, 731. [Google Scholar] [CrossRef]
- Macgregor, S.; Zhao, Z.Z.; Henders, A.; Nicholas, M.G.; Montgomery, G.W.; Visscher, P.M. Highly cost-efficient genome-wide association studies using DNA pools and dense SNP arrays. Nucleic Acids Res. 2008, 36, e35. [Google Scholar] [CrossRef]
- Type 2 Diabetes Knowledge Portal. Available online: https://t2d.hugeamp.org/ (accessed on 30 June 2022).
- Harmonizome: Integrated Knowledge about Genes and Proteins. Available online: https://maayanlab.cloud/Harmonizome/ (accessed on 30 June 2022).
- Network Analyst—Comprehensive Gene Expression Profiling and Network Visual Analytics. Available online: https://www.networkanalyst.ca/ (accessed on 30 June 2022).
- Chand, S.; McKnight, A.J.; Shabir, S.; Chan, W.; McCaughan, J.A.; Maxwell, A.P.; Harper, L.; Borrows, R. Analysis of single nucleotide polymorphisms implicate mTOR signalling in the development of new-onset diabetes after transplantation. BBA Clin. 2016, 5, 41–45. [Google Scholar] [CrossRef]
- Chen, Q.; Hao, W.; Xiao, C.; Wang, R.; Xu, X.; Lu, H.; Chen, W.; Deng, C.X. SIRT6 Is Essential for Adipocyte Differentiation by Regulating Mitotic Clonal Expansion. Cell Rep. 2017, 18, 3155–3166. [Google Scholar] [CrossRef]
- Del Rio-Moreno, M.; Luque, R.M.; Rangel-Zuniga, O.A.; Alors-Perez, E.; Alcala-Diaz, J.F.; Roncero-Ramos, I.; Camargo, A.; Gahete, M.D.; Lopez-Miranda, J.; Castano, J.P. Dietary Intervention Modulates the Expression of Splicing Machinery in Cardiovascular Patients at High Risk of Type 2 Diabetes Development: From the CORDIOPREV Study. Nutrients 2020, 12, 3528. [Google Scholar] [CrossRef] [PubMed]
- Goo, Y.H.; Son, S.H.; Yechoor, V.K.; Paul, A. Transcriptional Profiling of Foam Cells Reveals Induction of Guanylate-Binding Proteins Following Western Diet Acceleration of Atherosclerosis in the Absence of Global Changes in Inflammation. J. Am. Heart Assoc. 2016, 5, e002663. [Google Scholar] [CrossRef] [PubMed]
- Gurung, R.L.; FitzGerald, L.M.; Liu, E.; McComish, B.J.; Kaidonis, G.; Ridge, B.; Hewitt, A.W.; Vote, B.J.; Verma, N.; Craig, J.E.; et al. Identifying Genetic Biomarkers Predicting Response to Anti-Vascular Endothelial Growth Factor Injections in Diabetic Macular Edema. Int. J. Mol. Sci. 2022, 23, 4042. [Google Scholar] [CrossRef]
- Hebbar, P.; Abubaker, J.A.; Abu-Farha, M.; Alsmadi, O.; Elkum, N.; Alkayal, F.; John, S.E.; Channanath, A.; Iqbal, R.; Pitkaniemi, J.; et al. Genome-wide landscape establishes novel association signals for metabolic traits in the Arab population. Hum. Genet. 2021, 140, 505–528. [Google Scholar] [CrossRef] [PubMed]
- Hongbo, M.; Yanjiao, D.; Shuo, W.; Kun, S.; Yanjie, L.; Mengmeng, L. Podocyte RNF166 deficiency alleviates diabetic nephropathy by mitigating mitochondria impairment and apoptosis via regulation of CYLD signal. Biochem. Biophys. Res. Commun. 2021, 545, 46–53. [Google Scholar] [CrossRef]
- Huang, Q.; Deng, G.; Wei, R.; Wang, Q.; Zou, D.; Wei, J. Comprehensive Identification of Key Genes Involved in Development of Diabetes Mellitus-Related Atherogenesis Using Weighted Gene Correlation Network Analysis. Front. Cardiovasc. Med. 2020, 7, 580573. [Google Scholar] [CrossRef]
