Differential Methylation Profile in Fragile X Syndrome-Prone Offspring Mice after in Utero Exposure to Lactobacillus Reuteri
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mouse Models
2.2. DNA Methylation Array Processing and Mapping MethylationEPIC Primer to the Mouse Genome
2.3. Methylation Data Preprocessing
2.4. Identification of Differentially Methylated CpGs (DMs)
2.5. Pathway Analysis
2.6. Constructing a Functional Association Network for Some of the Proteins Involved in Brain Functions Using String
2.7. Western Blot Analysis
3. Results
3.1. Analysis
3.1.1. Mouse Models
3.1.2. Identification of Perfectly Mapping Probes to the Mouse Genome
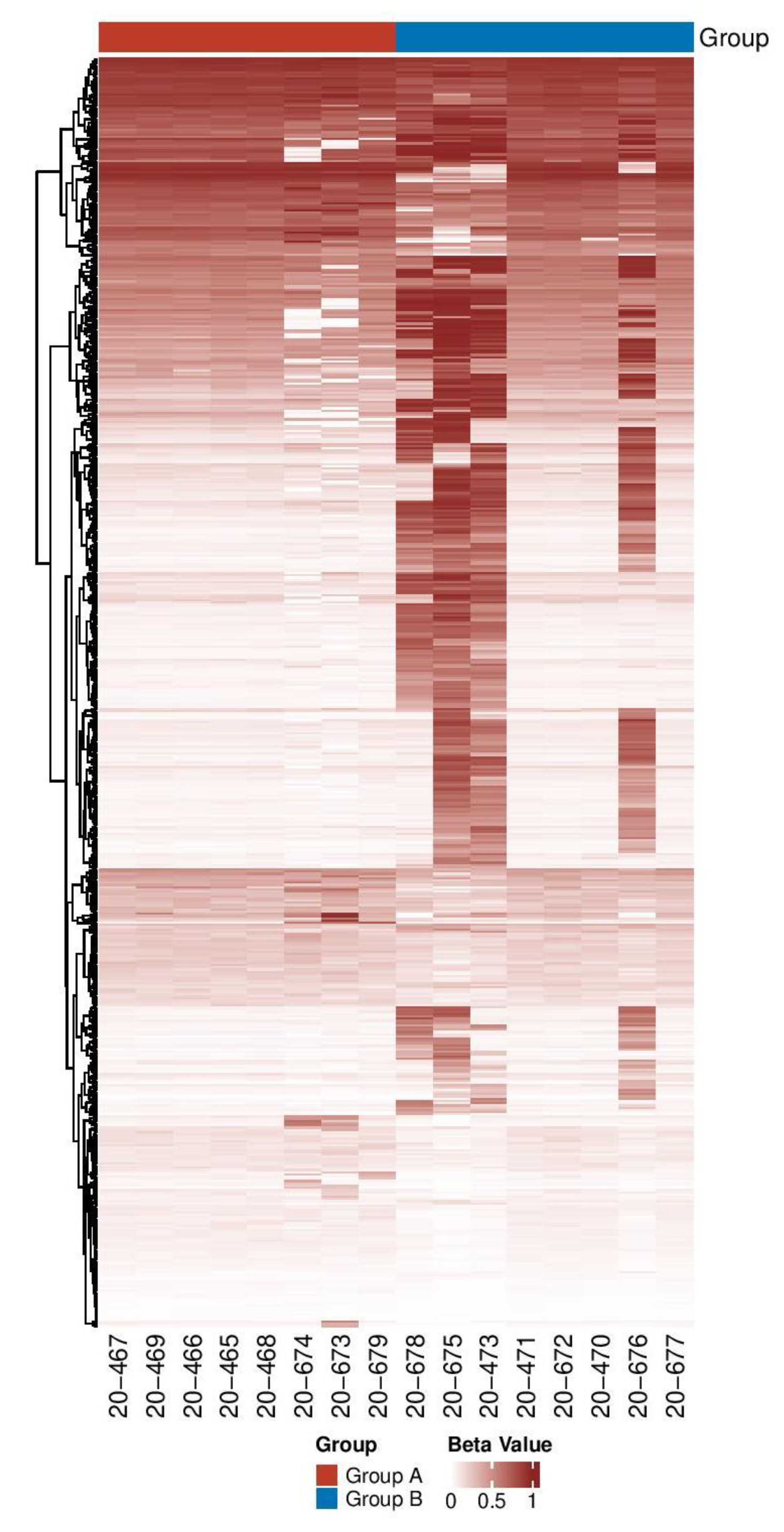
3.1.3. Identification and Distribution of DMs among the Two Groups
3.1.4. Differentially Methylated Genes in FXS-Like Mice Are Involved in Brain Functions
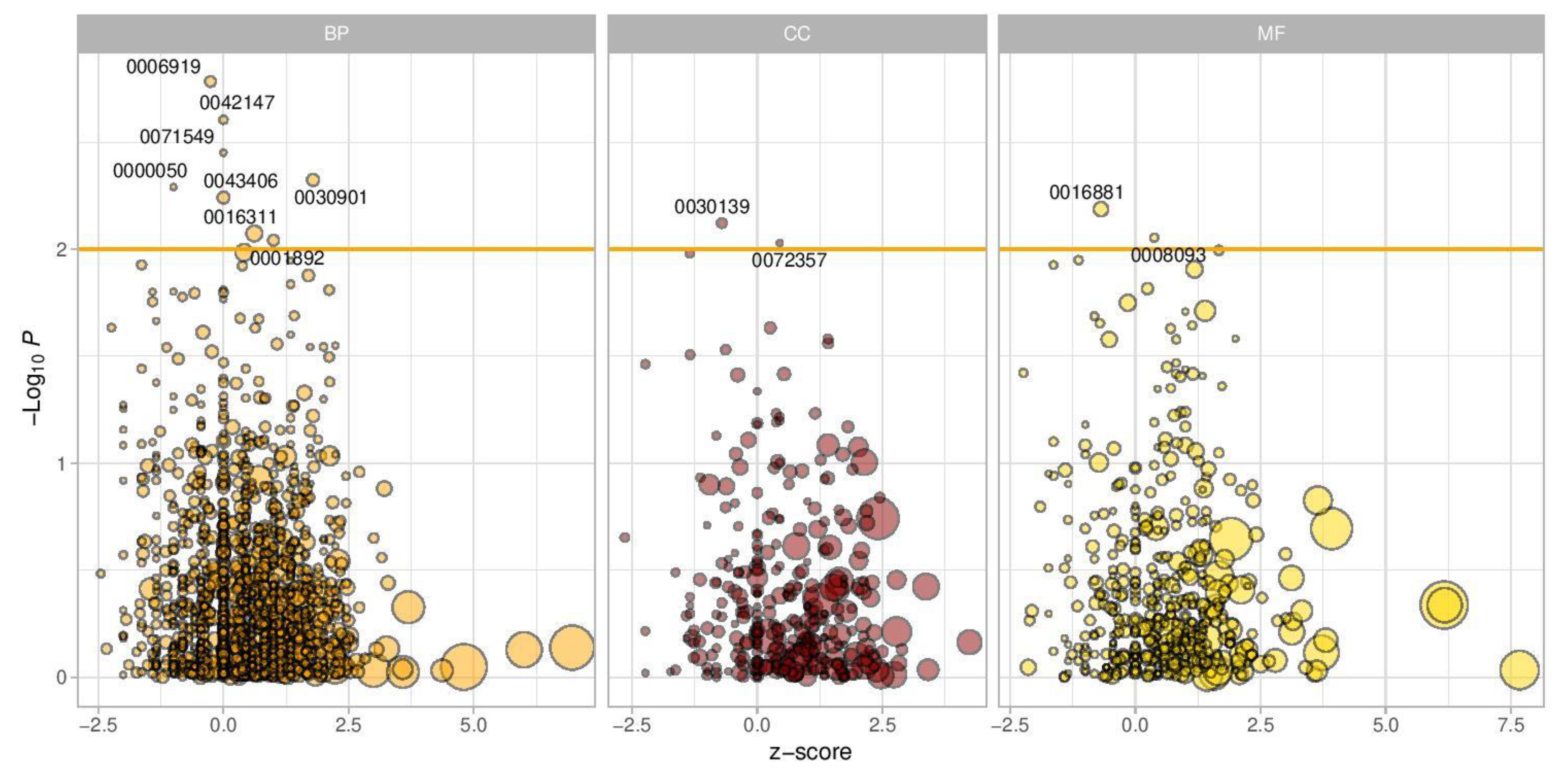
3.1.5. Genes and Pathway Enrichment Analysis
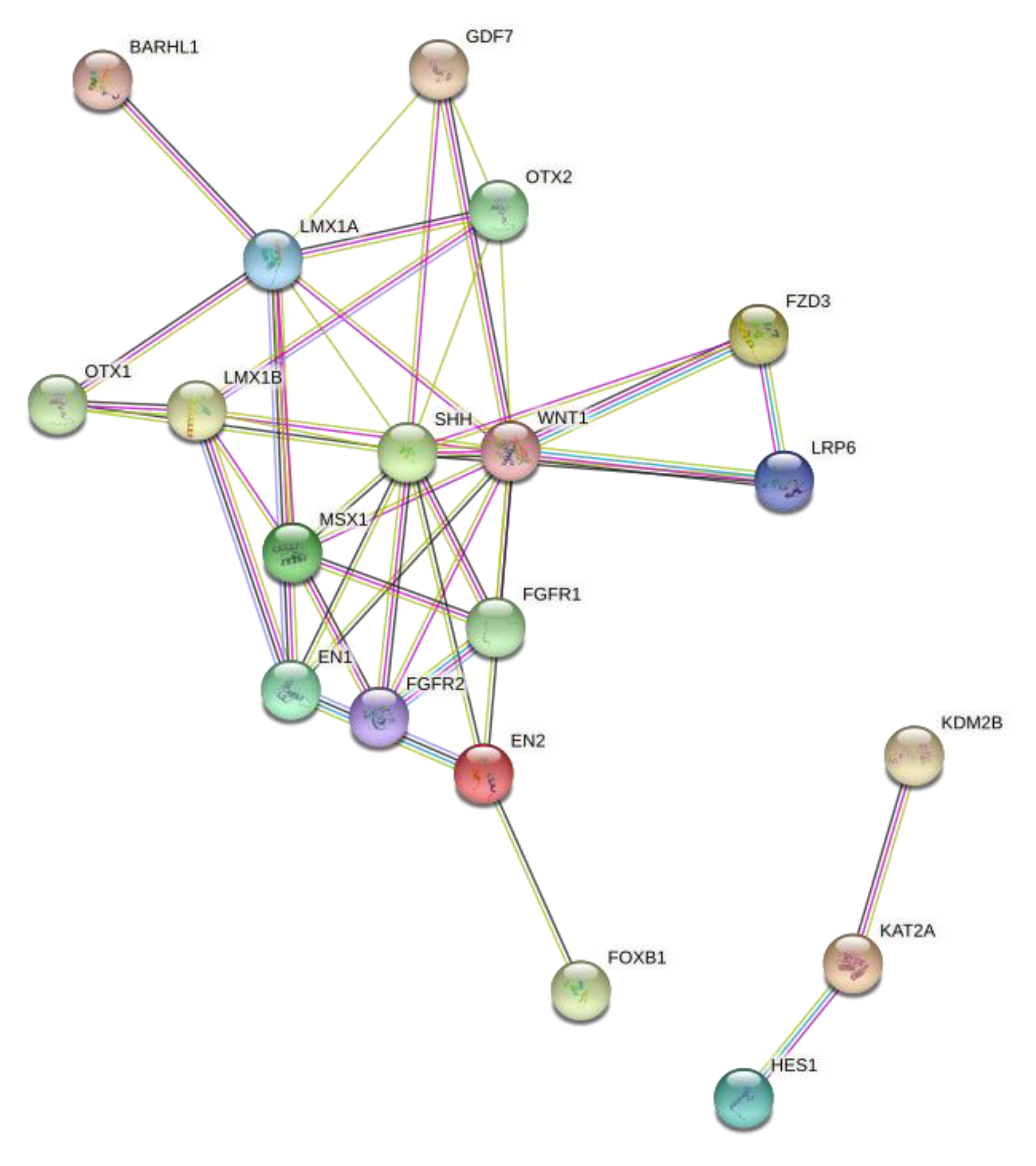
3.1.6. String-Based Protein–Protein Interaction Network Analysis
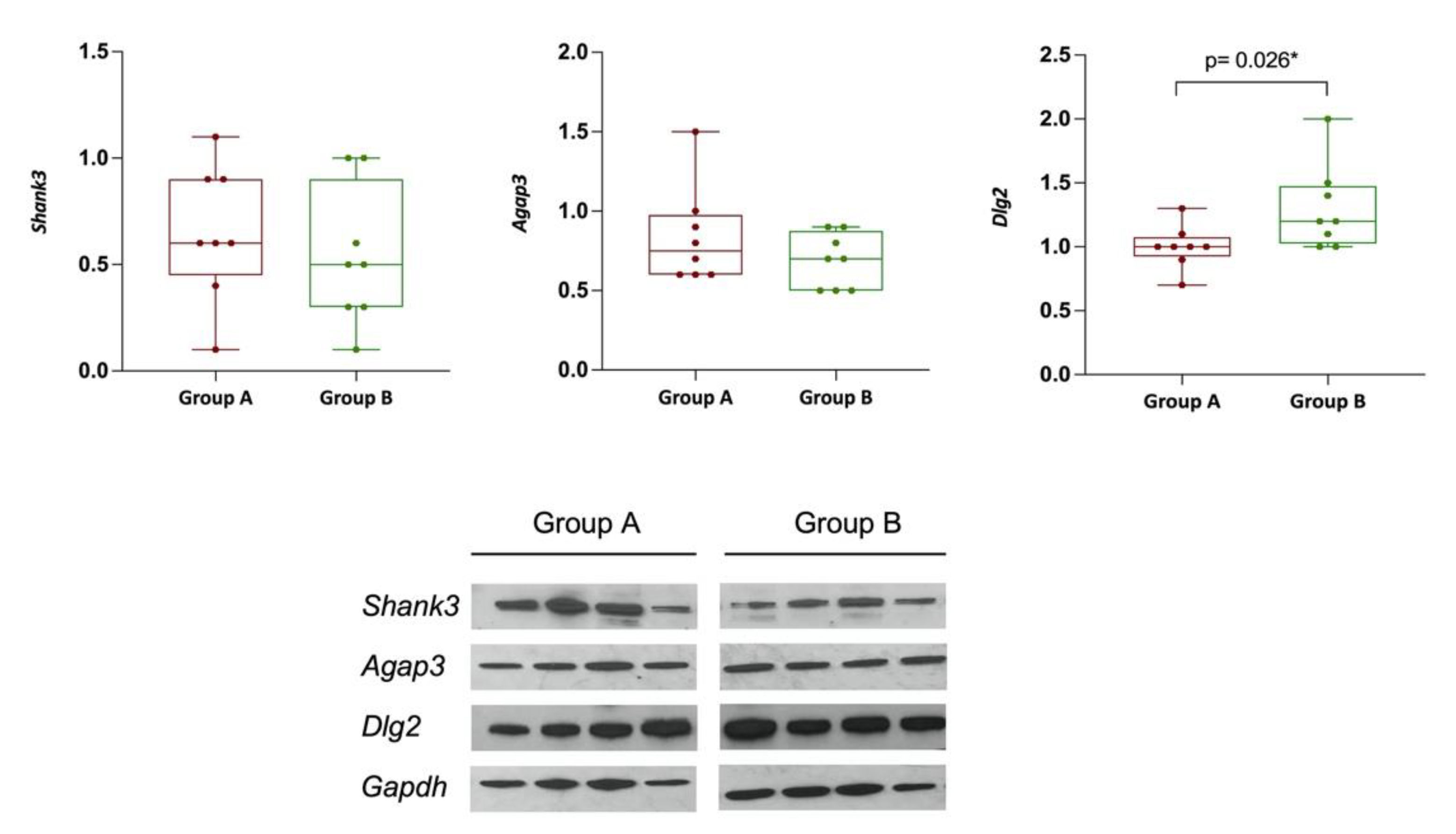
3.1.7. DLG2, SHANK3, and AGAP3 Protein Expression
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Rylaarsdam, L.; Guemez-Gamboa, A. Genetic Causes and Modifiers of Autism Spectrum Disorder. Front. Cell. Neurosci. 2019, 13, 385. [Google Scholar] [CrossRef] [PubMed]
- Bollati, V.; Baccarelli, A. Environmental epigenetics. Heredity 2010, 105, 105–112. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atladóttir, H.Ó.; Henriksen, T.B.; Schendel, D.E.; Parner, E.T. Autism After Infection, Febrile Episodes, and Antibiotic Use During Pregnancy: An Exploratory Study. Pediatrics 2012, 130, e1447–e1454. