The Effect of Selected Polymorphisms of the ACTN3, ACE, HIF1A and PPARA Genes on the Immediate Supercompensation Training Effect of Elite Slovak Endurance Runners and Football Players
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Genotype Frequency
3.2. Effect of ACTN3 Polymorphisms on the Supercompensation Effect at CJ10 in Experimental and Control Groups
3.3. The Effect of ACE Polymorphisms on the Supercompensation Effect in CJ10 in the Experimental and Control Groups
3.4. The Effect of HIF1A Polymorphisms on the Supercompensation Effect at CJ10 in the Experimental and Control Groups
3.5. The Effect of PPARA Polymorphisms on the Supercompensation Effect in CJ10 in the Experimental and Control Groups
3.6. The Combined Effect of Genes on Selected Parameters of the Supercompensation Effect in CJ10
4. Discussion
4.1. Genotype Frequency of ACTN3, ACE, HIF1A and PPARA Genes in Terms of Adaptation
4.2. Immediate Supercompensation Effect in CJ10 in Terms of Gene Polymorphisms
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Conflicts of Interest
References
- De Moor, M.H.; Spector, T.D.; Cherkas, L.F.; Falchi, M.; Hottenga, J.J.; Boomsma, D.I.; De Geus, E.J. Genome-wide linkage scan for athlete status in 700 British female DZ twin pairs. Twin Res. Hum. Genet. 2007, 10, 812–820. [Google Scholar] [CrossRef] [PubMed]
- Gaudino, C.; Canova, R.; Duca, M.; Silvaggi, N.; Gaudino, P. Optimizing Training and Performance. In Management of Track and Field Injures; Springer: Cham, Switzerland, 2022; pp. 349–362. [Google Scholar]
- Mukhopadhyay, K. Physiological basis of adaptation through super-compensation for better sporting result. Adv. Health Exerc. 2022, 1, 30–42. [Google Scholar]
- Zatsiorsky, V.M.; Kraemer, W.J.; Fry, A.C. Science and Practice of Strength Training; Human Kinetics: Champaign, IL, USA, 2020; ISBN 1492592005. [Google Scholar]
- Clark, R.A.; Bryant, A.L.; Reaburn, P. The acute effects of a single set of contrast preloading on a loaded countermovement jump training session. J. Strength Cond. Res. 2006, 20, 162–166. [Google Scholar] [PubMed]
- Coffey, V.G.; Hawley, J.A. The molecular bases of training adaptation. Sports Med. 2007, 37, 737–763. [Google Scholar] [CrossRef] [PubMed]
- Ahmetov, I.I.; Egorova, E.S.; Gabdrakhmanova, L.J.; Fedotovskaya, O.N. Genes and athletic performance: An update. Genet. Sports 2016, 61, 41–54. [Google Scholar]
- Pokrywka, A.; Kaliszewski, P.; Majorczyk, E.; Zembroń-Łacny, A. Review genes in sport and doping. Biol. Sport 2014, 30, 155–161. [Google Scholar] [CrossRef]
