Genetic Variation in DEAD-Box Helicase 20 as a Putative Marker of Recurrence in Propensity-Matched Colon Cancer Patients
Abstract
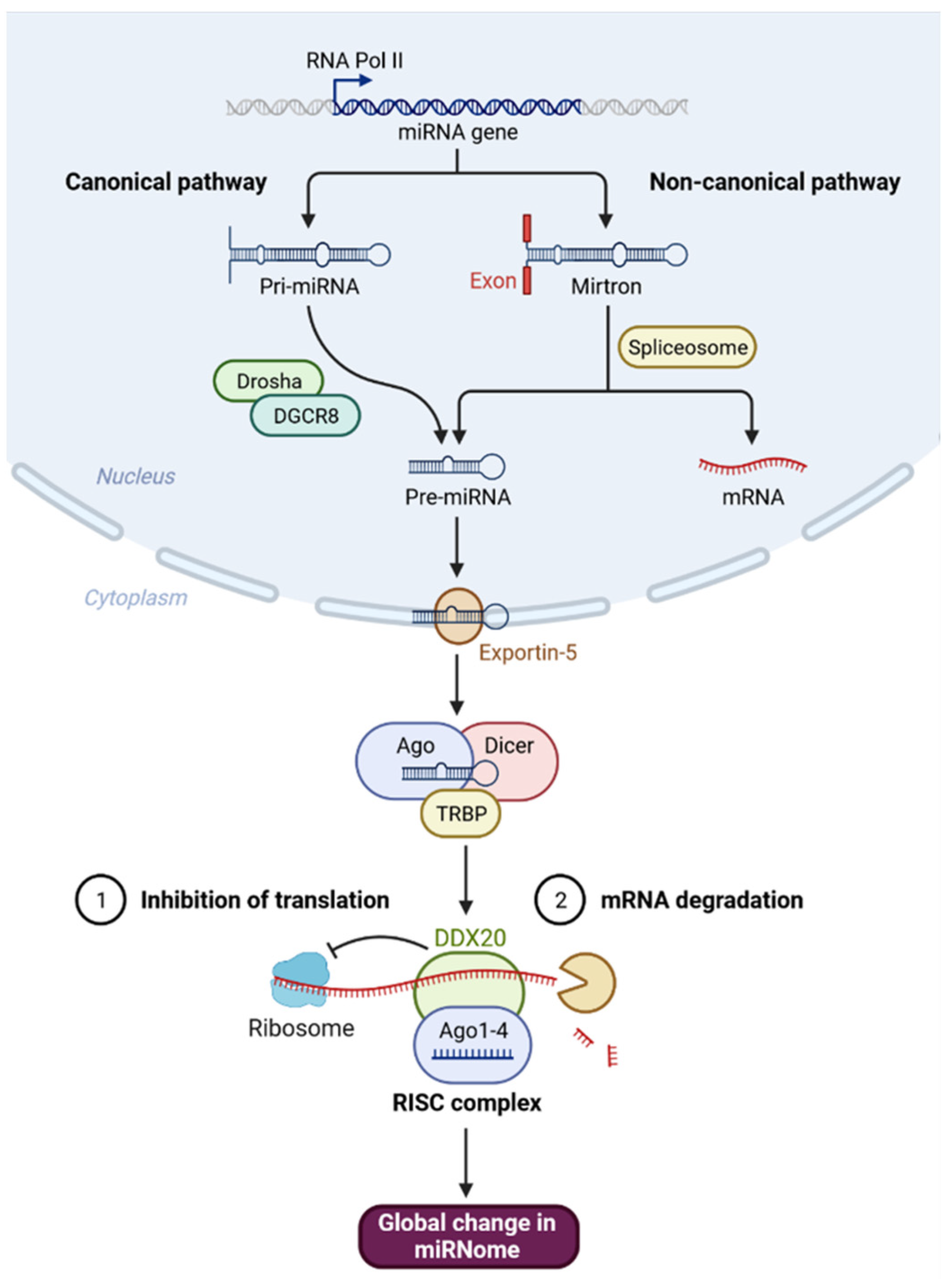
:1. Introduction
2. Materials and Methods
2.1. In Silico Data Analysis of DDX20 Gene and Protein
2.2. SNP Selection and Functional Variant Analysis
2.3. Study Subjects and Tissues
2.4. Assessment of Time-to-Event Endpoints
2.5. Propensity Scores Matched the Cohort
2.6. DDX20 rs197412 Variant Molecular Analysis
2.7. Meta-Analysis
2.8. Statistical Analysis
3. Results
3.1. In Silico Data Analysis
3.2. SNP Selection and Functional Prediction of the Consequence
3.3. Characteristics of Propensity-Matched Samples
3.4. DDX20 rs197412T/C: A Diagnostic and Prognostic Biomarker
3.5. Impact of Genotypes on Cancer Risk
3.6. Somatic Mutation Burden Analysis
3.7. DDX20 Variant Is a Poor Prognostic Marker
3.8. Meta-Analysis Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Ali, M.A.M. DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021, 296, 198352. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A.M. The DEAD-box protein family of RNA helicases: Sentinels for a myriad of cellular functions with emerging roles in tumorigenesis. Int. J. Clin. Oncol. 2021, 26, 795–825. [Google Scholar] [CrossRef] [PubMed]
- Andreou, A.Z.; Klostermeier, D. The DEAD-box helicase eIF4A: Paradigm or the odd one out? RNA Biol. 2013, 10, 19–32. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelhaleem, M. Do human RNA helicases have a role in cancer? Biochim. Biophys. Acta 2004, 1704, 37–46. [Google Scholar] [CrossRef]
- Fuller-Pace, F.V. DEAD box RNA helicase functions in cancer. RNA Biol. 2013, 10, 121–132. [Google Scholar] [CrossRef] [Green Version]
- Robert, F.; Pelletier, J. Perturbations of RNA helicases in cancer. Wiley Interdiscip. Rev. RNA 2013, 4, 333–349. [Google Scholar] [CrossRef]
- Charroux, B.; Pellizzoni, L.; Perkinson, R.A.; Shevchenko, A.; Mann, M.; Dreyfuss, G. Gemin3: A novel DEAD box protein that interacts with SMN, the spinal muscular atrophy gene product, and is a component of gems. J. Cell Biol. 1999, 147, 1181–1194. [Google Scholar] [CrossRef]
- Staněk, D.; Fox, A.H. Nuclear bodies: News insights into structure and function. Curr. Opin. Cell Biol. 2017, 46, 94–101. [Google Scholar] [CrossRef]
- Slaby, O.; Bienertova-Vasku, J.; Svoboda, M.; Vyzula, R. Genetic polymorphisms and microRNAs: New direction in molecular epidemiology of solid cancer. J. Cell. Mol. Med. 2012, 16, 8–21. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Du, Y.; Zhao, S.; Guo, Z. Single-nucleotide polymorphisms of microRNA processing machinery genes and risk of colorectal cancer. Onco Targets Ther. 2015, 8, 421–425. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Yang, H.; Lee, J.J.; Kim, E.; Lippman, S.M.; Khuri, F.R.; Spitz, M.R.; Lotan, R.; Hong, W.K.; Wu, X. MicroRNA-related genetic variations as predictors for risk of second primary tumor and/or recurrence in patients with early-stage head and neck cancer. Carcinogenesis 2010, 31, 2118–2123. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ye, Y.; Wang, K.K.; Gu, J.; Yang, H.; Lin, J.; Ajani, J.A.; Wu, X. Genetic variations in microRNA-related genes are novel susceptibility loci for esophageal cancer risk. Cancer Prev. Res. 2008, 1, 460–469. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourova-Flin, E.; Derakhshan, S.; Goudarzi, A.; Wang, T.; Vitte, A.L.; Chuffart, F.; Khochbin, S.; Rousseaux, S.; Aminishakib, P. The combined detection of Amphiregulin, Cyclin A1 and DDX20/Gemin3 expression predicts aggressive forms of oral squamous cell carcinoma. Br. J. Cancer 2021, 125, 1122–1134. [Google Scholar] [CrossRef]
- Turcot, V.; Lu, Y.; Highland, H.M.; Schurmann, C.; Justice, A.E.; Fine, R.S.; Bradfield, J.P.; Esko, T.; Giri, A.; Graff, M.; et al. Publisher Correction: Protein-altering variants associated with body mass index implicate pathways that control energy intake and expenditure in obesity. Nat. Genet. 2019, 51, 1191–1192. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clague, J.; Lippman, S.M.; Yang, H.; Hildebrandt, M.A.; Ye, Y.; Lee, J.J.; Wu, X. Genetic variation in MicroRNA genes and risk of oral premalignant lesions. Mol. Carcinog. 2010, 49, 183–189. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.; Diao, L.; Li, H.; Guo, Z. Single nucleotide polymorphisms of microRNA processing genes and outcome of non-Hodgkin's lymphoma. Onco Targets Ther. 2015, 8, 1735–1741. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, T.J.; Choquet, H.; Yin, J.; Banda, Y.; Kvale, M.N.; Glymour, M.; Schaefer, C.; Risch, N.; Jorgenson, E. A Large Multiethnic Genome-Wide Association Study of Adult Body Mass Index Identifies Novel Loci. Genetics 2018, 210, 499–515. [Google Scholar] [CrossRef] [Green Version]
- Ma, Y.; Yang, Y.; Wang, F.; Zhang, P.; Shi, C.; Zou, Y.; Qin, H. Obesity and risk of colorectal cancer: A systematic review of prospective studies. PLoS ONE 2013, 8, e53916. [Google Scholar] [CrossRef] [Green Version]
- Shaalan, A.A.M.; Mokhtar, S.H.; Ahmedah, H.T.; Almars, A.I.; Toraih, E.A.; Ibrahiem, A.T.; Fawzy, M.S.; Salem, M.A. Prognostic Value of LINC-ROR (rs1942347) Variant in Patients with Colon Cancer Harboring BRAF Mutation: A Propensity Score-Matched Analysis. Biomolecules 2022, 12, 569. [Google Scholar] [CrossRef]