- Jee, D.; Kang, S.; Park, S. Association of Age-Related Cataract Risk with High Polygenetic Risk Scores Involved in Galactose-Related Metabolism and Dietary Interactions. Lifestyle Genom. 2022, 15, 55–66. [Google Scholar] [CrossRef]
- Li, J.; Zhang, P.; Xia, Y. Study on CCDC69 interfering with the prognosis of patients with breast cancer through PPAR signal pathway. Eur. J. Histochem. EJH 2021, 65. [Google Scholar] [CrossRef]
- Li, X.Q.; Huang, T.Y. Notoginsenoside R1 alleviates high glucose-induced inflammation and oxidative stress in HUVECs via upregulating miR-147a. Kaohsiung J. Med. Sci. 2021, 37, 1101–1112. [Google Scholar] [CrossRef]
- Min, J.; Zeng, T.; Roux, M.; Lazar, D.; Chen, L.; Tudzarova, S. The Role of HIF1alpha-PFKFB3 Pathway in Diabetic Retinopathy. J. Clin. Endocrinol. Metab. 2021, 106, 2505–2519. [Google Scholar] [CrossRef]
- Ng, M.C.Y.; Graff, M.; Lu, Y.; Justice, A.E.; Mudgal, P.; Liu, C.T.; Young, K.; Yanek, L.R.; Feitosa, M.F.; Wojczynski, M.K.; et al. Discovery and fine-mapping of adiposity loci using high density imputation of genome-wide association studies in individuals of African ancestry: African Ancestry Anthropometry Genetics Consortium. PLoS Genet. 2017, 13, e1006719. [Google Scholar] [CrossRef] [PubMed]
- Qu, H.Q.; Qu, J.; Vaccaro, C.; Chang, X.; Mentch, F.; Li, J.; Mafra, F.; Nguyen, K.; Gonzalez, M.; March, M.; et al. Genetic Analysis for Type 1 Diabetes Genes in Juvenile Dermatomyositis Unveils Genetic Disease Overlap. Rheumatology 2022, keac100. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Perez, J.M.; Posadas-Sanchez, R.; Blachman-Braun, R.; Vargas-Alarcon, G.; Posadas-Romero, C.; Rodriguez-Cortes, A.A.; Lopez-Bautista, F.; Tovilla-Zarate, C.A.; Rojas-Toledo, E.X.; Borgonio-Cuadra, V.M.; et al. HHIPL-1 (rs2895811) gene polymorphism is associated with cardiovascular risk factors and cardiometabolic parameters in Mexicans patients with myocardial infarction. Gene 2018, 663, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Sajuthi, S.P.; Sharma, N.K.; Comeau, M.E.; Chou, J.W.; Bowden, D.W.; Freedman, B.I.; Langefeld, C.D.; Parks, J.S.; Das, S.K. Genetic regulation of adipose tissue transcript expression is involved in modulating serum triglyceride and HDL-cholesterol. Gene 2017, 632, 50–58. [Google Scholar] [CrossRef]
- Tang, G.Y.; Yu, P.; Zhang, C.; Deng, H.Y.; Lu, M.X.; Le, J.H. The Neuropeptide-Related HERC5/TAC1 Interactions May Be Associated with the Dysregulation of lncRNA GAS5 Expression in Gestational Diabetes Mellitus Exosomes. Dis. Markers 2022, 2022, 8075285. [Google Scholar] [CrossRef]
- Vivot, K.; Moulle, V.S.; Zarrouki, B.; Tremblay, C.; Mancini, A.D.; Maachi, H.; Ghislain, J.; Poitout, V. The regulator of G-protein signaling RGS16 promotes insulin secretion and β-cell proliferation in rodent and human islets. Mol. Metab. 2016, 5, 988–996. [Google Scholar] [CrossRef]
- Wang, L.; Lei, L.; Xu, T.; Wang, Y. GSTO1 regulates insulin biosynthesis in pancreatic β cells. Biochem. Biophys. Res. Commun. 2020, 524, 936–942. [Google Scholar] [CrossRef]
- Chen, J.; Sun, M.; Adeyemo, A.; Pirie, F.; Carstensen, T.; Pomilla, C.; Doumatey, A.P.; Chen, G.; Young, E.H.; Sandhu, M.; et al. Genome-wide association study of type 2 diabetes in Africa. Diabetologia 2019, 62, 1204–1211. [Google Scholar] [CrossRef]