- Lippi, G.; Longo, U.G.; Maffulli, N. Genetics and sports. Br. Med. Bull. 2010, 93, 27–47. [Google Scholar] [CrossRef]
- Puthucheary, Z.; Skipworth, J.R.; Rawal, J.; Loosemore, M.; Van Someren, K.; Montgomery, H.E. The ACE gene and human performance. Sports Med. 2011, 41, 433–448. [Google Scholar] [CrossRef]
- Ma, F.; Yang, Y.; Li, X.; Zhou, F.; Gao, C.; Li, M.; Gao, L. The association of sport performance with ACE and ACTN3 genetic polymorphisms: A systematic review and meta-analysis. PLoS ONE 2013, 8, e54685. [Google Scholar]
- Ahmetov, I.I.; Fedotovskaya, O.N. Current progress in sports genomics. Adv. Clin. Chem. 2015, 70, 247–314. [Google Scholar]
- Czarnik-Kwaśniak, J.; Kwaśniak, K.; Tabarkiewicz, J. How genetic predispositions may have impact on injury and success in sport. Eur. J. Clin. Exp. Med. 2018, 16, 366–375. [Google Scholar] [CrossRef]
- Zhang, B.; Tanaka, H.; Shono, N.; Miura, S.; Kiyonaga, A.; Shindo, M.; Saku, K. The I allele of the angiotensin-converting enzyme gene is associated with an increased percentage of slow-twitch type I fibers in human skeletal muscle. Clin. Genet. 2003, 63, 139–144. [Google Scholar] [CrossRef]
- Goh, K.P.; Chew, K.; Koh, A.; Guan, M.; Wong, Y.S.; Sum, C.F. The relationship between ACE gene ID polymorphism and aerobic capacity in Asian rugby players. Singap. Med. J. 2009, 50, 997. [Google Scholar]
- Bueno, S.; Pasqua, L.A.; de Araujo, G.; Eduardo Lima-Silva, A.; Bertuzzi, R. The association of ACE genotypes on cardiorespiratory variables related to physical fitness in healthy men. PLoS ONE 2016, 11, e0165310. [Google Scholar]
- Dionísio, T.J.; Thiengo, C.R.; Brozoski, D.T.; Dionísio, E.J.; Talamoni, G.A.; Silva, R.B.; Garlet, G.P.; Santos, C.F.; Amaral, S.L. The influence of genetic polymorphisms on performance and cardiac and hemodynamic parameters among Brazilian soccer players. Appl. Physiol. Nutr. Metab. 2017, 42, 596–604. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Fedotovskaya, O.N. Sports genomics: Current state of knowledge and future directions. Cell. Mol. Exerc. Physiol. 2012, 1, e1. [Google Scholar] [CrossRef]
- Orysiak, J.; Mazur-Rózycka, J.; Busko, K.; Gajewski, J.; Szczepanska, B.; Malczewska-Lenczowska, J. Individual and combined influence of ACE and ACTN3 genes on muscle phenotypes in Polish athletes. J. Strength Cond. Res. 2018, 32, 2776–2782. [Google Scholar] [CrossRef] [PubMed]
- Šećerović, A.; Gurbeta, L.; Omanović-Mikličanin, E.; Badnjević, A. Genotype Association with Sport Activity: The Impact of ACE and ACTN3 Gene Polymorphism on Athletic Performance. Int. J. Eng. Res. Technol. 2017, 6, 859–863. [Google Scholar]