- Amin, M.B.; Greene, F.L.; Edge, S.B.; Compton, C.C.; Gershenwald, J.E.; Brookland, R.K.; Meyer, L.; Gress, D.M.; Byrd, D.R.; Winchester, D.P. The Eighth Edition AJCC Cancer Staging Manual: Continuing to build a bridge from a population-based to a more "personalized" approach to cancer staging. CA Cancer J. Clin. 2017, 67, 93–99. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, Y.; Zhao, Y.; Guo, Z. Single-nucleotide polymorphisms of microRNA processing machinery genes are associated with risk for gastric cancer. Onco Targets Ther. 2015, 8, 567–571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roy, R.; De Sarkar, N.; Ghose, S.; Paul, R.R.; Pal, M.; Bhattacharya, C.; Chowdhury, S.K.; Ghosh, S.; Roy, B. Genetic variations at microRNA and processing genes and risk of oral cancer. Tumour. Biol. 2014, 35, 3409–3414. [Google Scholar] [CrossRef] [PubMed]
- Zhu, W.; Zhao, J.; He, J.; Qi, D.; Wang, L.; Ma, X.; Liu, P. Genetic variants in the MicroRNA biosynthetic pathway Gemin3 and Gemin4 are associated with a risk of cancer: A meta-analysis. PeerJ 2016, 4, e1724. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, Y.; Chen, J.; Wu, J.; Hu, Z.; Qin, Z.; Liu, X.; Guan, X.; Wang, Y.; Han, J.; Jiang, T.; et al. Evaluation of genetic variants in microRNA biosynthesis genes and risk of breast cancer in Chinese women. Int. J. Cancer 2013, 133, 2216–2224. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.S.; Choi, Y.Y.; Jin, G.; Kang, H.G.; Choi, J.E.; Jeon, H.S.; Lee, W.K.; Kim, D.S.; Kim, C.H.; Kim, Y.J.; et al. Association of a common AGO1 variant with lung cancer risk: A two-stage case-control study. Mol. Carcinog. 2010, 49, 913–921. [Google Scholar] [CrossRef]
- Horikawa, Y.; Wood, C.G.; Yang, H.; Zhao, H.; Ye, Y.; Gu, J.; Lin, J.; Habuchi, T.; Wu, X. Single nucleotide polymorphisms of microRNA machinery genes modify the risk of renal cell carcinoma. Clin. Cancer Res. 2008, 14, 7956–7962. [Google Scholar] [CrossRef] [Green Version]
- Sawicki, T.; Ruszkowska, M.; Danielewicz, A.; Niedźwiedzka, E.; Arłukowicz, T.; Przybyłowicz, K.E. A Review of Colorectal Cancer in Terms of Epidemiology, Risk Factors, Development, Symptoms and Diagnosis. Cancers 2021, 13, 2025. [Google Scholar] [CrossRef]
- Ferlizza, E.; Solmi, R.; Sgarzi, M.; Ricciardiello, L.; Lauriola, M. The Roadmap of Colorectal Cancer Screening. Cancers 2021, 13, 1101. [Google Scholar] [CrossRef]
- Lin, J.; Horikawa, Y.; Tamboli, P.; Clague, J.; Wood, C.G.; Wu, X. Genetic variations in microRNA-related genes are associated with survival and recurrence in patients with renal cell carcinoma. Carcinogenesis 2010, 31, 1805–1812. [Google Scholar] [CrossRef] [Green Version]
- Faluyi, O.O.; Eng, L.; Qiu, X.; Che, J.; Zhang, Q.; Cheng, D.; Ying, N.; Tse, A.; Kuang, Q.; Dodbiba, L.; et al. Validation of microRNA pathway polymorphisms in esophageal adenocarcinoma survival. Cancer Med. 2017, 6, 361–373. [Google Scholar] [CrossRef] [Green Version]