- Tabassum, R.; Chauhan, G.; Dwivedi, O.P.; Mahajan, A.; Jaiswal, A.; Kaur, I.; Bandesh, K.; Singh, T.; Mathai, B.J.; Pandey, Y.; et al. Genome-wide association study for type 2 diabetes in Indians identifies a new susceptibility locus at 2q21. Diabetes 2013, 62, 977–986. [Google Scholar] [CrossRef]
- Saxena, R.; Saleheen, D.; Been, L.F.; Garavito, M.L.; Braun, T.; Bjonnes, A.; Young, R.; Ho, W.K.; Rasheed, A.; Frossard, P.; et al. Genome-wide association study identifies a novel locus contributing to type 2 diabetes susceptibility in Sikhs of Punjabi origin from India. Diabetes 2013, 62, 1746–1755. [Google Scholar] [CrossRef]
- Venkatesan, V.; Lopez-Alvarenga, J.C.; Arya, R.; Ramu, D.; Koshy, T.; Ravichandran, U.; Ponnala, A.R.; Sharma, S.K.; Lodha, S.; Sharma, K.K.; et al. Burden of Type 2 Diabetes and Associated Cardiometabolic Traits and Their Heritability Estimates in Endogamous Ethnic Groups of India: Findings From the INDIGENIUS Consortium. Front. Endocrinol. 2022, 13, 847692. [Google Scholar] [CrossRef] [PubMed]
- Shpakov, A.O.; Derkach, K.V.; Berstein, L.M. Brain signaling systems in the Type 2 diabetes and metabolic syndrome: Promising target to treat and prevent these diseases. Future Sci. OA 2015, 1, FSO25. [Google Scholar] [CrossRef] [PubMed]
- Hao, K.; Di Narzo, A.F.; Ho, L.; Luo, W.; Li, S.; Chen, R.; Li, T.; Dubner, L.; Pasinetti, G.M. Shared genetic etiology underlying Alzheimer’s disease and type 2 diabetes. Mol. Asp. Med. 2015, 43–44, 66–76. [Google Scholar] [CrossRef] [PubMed]
- Rahimi, M.; Vinciguerra, M.; Daghighi, M.; Ozcan, B.; Akbarkhanzadeh, V.; Sheedfar, F.; Amini, M.; Mazza, T.; Pazienza, V.; Motazacker, M.M.; et al. Age-related obesity and type 2 diabetes dysregulate neuronal associated genes and proteins in humans. Oncotarget 2015, 6, 29818–29832. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Santulli, G.; Pagano, G.; Sardu, C.; Xie, W.; Reiken, S.; D’Ascia, S.L.; Cannone, M.; Marziliano, N.; Trimarco, B.; Guise, T.A.; et al. Calcium release channel RyR2 regulates insulin release and glucose homeostasis. J. Clin. Investig. 2015, 125, 1968–1978. [Google Scholar] [CrossRef] [PubMed]
- Bruno, E.; Manoukian, S.; Venturelli, E.; Oliverio, A.; Rovera, F.; Iula, G.; Morelli, D.; Peissel, B.; Azzolini, J.; Roveda, E.; et al. Adherence to Mediterranean Diet and Metabolic Syndrome in BRCA Mutation Carriers. Integr. Cancer Ther. 2018, 17, 153–160. [Google Scholar] [CrossRef]
- Yin, Q.; Ni, Q.; Wang, Y.; Zhang, H.; Li, W.; Nie, A.; Wang, S.; Gu, Y.; Wang, Q.; Ning, G. Raptor determines β-cell identity and plasticity independent of hyperglycemia in mice. Nat. Commun. 2020, 11, 2538. [Google Scholar] [CrossRef]
- Cras-Meneur, C.; Conlon, M.; Zhang, Y.; Pasca Di Magliano, M.; Bernal-Mizrachi, E. Early pancreatic islet fate and maturation is controlled through RBP-Jkappa. Sci. Rep. 2016, 6, 26874. [Google Scholar] [CrossRef]
- Bonal, C.; Herrera, P.L. Understanding the extrinsic and intrinsic signals involved in pancreas and β-cell development: From endoderm to β cells. Curr. Opin. Organ Transplant. 2007, 12, 40–48. [Google Scholar] [CrossRef]