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.S.; Sourander, A.; Hinkka-Yli-Salomäki, S.; McKeague, I.W.; Sundvall, J.; Surcel, H.-M. Elevated maternal C-reactive protein and autism in a national birth cohort. Mol. Psychiatry 2014, 19, 259–264. [Google Scholar] [CrossRef]
- Cornish, K.M.; Kraan, C.M.; Bui, Q.M.; Bellgrove, M.A.; Metcalfe, S.A.; Trollor, J.N.; Hocking, D.R.; Slater, H.R.; Inaba, Y.; Li, X.; et al. Novel methylation markers of the dysexecutive-psychiatric phenotype in FMR1 premutation women. Neurology 2015, 84, 1631–1638. [Google Scholar] [CrossRef] [Green Version]
- Lu, J.; Gao, F.-H. Role and molecular mechanism of heterogeneous nuclear ribonucleoprotein K in tumor development and progression. Biomed. Rep. 2016, 4, 657–663. [Google Scholar] [CrossRef] [Green Version]
- Paul, B.; Barnes, S.; Demark-Wahnefried, W.; Morrow, C.D.; Salvador, C.; Skibola, C.F.; Tollefsbol, T.O. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenet. 2015, 7, 112. [Google Scholar] [CrossRef] [Green Version]
- Lee, H.-S. The interaction between gut microbiome and nutrients on development of human disease through epigenetic mechanisms. Genom. Inform. 2019, 17, e24. [Google Scholar] [CrossRef] [PubMed]
- Schächtle, M.A.; Rosshart, S.P. The Microbiota-Gut-Brain Axis in Health and Disease and Its Implications for Translational Research. Front. Cell. Neurosci. 2021, 15, 698172. [Google Scholar] [CrossRef]
- Zheng, D.; Liwinski, T.; Elinav, E. Interaction between microbiota and immunity in health and disease. Cell Res. 2020, 30, 492–506. [Google Scholar] [CrossRef]