- Lin, M.; Gu, J.; Eng, C.; Ellis, L.M.; Hildebrandt, M.A.; Lin, J.; Huang, M.; Calin, G.A.; Wang, D.; Dubois, R.N.; et al. Genetic polymorphisms in MicroRNA-related genes as predictors of clinical outcomes in colorectal adenocarcinoma patients. Clin. Cancer Res. 2012, 18, 3982–3991. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Savas, S.; Xu, J.; Werdyani, S.; Shestopaloff, K.; Dicks, E.; Green, J.; Parfrey, P.; Green, R.; Xu, W. A Survival Association Study of 102 Polymorphisms Previously Associated with Survival Outcomes in Colorectal Cancer. Biomed. Res. Int. 2015, 2015, 968743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, H.C.; Kim, J.G.; Chae, Y.S.; Sohn, S.K.; Kang, B.W.; Moon, J.H.; Jeon, S.W.; Lee, M.H.; Lim, K.H.; Park, J.Y.; et al. Prognostic impact of microRNA-related gene polymorphisms on survival of patients with colorectal cancer. J. Cancer Res. Clin. Oncol. 2010, 136, 1073–1078. [Google Scholar] [CrossRef] [PubMed]
- Boni, V.; Zarate, R.; Villa, J.C.; Bandrés, E.; Gomez, M.A.; Maiello, E.; Garcia-Foncillas, J.; Aranda, E. Role of primary miRNA polymorphic variants in metastatic colon cancer patients treated with 5-fluorouracil and irinotecan. Pharm. J. 2011, 11, 429–436. [Google Scholar] [CrossRef] [PubMed]
- Ke, H.L.; Chen, M.; Ye, Y.; Hildebrandt, M.A.; Wu, W.J.; Wei, H.; Huang, M.; Chang, D.W.; Dinney, C.P.; Wu, X. Genetic variations in micro-RNA biogenesis genes and clinical outcomes in non-muscle-invasive bladder cancer. Carcinogenesis 2013, 34, 1006–1011. [Google Scholar] [CrossRef] [Green Version]
- Liu, S.; An, J.; Lin, J.; Liu, Y.; Bao, L.; Zhang, W.; Zhao, J.J. Single nucleotide polymorphisms of microRNA processing machinery genes and outcome of hepatocellular carcinoma. PLoS ONE 2014, 9, e92791. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Dinney, C.P.; Ye, Y.; Zhu, Y.; Grossman, H.B.; Wu, X. Evaluation of genetic variants in microRNA-related genes and risk of bladder cancer. Cancer Res. 2008, 68, 2530–2537. [Google Scholar] [CrossRef] [Green Version]







| Variable | Total (N = 122) | Primary (N = 61) | Recurrent (N = 61) | p-Value | |
|---|---|---|---|---|---|
| Age (y) | ≤60 | 77 (63.1) | 41 (67.2) | 36 (59) | 0.45 |
| >60 | 45 (36.9) | 20 (32.8) | 25 (41) | ||
| Sex | Male | 82 (67.2) | 42 (68.9) | 40 (65.6) | 0.85 |
| Female | 40 (32.8) | 19 (31.1) | 21 (34.4) | ||
| Location | Right | 67 (54.9) | 35 (57.4) | 32 (52.5) | 0.49 |
| Transverse | 7 (5.7) | 2 (3.3) | 5 (8.2) | ||
| Left | 48 (39.3) | 24 (39.3) | 24 (39.3) | ||
| Type | Adenocarcinoma | 84 (68.9) | 41 (67.2) | 43 (70.5) | 0.75 |
| Mucinous | 16 (13.1) | 7 (11.5) | 9 (14.8) | ||
| Signet cell | 18 (14.8) | 11 (18) | 7 (11.5) | ||
| Undifferentiated | 4 (3.3) | 2 (3.3) | 2 (3.3) | ||
| Grade | G1/2 | 83 (68) | 41 (67.2) | 42 (68.9) | 0.84 |