- Goth, L.; Nagy, T.; Kosa, Z.; Fejes, Z.; Bhattoa, H.P.; Paragh, G.; Kaplar, M. Effects of rs769217 and rs1001179 polymorphisms of catalase gene on blood catalase, carbohydrate and lipid biomarkers in diabetes mellitus. Free Radic. Res. 2012, 46, 1249–1257. [Google Scholar] [CrossRef]
- Flekac, M.; Skrha, J.; Hilgertova, J.; Lacinova, Z.; Jarolimkova, M. Gene polymorphisms of superoxide dismutases and catalase in diabetes mellitus. BMC Med. Genet. 2008, 9, 30. [Google Scholar] [CrossRef] [PubMed]
- Musicant, A.M.; Parag-Sharma, K.; Gong, W.; Sengupta, M.; Chatterjee, A.; Henry, E.C.; Tsai, Y.H.; Hayward, M.C.; Sheth, S.; Betancourt, R.; et al. CRTC1/MAML2 directs a PGC-1alpha-IGF-1 circuit that confers vulnerability to PPARgamma inhibition. Cell Rep. 2021, 34, 108768. [Google Scholar] [CrossRef] [PubMed]
- White, P.; May, C.L.; Lamounier, R.N.; Brestelli, J.E.; Kaestner, K.H. Defining pancreatic endocrine precursors and their descendants. Diabetes 2008, 57, 654–668. [Google Scholar] [CrossRef] [PubMed]
- Macgregor, S. Most pooling variation in array-based DNA pooling is attributable to array error rather than pool construction error. Eur. J. Hum. Genet. 2007, 15, 501–504. [Google Scholar] [CrossRef] [PubMed][Green Version]
| Pool Id | T2D | CO | HTN | DL | n | % |
|---|---|---|---|---|---|---|
| 1 | No | No | No | No | 190 | 13.55 |
| 2 | No | No | No | Yes | 49 | 3.50 |
| 3 | No | No | Yes | No | 65 | 4.64 |
| 4 | No | No | Yes | Yes | 30 | 2.14 |
| 5 | No | Yes | No | No | 230 | 16.41 |
| 6 | No | Yes | No | Yes | 90 | 6.42 |
| 7 | No | Yes | Yes | No | 224 | 15.98 |
| 8 | No | Yes | Yes | Yes | 106 | 7.56 |
| 9 | Yes | No | No | No | 22 | 1.57 |
| Yes | No | No | Yes | 7 | 0.50 | |
| 10 | Yes | No | Yes | No | 19 | 1.36 |
| Yes | No | Yes | Yes | 11 | 0.78 | |
| 11 | Yes | Yes | No | No | 38 | 2.71 |
| 12 | Yes | Yes | No | Yes | 34 | 2.43 |
| 13 | Yes | Yes | Yes | No | 179 | 12.77 |
| 14 | Yes | Yes | Yes | Yes | 108 | 7.70 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pipal, K.V.; Mamtani, M.; Patel, A.A.; Jaiswal, S.G.; Jaisinghani, M.T.; Kulkarni, H. Susceptibility Loci for Type 2 Diabetes in the Ethnically Endogamous Indian Sindhi Population: A Pooled Blood Genome-Wide Association Study. Genes 2022, 13, 1298. https://doi.org/10.3390/genes13081298
Pipal KV, Mamtani M, Patel AA, Jaiswal SG, Jaisinghani MT, Kulkarni H. Susceptibility Loci for Type 2 Diabetes in the Ethnically Endogamous Indian Sindhi Population: A Pooled Blood Genome-Wide Association Study. Genes. 2022; 13(8):1298. https://doi.org/10.3390/genes13081298
Chicago/Turabian StylePipal, Kanchan V., Manju Mamtani, Ashwini A. Patel, Sujeet G. Jaiswal, Manisha T. Jaisinghani, and Hemant Kulkarni. 2022. "Susceptibility Loci for Type 2 Diabetes in the Ethnically Endogamous Indian Sindhi Population: A Pooled Blood Genome-Wide Association Study" Genes 13, no. 8: 1298. https://doi.org/10.3390/genes13081298
APA StylePipal, K. V., Mamtani, M., Patel, A. A., Jaiswal, S. G., Jaisinghani, M. T., & Kulkarni, H. (2022). Susceptibility Loci for Type 2 Diabetes in the Ethnically Endogamous Indian Sindhi Population: A Pooled Blood Genome-Wide Association Study. Genes, 13(8), 1298. https://doi.org/10.3390/genes13081298