- Jacob, Y.; Spiteri, T.; Hart, N.H.; Anderton, R.S. The potential role of genetic markers in talent identification and athlete assessment in elite sport. Sports 2018, 6, 88. [Google Scholar] [CrossRef]
- John, R.; Dhillon, M.S.; Dhillon, S. Genetics and the elite athlete: Our understanding in 2020. Indian J. Orthop. 2020, 54, 256–263. [Google Scholar] [CrossRef]
- Williams, A.G.; Day, S.H.; Folland, J.P.; Gohlke, P.; Dhamrait, S.; Montgomery, H.E. Circulating angiotensin converting enzyme activity is correlated with muscle strength. Med. Sci. Sports Exerc. 2005, 37, 944–948. [Google Scholar] [PubMed]
- Charbonneau, D.E.; Hanson, E.D.; Ludlow, A.T.; Delmonico, M.J.; Hurley, B.F.; Roth, S.M. ACE genotype and the muscle hypertrophic and strength responses to strength training. Med. Sci. Sports Exerc. 2008, 40, 677. [Google Scholar] [CrossRef] [PubMed]
- Eynon, N.; Hanson, E.D.; Lucia, A.; Houweling, P.J.; Garton, F.; North, K.N.; Bishop, D.J. Genes for elite power and sprint performance: ACTN3 leads the way. Sports Med. 2013, 43, 803–817. [Google Scholar] [CrossRef]
- Yang, N.; MacArthur, D.G.; Gulbin, J.P.; Hahn, A.G.; Beggs, A.H.; Easteal, S.; North, K. ACTN3 genotype is associated with human elite athletic performance. Am. J. Hum. Genet. 2003, 73, 627–631. [Google Scholar] [CrossRef]
- Silva, M.S.; Bolani, W.; Alves, C.R.; Biagi, D.G.; Lemos, J.R.; da Silva, J.L.; de Oliveira, P.A.; Alves, G.B.; de Oliveira, E.M.; Negrão, C.E.; et al. Elimination of influences of the ACTN3 R577X variant on oxygen uptake by endurance training in healthy individuals. Int. J. Sports Physiol. Perform. 2015, 10, 636–641. [Google Scholar] [CrossRef] [PubMed]
- Pickering, C.; Kiely, J. ACTN3: More than just a gene for speed. Front. Physiol. 2017, 8, 1080. [Google Scholar] [CrossRef]
- North, K. Why is α-actinin-3 deficiency so common in the general population? The evolution of athletic performance. Twin Res. Hum. Genet. 2008, 11, 384–394. [Google Scholar] [CrossRef]
- Vincent, B.; De Bock, K.; Ramaekers, M.; Van den Eede, E.; Van Leemputte, M.; Hespel, P.; Thomis, M.A. ACTN3 (R577X) genotype is associated with fiber type distribution. Physiol. Genom. 2007, 32, 58–63. [Google Scholar] [CrossRef] [PubMed]
- Ahmetov, I.I.; Gavrilov, D.N.; Astratenkova, I.V.; Druzhevskaya, A.M.; Malinin, A.V.; Romanova, E.E.; Rogozkin, V.A. The association of ACE, ACTN3 and PPARA gene variants with strength phenotypes in middle school-age children. J. Physiol. Sci. 2013, 63, 79–85. [Google Scholar] [CrossRef] [PubMed]