- Suganya, K.; Koo, B.-S. Gut–Brain Axis: Role of Gut Microbiota on Neurological Disorders and How Probiotics/Prebiotics Beneficially Modulate Microbial and Immune Pathways to Improve Brain Functions. Int. J. Mol. Sci. 2020, 21, 7551. [Google Scholar] [CrossRef] [PubMed]
- Choi, G.B.; Yim, Y.S.; Kim, V.; Hoeffer, C.A.; Littman, D.R.; Huh, J.R. The maternal interleukin-17a pathway in mice promotes autismlike phenotypes in offspring. Science 2016, 351, 933–939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef]
- Szkaradkiewicz, A.K.; Stopa, J.; Karpiński, T.M. Effect of Oral Administration Involving a Probiotic Strain of Lactobacillus reuteri on Pro-Inflammatory Cytokine Response in Patients with Chronic Periodontitis. Arch. Immunol. Ther. Exp. 2014, 62, 495–500. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, S.W.; Hessl, D.; Goodlin-Jones, B.; Ferranti, J.; Bacalman, S.; Barbato, I.; Tassone, F.; Hagerman, P.J.; Herman, K.; Hagerman, R.J. Autism Profiles of Males With Fragile X Syndrome. Am. J. Ment. Retard. 2008, 113, 427–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wheeler, A.C.; Mussey, J.; Villagomez, A.; Bishop, E.; Raspa, M.; Edwards, A.; Bodfish, J.; Bann, C.; Bailey, D.B. DSM-5 Changes and the Prevalence of Parent-Reported Autism Spectrum Symptoms in Fragile X Syndrome. J. Autism Dev. Disord. 2015, 45, 816–829. [Google Scholar] [CrossRef]
- Verheij, C.; Bakker, C.; De Graaff, E.; Keulemans, J.; Willemsen, R.; Verkerk, A.J.M.H.; Galjaard, H.; Reuser, A. Characterization and localization of the FMR-1 gene product associated with fragile X syndrome. Nature 1993, 363, 722–724. [Google Scholar] [CrossRef]