| G3 | 39 (32) | 20 (32.8) | 19 (31.1) | ||
| T stage | T1/2 | 65 (53.3) | 32 (52.5) | 33 (54.1) | 0.85 |
| T3/4 | 57 (46.7) | 29 (47.5) | 28 (45.9) | ||
| N stage | N0 | 45 (36.9) | 20 (32.8) | 25 (41) | 0.45 |
| N1 | 77 (63.1) | 41 (67.2) | 36 (59) | ||
| M stage | M0 | 98 (80.3) | 49 (80.3) | 49 (80.3) | 0.82 |
| M1 | 24 (19.7) | 12 (19.7) | 12 (19.7) | ||
| Lymphovascular invasion | No | 82 (67.2) | 41 (67.2) | 41 (67.2) | 0.85 |
| Yes | 40 (32.8) | 20 (32.8) | 20 (32.8) | ||
| Duke’s stage | A/B | 63 (51.6) | 31 (50.8) | 32 (52.5) | 0.85 |
| C/D | 59 (48.4) | 30 (49.2) | 29 (47.5) | ||
| BRAF mutation | Wild type | 86 (70.5) | 42 (68.9) | 44 (72.1) | 0.84 |
| Mutant | 36 (29.5) | 19 (31.1) | 17 (27.9) | ||
| Relapse | No | 83 (68) | 44 (72.1) | 39 (63.9) | 0.43 |
| Yes | 39 (32) | 17 (27.9) | 22 (36.1) | ||
| Mortality | Survived | 90 (73.8) | 49 (80.3) | 41 (67.2) | 0.15 |
| Died | 32 (26.2) | 12 (19.7) | 20 (32.8) | ||
| Disease-free survival | Prolonged > 2y | 46 (39) | 21 (36.2) | 25 (41.7) | 0.57 |
| Short ≤ 2y | 72 (61) | 37 (63.8) | 35 (58.3) | ||
| Overall survival | Prolonged > 2y | 61 (52.6) | 30 (51.7) | 31 (53.4) | 0.85 |
| Short ≤ 2y | 55 (47.4) | 28 (48.3) | 27 (46.6) | ||
| Frequencies | Total (N = 244) | Control Samples (N = 122) | Tumor Samples (N = 122) | p-Value | Primary (N = 61) | Recurrent (N = 61) | p-Value |
|---|---|---|---|---|---|---|---|
| Alleles | |||||||
| C | 254 (52) | 157 (64.3) | 97 (39.8) | <0.001 | 64 (52.5) | 33 (27) | <0.001 |
| T | 234 (48) | 87 (35.7) | 147 (60.2) | 58 (47.5) | 89 (73) | ||
| Genotypes | |||||||
| C/C | 71 (29.1) | 50 (41.0) | 21 (17.2) | <0.001 | 17 (27.9) | 4 (6.6) | <0.001 |
| C/T | 112 (45.9) | 57 (46.7) | 55 (45.1) | 30 (49.2) | 25 (41) | ||
| T/T | 61 (25.0) | 15 (12.3) | 46 (37.7) | 14 (23) | 32 (52.5) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hobani, Y.H.; Almars, A.I.; Alelwani, W.; Toraih, E.A.; Nemr, N.A.; Shaalan, A.A.M.; Fawzy, M.S.; Attallah, S.M. Genetic Variation in DEAD-Box Helicase 20 as a Putative Marker of Recurrence in Propensity-Matched Colon Cancer Patients. Genes 2022, 13, 1404. https://doi.org/10.3390/genes13081404
Hobani YH, Almars AI, Alelwani W, Toraih EA, Nemr NA, Shaalan AAM, Fawzy MS, Attallah SM. Genetic Variation in DEAD-Box Helicase 20 as a Putative Marker of Recurrence in Propensity-Matched Colon Cancer Patients. Genes. 2022; 13(8):1404. https://doi.org/10.3390/genes13081404
Chicago/Turabian StyleHobani, Yahya H., Amany I. Almars, Walla Alelwani, Eman A. Toraih, Nader A. Nemr, Aly A. M. Shaalan, Manal S. Fawzy, and Samy M. Attallah. 2022. "Genetic Variation in DEAD-Box Helicase 20 as a Putative Marker of Recurrence in Propensity-Matched Colon Cancer Patients" Genes 13, no. 8: 1404. https://doi.org/10.3390/genes13081404