- Pereira, A.; Costa, A.M.; Izquierdo, M.; Silva, A.J.; Bastos, E.; Marques, M.C. ACE I/D and ACTN3 R/X polymorphisms as potential factors in modulating exercise-related phenotypes in older women in response to a muscle power training stimuli. Age 2013, 35, 1949–1959. [Google Scholar] [CrossRef]
- Lopez-Leon, S.; Tuvblad, C.; Forero, D.A. Sports genetics: The PPARA gene and athletes’ high ability in endurance sports. A systematic review and meta-analysis. Biol. Sport 2016, 33, 3–6. [Google Scholar]
- Russell, A.P.; Feilchenfeldt, J.; Schreiber, S.; Praz, M.; Crettenand, A.; Gobelet, C.; Meier, C.A.; Bell, D.R.; Kralli, A.; Giacobino, J.P.; et al. Endurance training in humans leads to fiber type-specific increases in levels of peroxisome proliferator-activated receptor-γ coactivator-1 and peroxisome proliferator-activated receptor-α in skeletal muscle. Diabetes 2003, 52, 2874–2881. [Google Scholar] [CrossRef] [PubMed]
- Petr, M.; Stastny, P.; Zajac, A.; Tufano, J.J.; Maciejewska-Skrendo, A. The role of peroxisome proliferator-activated receptors and their transcriptional coactivators gene variations in human trainability: A systematic review. Int. J. Mol. Sci. 2018, 19, 1472. [Google Scholar] [CrossRef] [PubMed]
- Ahmetov, I.I.; Williams, A.G.; Popov, D.V.; Lyubaeva, E.V.; Hakimullina, A.M.; Fedotovskaya, O.N.; Mozhayskaya, I.A.; Vinogradova, O.L.; Astratenkova, I.V.; Montgomery, H.E.; et al. The combined impact of metabolic gene polymorphisms on elite endurance athlete status and related phenotypes. Hum. Genet. 2009, 126, 751–761. [Google Scholar] [CrossRef] [PubMed]
- Tural, E.; Kara, N.; Agaoglu, S.A.; Elbistan, M.; Tasmektepligil, M.Y.; Imamoglu, O. PPAR-α and PPARGC1A gene variants have strong effects on aerobic performance of Turkish elite endurance athletes. Mol. Biol. Rep. 2014, 41, 5799–5804. [Google Scholar] [CrossRef]
- Akhmetov, I.I.; Popov, D.V.; Mozhaĭskaia, I.A.; Missina, S.S.; Astratenkova, I.V.; Vinogradova, O.L.; Rogozkin, V.A. Association of regulatory genes polymorphisms with aerobic and anaerobic performance of athletes. Ross. Fiziol. Zhurnal Im. IM Sechenova 2007, 93, 837–843. [Google Scholar]
- Ginevičienė, V.; Žavoronkova, R.; Milašius, K. Association of PPARA gene variant with sprint and power ability of Lithuanian elite athletes. In Sporto Mokslas = Sport Science; Vytauto Didžiojo Universiteto Švietimo Akademija: Kaunas, Lithuania; Lietuvos Olimpinė Akademija: Vilnius, Lithuania, 2020; p. 1. [Google Scholar]
- Ginevičienė, V.; Pranckevičienė, E.; Milašius, K.; Kučinskas, V. Relating fitness phenotypes to genotypes in Lithuanian elite athletes. Acta Med. Litu. 2010, 17, 1–10. [Google Scholar]