- Kraan, C.M.; Hocking, D.R.; Georgiou-Karistianis, N.; Metcalfe, S.A.; Archibald, A.D.; Fielding, J.; Trollor, J.; Bradshaw, J.L.; Cohen, J.; Cornish, K.M. Age and CGG-repeat length are associated with neuromotor impairments in at-risk females with the FMR1 premutation. Neurobiol. Aging 2014, 35, 2179.e7–2179.e13. [Google Scholar] [CrossRef]
- Poutahidis, T.; Varian, B.J.; Levkovich, T.; Lakritz, J.R.; Mirabal, S.; Kwok, C.; Ibrahim, Y.M.; Kearney, S.M.; Chatzigiagkos, A.; Alm, E.J.; et al. Dietary Microbes Modulate Transgenerational Cancer Risk. Cancer Res. 2015, 75, 1197–1204. [Google Scholar] [CrossRef] [Green Version]
- Gujar, H.; Liang, J.W.; Wong, N.C.; Mozhui, K. Profiling DNA methylation differences between inbred mouse strains on the Illumina Human Infinium MethylationEPIC microarray. PLoS ONE 2018, 13, e0193496. [Google Scholar] [CrossRef] [Green Version]
- Needhamsen, M.; Ewing, E.; Lund, H.; Gomez-Cabrero, D.; Harris, R.A.; Kular, L.; Jagodic, M. Usability of human Infinium MethylationEPIC BeadChip for mouse DNA methylation studies. BMC Bioinform. 2017, 18, 486. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fortin, J.P.; Triche, T.J.; Hansen, K.D. Preprocessing, normalization and integration of the Illumina HumanMethylationEPIC array with minfi. Bioinformatics 2017, 33, 558–560. [Google Scholar] [CrossRef] [PubMed]
- Du, P.; Zhang, X.; Huang, C.-C.; Jafari, N.; Kibbe, A.W.; Hou, L.; Lin, S.M. Comparison of Beta-value and M-value methods for quantifying methylation levels by microarray analysis. BMC Bioinform. 2010, 11, 587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Classics in the History of Psychology—Fisher (1925) Index. Available online: https://psychclassics.yorku.ca/Fisher/Methods/index.htm (accessed on 1 October 2021).