- Petr, M.; Šťastný, P.; Pecha, O.; Šteffl, M.; Šeda, O.; Kohlíková, E. PPARA intron polymorphism associated with power performance in 30-s anaerobic Wingate Test. PLoS ONE 2014, 9, e107171. [Google Scholar] [CrossRef]
- Alvarez-Romero, J.; Voisin, S.; Eynon, N.; Hiam, D. Mapping robust genetic variants associated with exercise responses. Int. J. Sports Med. 2021, 42, 3–18. [Google Scholar] [CrossRef]
- Lysiak, J.J.; Kirby, J.L.; Tremblay, J.J.; Woodson, R.I.; Reardon, M.A.; Palmer, L.A.; Turner, T.T. Hypoxia-inducible factor-1α is constitutively expressed in murine Leydig cells and regulates 3β-hydroxysteroid dehydrogenase type 1 promoter activity. J. Androl. 2009, 30, 146–156. [Google Scholar] [CrossRef]
- Ahmetov, I.I.; Hakimullina, A.M.; Lyubaeva, E.V.; Vinogradova, O.L.; Rogozkin, V.A. Effect of HIF1A gene polymorphism on human muscle performance. Bull. Exp. Biol. Med. 2008, 146, 351–353. [Google Scholar] [CrossRef] [PubMed]
- Vogt, M.; Puntschart, A.; Geiser, J.; Zuleger, C.; Billeter, R.; Hoppeler, H. Molecular adaptations in human skeletal muscle to endurance training under simulated hypoxic conditions. J. Appl. Physiol. 2001, 91, 173–182. [Google Scholar] [CrossRef] [PubMed]
- Cieszczyk, P.; Eider, J.; Arczewska, A.; Ostanek, M.; Leonska-Duniec, A.; Sawczyn, S.; Ficek, K.; Jascaniene, N.; Kotarska, K.; Sygit, K. The HIF1A gene Pro582Ser polymorphism in polish power-orientated athletes. Biol. Sport 2011, 28, 111. [Google Scholar] [CrossRef]
- Wiener, C.M.; Booth, G.; Semenza, G.L. In vivoexpression of mRNAs encoding hypoxia-inducible factor 1. Biochem. Biophys. Res. Commun. 1996, 225, 485–488. [Google Scholar] [CrossRef] [PubMed]
- Hoppeler, H.; Baum, O.; Lurman, G.; Mueller, M. Molecular mechanisms of muscle plasticity with exercise. Compr. Physiol. 2011, 1, 1383–1412. [Google Scholar]
- Drozdovska, S.B.; Dosenko, V.E.; Ahmetov, I.I.; Ilyin, V. The association of gene polymorphisms with athlete status in Ukrainians. Biol. Sport 2013, 30, 163–167. [Google Scholar] [CrossRef] [PubMed]
- Gabbasov, R.T.; Arkhipova, A.A.; Borisova, A.V.; Hakimullina, A.M.; Kuznetsova, A.V.; Williams, A.G.; Day, S.H.; Ahmetov, I.I. The HIF1A gene Pro582Ser polymorphism in Russian strength athletes. J. Strength Cond. Res. 2013, 27, 2055–2058. [Google Scholar] [CrossRef]
- Maciejewska-Skrendo, A.; Cięszczyk, P.; Chycki, J.; Sawczuk, M.; Smółka, W. Genetic markers associated with power athlete status. J. Hum. Kinet. 2019, 68, 17–36. [Google Scholar] [CrossRef]