- Lai, L.; Hennessey, J.; Bares, V.; Son, E.W.; Ban, Y.; Wang, W.; Qi, J.; Jiang, G.; Liberzon, A.; Ge, S.X. GSKB: A gene set database for pathway analysis in mouse. bioRxiv 2016, 082511. [Google Scholar] [CrossRef] [Green Version]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P.; et al. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Prieto, C.A.; Álvarez-Errico, D.; Musulen, E.; Bueno-Costa, A.; Vazquez, B.N.; Vaquero, A.; Esteller, M. Validation of a DNA methylation microarray for 285,000 CpG sites in the mouse genome. Epigenetics 2022, 1–9. [Google Scholar] [CrossRef]
- Oku, Y.; Huganir, R.L. AGAP3 and Arf6 Regulate Trafficking of AMPA Receptors and Synaptic Plasticity. J. Neurosci. 2013, 33, 12586–12598. [Google Scholar] [CrossRef] [Green Version]
- Gross, C.; Chang, C.-W.; Kelly, S.M.; Bhattacharya, A.; McBride, S.M.; Danielson, S.W.; Jiang, M.Q.; Chan, C.B.; Ye, K.; Gibson, J.R.; et al. Increased Expression of the PI3K Enhancer PIKE Mediates Deficits in Synaptic Plasticity and Behavior in Fragile X Syndrome. Cell Rep. 2016, 11, 727–736. [Google Scholar] [CrossRef] [Green Version]
- Manuel, M.N.; Mi, D.; Mason, J.O.; Price, D.J. Regulation of cerebral cortical neurogenesis by the Pax6 transcription factor. Front. Cell. Neurosci. 2015, 9, 70. [Google Scholar] [CrossRef] [Green Version]
- Jones, K.A.; Luo, Y.; Dukes-Rimsky, L.; Srivastava, D.P.; Koul-Tewari, R.; Russell, T.A.; Shapiro, L.P.; Srivastava, A.K.; Penzes, P. Neurodevelopmental disorder-associated ZBTB20 gene variants affect dendritic and synaptic structure. PLoS ONE 2018, 13, e0203760. [Google Scholar] [CrossRef]
- Kantarci, H.; Edlund, R.K.; Groves, A.K.; Riley, B.B. Tfap2a Promotes Specification and Maturation of Neurons in the Inner Ear through Modulation of Bmp, Fgf and Notch Signaling. PLoS Genet. 2015, 11, e1005037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, J.; Liu, Y.; Baudry, M.; Bi, X. SK2 channel regulation of neuronal excitability, synaptic transmission, and brain rhythmic activity in health and diseases. Biochim. Biophys. Acta Mol. Cell Res. 2020, 1867, 118834. [Google Scholar] [CrossRef] [PubMed]
- Favaro, P.D.; Huang, X.; Hosang, L.; Stodieck, S.; Cui, L.; Liu, Y.-Z.; Engelhardt, K.-A.; Schmitz, F.; Dong, Y.; Löwel, S.; et al. An opposing function of paralogs in balancing developmental synapse maturation. PLOS Biol. 2018, 16, e2006838. [Google Scholar] [CrossRef] [PubMed]
- Yoo, T.; Kim, S.G.; Yang, S.H.; Kim, H.; Kim, E.; Kim, S.Y. A DLG2 deficiency in mice leads to reduced sociability and increased repetitive behavior accompanied by aberrant synaptic transmission in the dorsal striatum. Mol. Autism 2020, 11, 19. [Google Scholar] [CrossRef]
- Ali, S.; Hoven, A.; Dress, R.J.; Schaal, H.; Alferink, J.; Scheu, S. Identification of a novel Dlg2 isoform differentially expressed in IFNβ-producing plasmacytoid dendritic cells. BMC Genom. 2018, 19, 194. [Google Scholar] [CrossRef]
- Dillon, G.M.; Tyler, W.A.; Omuro, K.C.; Kambouris, J.; Tyminski, C.; Henry, S.; Haydar, T.; Beffert, U.; Ho, A. CLASP2 Links Reelin to the Cytoskeleton during Neocortical Development. Neuron 2017, 93, 1344–1358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, X.; Hou, D.; Jiang, W.; Zhang, C. Intercellular protein–protein interactions at synapses. Protein Cell 2014, 5, 420–444. [Google Scholar] [CrossRef] [Green Version]
- Meyer, R.; Begemann, M.; Demuth, S.; Kraft, F.; Dey, D.; Schüler, H.; Busse, S.; Häusler, M.; Zerres, K.; Kurth, I.; et al. Inherited cases of CNOT3 -associated intellectual developmental disorder with speech delay, autism, and dysmorphic facies. Clin. Genet. 2020, 98, 408–412. [Google Scholar] [CrossRef]
- Zhang, Q.; Peng, C.; Song, J.; Zhang, Y.; Chen, J.; Song, Z.; Shou, X.; Ma, Z.; Peng, H.; Jian, X.; et al. Germline Mutations in CDH23, Encoding Cadherin-Related 23, Are Associated with Both Familial and Sporadic Pituitary Adenomas. Am. J. Hum. Genet. 2017, 100, 817–823. [Google Scholar] [CrossRef] [Green Version]
- Zhu, L.; Wang, X.; Li, X.-L.; Towers, A.; Cao, X.; Wang, P.; Bowman, R.; Yang, H.; Goldstein, J.; Li, Y.-J.; et al. Epigenetic dysregulation of SHANK3 in brain tissues from individuals with autism spectrum disorders. Hum. Mol. Genet. 2014, 23, 1563–1578. [Google Scholar] [CrossRef] [Green Version]
- Kovács, R.A.; Vadászi, H.; Bulyáki, É.; Török, G.; Tóth, V.; Mátyás, D.; Kun, J.; Hunyadi-Gulyás, É.; Fedor, F.Z.; Csincsi, Á.; et al. Identification of Neuronal Pentraxins as Synaptic Binding Partners of C1q and the Involvement of NP1 in Synaptic Pruning in Adult Mice. Front. Immunol. 2021, 11, 599771. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Ryan, T.A. Synaptic Vesicle Recycling at CNS Synapses without AP-2. J. Neurosci. 2009, 29, 3865–3874. [Google Scholar] [CrossRef] [PubMed]
- Mohn, J.L.; Alexander, J.; Pirone, A.; Palka, C.D.; Lee, S.-Y.; Mebane, L.; Haydon, P.G.; Jacob, M.H. Adenomatous polyposis coli protein deletion leads to cognitive and autism-like disabilities. Mol. Psychiatry 2014, 19, 1133–1142. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Carosso, G.A.; Boukas, L.; Augustin, J.J.; Nguyen, H.N. Transcriptional suppression from KMT2D loss disrupts cell cycle and hypoxic responses in neurodevelopmental models of Kabuki syndrome. bioRxiv 2018, 484410. Available online: https://www.biorxiv.org/content/early/2018/12/02/484410 (accessed on 17 June 2022).
- Fatima, A.; Hoeber, J.; Schuster, J.; Koshimizu, E.; Maya-Gonzalez, C.; Keren, B.; Mignot, C.; Akram, T.; Ali, Z.; Miyatake, S.; et al. Monoallelic and bi-allelic variants in NCDN cause neurodevelopmental delay, intellectual disability, and epilepsy. Am. J. Hum. Genet. 2021, 108, 739–748. [Google Scholar] [CrossRef] [PubMed]
- Tian, W.T.; Liu, L.-H.; Zhou, H.-Y.; Zhang, C.; Zhan, F.-X.; Zhu, Z.-Y.; Chen, S.-D.; Luan, X.-H.; Cao, L. New phenotype of DCTN1-related spectrum: Early-onset dHMN plus congenital foot deformity. Ann. Clin. Transl. Neurol. 2020, 7, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Haldeman-Englert, C.R.; Hurst, A.C.; Levine, M.A. Disorders of GNAS Inactivation. GeneReviews®. 2017. Available online: https://www.ncbi.nlm.nih.gov/books/NBK459117/ (accessed on 6 October 2021).
- Mulhern, M.S.; Stumpel, C.; Stong, N.; Brunner, H.G.; Bier, L.; Lippa, N.; Riviello, J.; Rouhl, R.P.W.; Kempers, M.; Pfundt, R.; et al. NBEA: Developmental disease gene with early generalized epilepsy phenotypes. Ann. Neurol. 2018, 84, 788–795. [Google Scholar] [CrossRef] [Green Version]
- Megarbane, A.; Noguier, F.; Stora, S.; Manchon, L.; Mircher, C.; Bruno, R.; Dorison, N.; Pierrat, F.; Réthoré, M.-O.; Trentin, B.; et al. The intellectual disability of trisomy 21: Differences in gene expression in a case series of patients with lower and higher IQ. Eur. J. Hum. Genet. 2013, 21, 1253–1259. [Google Scholar] [CrossRef] [Green Version]
- Piper, M.; Dawson, A.-L.S.; Lindwall, C.; Barry, G.; Plachez, C.; Richards, L.J. Emx and Nfi Genes Regulate Cortical Development and Axon Guidance in the Telencephalon. Cortical Dev. Genes Genet. Abnorm. 2008, 288, 230–245. [Google Scholar] [CrossRef]
- Arnold, M.; Cross, R.; Singleton, K.S.; Zlatic, S.; Chapleau, C.; Mullin, A.P.; Rolle, I.; Moore, C.C.; Theibert, A.; Pozzo-Miller, L.; et al. The Endosome Localized Arf-GAP AGAP1 Modulates Dendritic Spine Morphology Downstream of the Neurodevelopmental Disorder Factor Dysbindin. Front. Cell. Neurosci. 2016, 10, 218. [Google Scholar] [CrossRef] [Green Version]
- Marsh, A.P.L.; Heron, D.; Edwards, T.; Quartier, A.; Galea, C.; Nava, C.; Rastetter, A.; Moutard, M.-L.; Anderson, V.; Bitoun, P.; et al. Mutations in DCC cause isolated agenesis of the corpus callosum with incomplete penetrance. Nat. Genet. 2017, 49, 511–514. [Google Scholar] [CrossRef] [PubMed]
- Cerdó, T.; García-Valdés, L.; Altmäe, S.; Ruíz, A.; Suárez, A.; Campoy, C. Role of microbiota function during early life on child’s neurodevelopment. Trends Food Sci. Technol. 2016, 57, 273–288. [Google Scholar] [CrossRef]
- Laker, R.C.; Wlodek, M.E.; Connelly, J.; Yan, Z. Epigenetic origins of metabolic disease: The impact of the maternal condition to the offspring epigenome and later health consequences. Food Sci. Hum. Wellness 2013, 2, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Borre, Y.E.; O’Keeffe, G.W.; Clarke, G.; Stanton, C.; Dinan, T.G.; Cryan, J.F. Microbiota and neurodevelopmental windows: Implications for brain disorders. Trends Mol. Med. 2014, 20, 509–518. [Google Scholar] [CrossRef]
- Hornig, M.; Bresnahan, M.A.; Che, X.; Schultz, A.F.; Ukaigwe, J.E.; Eddy, M.L.; Hirtz, D.; Gunnes, N.; Lie, K.K.; Magnus, P.; et al. Prenatal fever and autism risk. Mol. Psychiatry 2018, 23, 759–766. [Google Scholar] [CrossRef]
- Brown, A.S.; Conway, F. Maternal immune activation and related factors in the risk of offspring psychiatric disorders. Front. Psychiatry 2019, 10, 430. [Google Scholar] [CrossRef]
- Paraschivescu, C.; Barbosa, S.; Lorivel, T.; Glaichenhaus, N.; Davidovic, L. Cytokine changes associated with the maternal immune activation (MIA) model of autism: A penalized regression approach. PLoS ONE 2020, 15, e0231609. [Google Scholar] [CrossRef]
- Patel, S.; Dale, R.C.; Rose, D.; Heath, B.; Nordahl, C.W.; Rogers, S.; Guastella, A.J.; Ashwood, P. Maternal immune conditions are increased in males with autism spectrum disorders and are associated with behavioural and emotional but not cognitive co-morbidity. Transl. Psychiatry 2020, 10, 286. [Google Scholar] [CrossRef]
- Wang, X.; Yang, J.; Zhang, H.; Yu, J.; Yao, Z. Oral probiotic administration during pregnancy prevents autism-related behaviors in offspring induced by maternal immune activation via anti-inflammation in mice. Autism Res. 2019, 12, 576–588. [Google Scholar] [CrossRef]
- Buffington, S.A.; Dooling, S.W.; Sgritta, M.; Noecker, C.; Murillo, O.D.; Felice, D.F.; Turnbaugh, P.J.; Costa-Mattioli, M. Dissecting the contribution of host genetics and the microbiome in complex behaviors. Cell 2021, 184, 1740–1756.e16. [Google Scholar] [CrossRef]
- Vuong, H.E.; Hsiao, E.Y. Gut Microbes Join the Social Network. Neuron 2019, 101, 196–198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sgritta, M.; Dooling, S.W.; Buffington, S.A.; Momin, E.N.; Francis, M.B.; Britton, R.A.; Costa-Mattioli, M. Mechanisms Underlying Microbial-Mediated Changes in Social Behavior in Mouse Models of Autism Spectrum Disorder. Neuron 2019, 101, 246–259.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schilling, E.; El Chartouni, C.; Rehli, M. Allele-specific DNA methylation in mouse strains is mainly determined by cis-acting sequences. Genome Res. 2009, 19, 2028–2035. [Google Scholar] [CrossRef] [Green Version]
- Agarwal, A.K.; Tunison, K.; Dalal, J.S.; Nagamma, S.S.; Hamra, F.K.; Sankella, S.; Shao, X.; Auchus, R.J.; Garg, A. Metabolic, Reproductive, and Neurologic Abnormalities in Agpat1-Null Mice. Endocrinology 2017, 158, 3954–3973. [Google Scholar] [CrossRef] [Green Version]
- Jansen, S.; Van Der Werf, I.M.; Innes, A.M.; Afenjar, A.; Agrawal, P.B.; Anderson, I.J.; Atwal, P.S.; Van Binsbergen, E.; Boogaard, M.-J.V.D.; Castiglia, L.; et al. De novo variants in FBXO11 cause a syndromic form of intellectual disability with behavioral problems and dysmorphisms. Eur. J. Hum. Genet. 2019, 27, 738–746. [Google Scholar] [CrossRef] [PubMed]
- Yoshizaki, K.; Furuse, T.; Kimura, R.; Tucci, V.; Kaneda, H.; Wakana, S.; Osumi, N. Paternal Aging Affects Behavior in Pax6 Mutant Mice: A Gene/Environment Interaction in Understanding Neurodevelopmental Disorders. PLoS ONE 2016, 11, e0166665. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, A.H.; Rasmussen, H.B.; Silahtaroglu, A. The DLGAP family: Neuronal expression, function and role in brain disorders. Mol. Brain 2017, 10, 43. [Google Scholar] [CrossRef] [Green Version]
- Moessner, R.; Marshall, C.R.; Sutcliffe, J.S.; Skaug, J.; Pinto, D.; Vincent, J.; Zwaigenbaum, L.; Fernandez, B.; Roberts, W.; Szatmari, P.; et al. Contribution of SHANK3 Mutations to Autism Spectrum Disorder. Am. J. Hum. Genet. 2007, 81, 1289–1297. [Google Scholar] [CrossRef] [Green Version]
- Phelan, K.; McDermid, H.E. The 22q13.3 Deletion Syndrome (Phelan-McDermid Syndrome). Mol. Syndr. 2011, 2, 186–201. [Google Scholar] [CrossRef] [Green Version]
- Soorya, L.; Kolevzon, A.; Zweifach, J.; Lim, T.; Dobry, Y.; Schwartz, L.; Frank, Y.; Wang, A.T.; Cai, G.; Parkhomenko, E.; et al. Prospective investigation of autism and genotype-phenotype correlations in 22q13 deletion syndrome and SHANK3 deficiency. Mol. Autism 2013, 4, 18. [Google Scholar] [CrossRef] [Green Version]
| Gene | p-Value | Me Status | Function/Implication |
|---|---|---|---|
| Agap3 * | 0.008 | Hypermethylated | Regulates NMDA receptor-mediated Ras/ERK and Arf6 signaling pathways in LTP [28,29] |
| Pax6 * | 0.004 | Hypermethylated | Regulates cortical progenitor proliferation, neurogenesis, and formation of cortical layers [30] |
| Zbtb20 * | 0.01 | Hypermethylated | Plays a role in dendritic and synaptic structure [31] |
| Tfap2a | 0.007 | Hypermethylated | Promotes specification and maturation of neurons [32] |
| Kcnn2 | 0.02 | Hypermethylated | Plays a role in synaptic transmission [33] |
| Dlg2 * | 0.01 | Hypermethylated | A DLG2 deficiency in mice leads to reduced sociability and increased repetitive behavior accompanied by aberrant synaptic transmission [34,35,36] |
| Clasp2 * | 0.03 | Hypermethylated | Plays an important role in the reelin signaling pathway [37] |
| Dlgap3 * | 0.03 | Hypomethylated | Important for protein–protein interactions at synapses and transmission across chemical synapses. Implicated in body dysmorphic disorders [38] |
| CNot3 * | 0.04 | Hypomethylated | Diseases associated with CNOT3 include intellectual developmental disorder with speech delay, autism, and dysmorphic facies [39] |
| Cdh23 | 0.05 | Hypomethylated | Implicated in pituitary adenoma, deafness [40] |
| Shank3 * | 0.01 | Hypermethylated | Implicated in schizophrenia, ASD, and other neurological disorders [41]. |
| Abl1 | 0.03 | Hypermethylated | Important for assembly and remodeling of synapses [42] |
| Ap2a1 | 0.01 | Hypomethylated | AP-2 seems to play a role in the recycling of synaptic vesicle membranes [43] |
| APC | 0.02 | Hypomethylated | Regulates synaptic adhesion and signal transduction pathways critical for normal cognition and behavior [44] |
| Kmt2d * | 0.03 | Hypermethylated | Promotes transcriptional activation and its loss causes the intellectual disability disorder Kabuki syndrome 1 (KS1) [45] |
| Ncdn | 0.02 | Hypermethylated | Neurodevelopmental delay, intellectual disability, and epilepsy [46] |
| Dctna | 0.03 | Hypermethylated | Stabilizing neuron cytoskeleton [47] |
| Gnas * | 0.04 | Hypermethylated | Implicated in variable degrees of intellectual disability and developmental delay [48]. |
| Nbea * | 0.04 | Hypermethylated | Developmental disease gene with early generalized epilepsy phenotypes [49]. |
| Gart | 0.04 | Hypermethylated | Overexpression improves the IQ [50] |
| Emx2 | 0.05 | Hypermethylated | Schizencephaly, CNS, tumorigenesis [51] |
| Agap1 * | 0.03 | Hypermethylated | Dendritic spines, neuron endosome trafficking, neurodevelopmental disorders [52] |
| Dcc * | 0.04 | Hypermethylated | Mirror movement, gaze palsy, impaired intellectual disability [53] |
| Description | Number of Genes | Z-Score | p-Value | Genes |
|---|---|---|---|---|
| GO_BP_MM_MIDBRAIN_DEVELOPMENT | 22 | 1.788854382 | 0.004 | Barhl1;Msx1;Kdm2b;Lrp6;Foxb1;Otx1;Otx2;Fgfr2;En2;Lmx1b;En1;Aplp2;Lmx1a;Rfx4;Gdf7;Hes1;Fgfr1;Shh;Fzd3;Tal2;Kat2a;Wnt1 |
| GO_BP_MM_POSITIVE_REGULATION_OF_NEURON_APOPTOTIC_PROCESS | 19 | 1.697749375 | 0.01 | Tfap2a;Cdk5;Map3k11;Nf1;Tfap2b;Bax;Hrk;Jun;Cdk5r1;Ptprf;Map2k7;Trp53;Ascl1;Agrn;Srpk2;Tgfb2;Nr3c1;Epha7;Ube2m |
| GO_BP_MM_NEGATIVE_REGULATION_OF_NEURON_PROJECTION_DEVELOPMENT | 16 | 1.069044968 | 0.02 | Pafah1b1; Cbfa2t2;Bcl11a;Gfap;Ntm;Dpysl3;Runx1t1;Vim;Inppl1;Gfi1;Fkbp4;Trpv4;Itm2c;Inpp5j;Lpar1;Rtn4 |
| GO_BP_MM_CEREBRAL_CORTEX_NEURON_DIFFERENTIATION | 6 | 0.447213595 | 0.04 | Pafah1b1;Nkx2-1;Id4;Lhx6;Nr2e1;Pex5 |
| GO_BP_MM_NEUROMUSCULAR_JUNCTION_DEVELOPMENT | 14 | –0.632455532 | 0.05 | Cacnb4;Fgfr2;Cacna1s;Col4a1;Cacna2d2;Pdzrn3;Cacng2;Lamb2;Agrn;Pak1; Dvl1;Erbb2;Tnc;Ky |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
AlOlaby, R.R.; Zafarullah, M.; Barboza, M.; Peng, G.; Varian, B.J.; Erdman, S.E.; Lebrilla, C.; Tassone, F. Differential Methylation Profile in Fragile X Syndrome-Prone Offspring Mice after in Utero Exposure to Lactobacillus Reuteri. Genes 2022, 13, 1300. https://doi.org/10.3390/genes13081300
AlOlaby RR, Zafarullah M, Barboza M, Peng G, Varian BJ, Erdman SE, Lebrilla C, Tassone F. Differential Methylation Profile in Fragile X Syndrome-Prone Offspring Mice after in Utero Exposure to Lactobacillus Reuteri. Genes. 2022; 13(8):1300. https://doi.org/10.3390/genes13081300
Chicago/Turabian StyleAlOlaby, Reem R., Marwa Zafarullah, Mariana Barboza, Gang Peng, Bernard J. Varian, Susan E. Erdman, Carlito Lebrilla, and Flora Tassone. 2022. "Differential Methylation Profile in Fragile X Syndrome-Prone Offspring Mice after in Utero Exposure to Lactobacillus Reuteri" Genes 13, no. 8: 1300. https://doi.org/10.3390/genes13081300
APA StyleAlOlaby, R. R., Zafarullah, M., Barboza, M., Peng, G., Varian, B. J., Erdman, S. E., Lebrilla, C., & Tassone, F. (2022). Differential Methylation Profile in Fragile X Syndrome-Prone Offspring Mice after in Utero Exposure to Lactobacillus Reuteri. Genes, 13(8), 1300. https://doi.org/10.3390/genes13081300