- Tiret, L.; Rigat, B.; Visvikis, S.; Breda, C.; Corvol, P.; Cambien, F.; Soubrier, F. Evidence, from combined segregation and linkage analysis, that a variant of the angiotensin I-converting enzyme (ACE) gene controls plasma ACE levels. Am. J. Hum. Genet. 1992, 51, 197. [Google Scholar]
- Flavell, D.M.; Jamshidi, Y.; Hawe, E.; Pineda Torra, I.; Taskinen, M.R.; Frick, M.H.; Nieminen, M.S.; Kesäniemi, Y.A.; Pasternack, A.; Staels, B.; et al. Peroxisome proliferator-activated receptor α gene variants influence progression of coronary atherosclerosis and risk of coronary artery disease. Circulation 2002, 105, 1440–1445. [Google Scholar] [CrossRef] [PubMed]
- Zemková, E.; Hamar, D. JUMP ERGOMETER IN SPORT PERFORMANCE TESTING. Acta Univ. Palacki. Olomuc. Gymnica 2005, 35, 7–16. [Google Scholar]
- Turner, A.; Comfort, P. Advanced Strength and Conditioning: An Evidence-Based Approach; Routledge: London, UK, 2022; ISBN 9780367491352. [Google Scholar]
- Bosquet, L.; Berryman, N.; Dupuy, O. A comparison of 2 optical timing systems designed to measure flight time and contact time during jumping and hopping. J. Strength Cond. Res. 2009, 23, 2660–2665. [Google Scholar] [CrossRef] [PubMed]
- Glatthorn, J.F.; Gouge, S.; Nussbaumer, S.; Stauffacher, S.; Impellizzeri, F.M.; Maffiuletti, N.A. Validity and reliability of Optojump photoelectric cells for estimating vertical jump height. J. Strength Cond. Res. 2011, 25, 556–560. [Google Scholar] [CrossRef]
- Kim, H.Y. Analysis of variance (ANOVA) comparing means of more than two groups. Restor. Dent. Endod. 2014, 39, 74–77. [Google Scholar] [CrossRef]
- Kaufmann, J.; Schering, A.G. Analysis of variance ANOVA. In Wiley Encyclopedia of Clinical Trials; John Wiley & Sons: Hoboken, NJ, USA, 2007. [Google Scholar]
- Gineviciene, V.; Jakaitiene, A.; Tubelis, L.; Kucinskas, V. Variation in the ACE, PPARGC1A and PPARA genes in Lithuanian football players. Eur. J. Sport Sci. 2014, 14 (Suppl. S1), S289–S295. [Google Scholar] [CrossRef]
- McAuley, A.B.; Hughes, D.C.; Tsaprouni, L.G.; Varley, I.; Suraci, B.; Roos, T.R.; Herbert, A.J.; Kelly, A.L. The association of the ACTN3 R577X and ACE I/D polymorphisms with athlete status in football: A systematic review and meta-analysis. J. Sports Sci. 2021, 39, 200–211. [Google Scholar] [CrossRef]
- Pimjan, L.; Ongvarrasopone, C.; Chantratita, W.; Polpramool, C.; Cherdrungsi, P.; Bangrak, P.; Yimlamai, T. A study on ACE, ACTN3, and VDR genes polymorphism in Thai weightlifters. Walailak J. Sci. Technol. WJST 2018, 15, 609–626. [Google Scholar] [CrossRef]
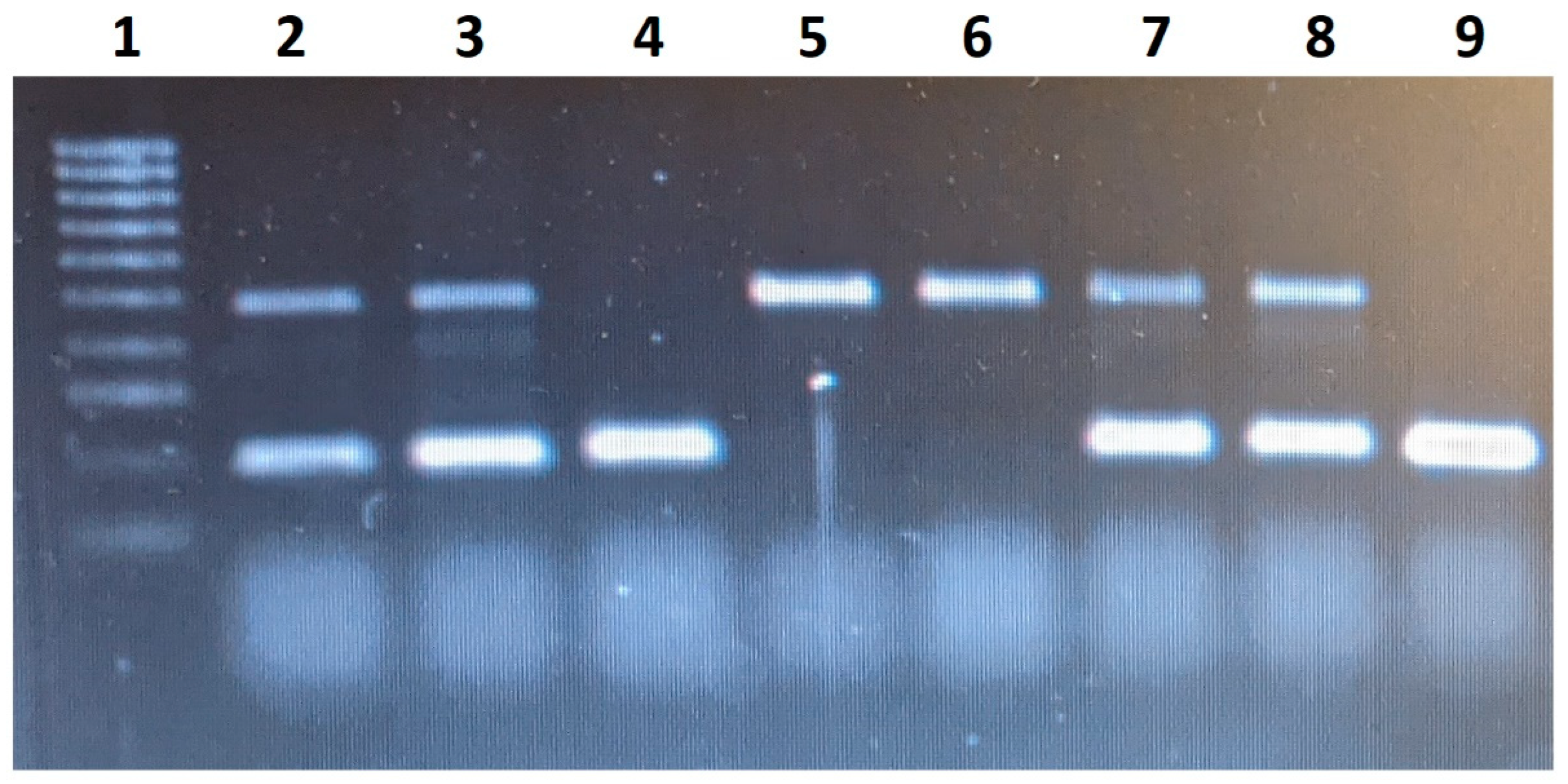
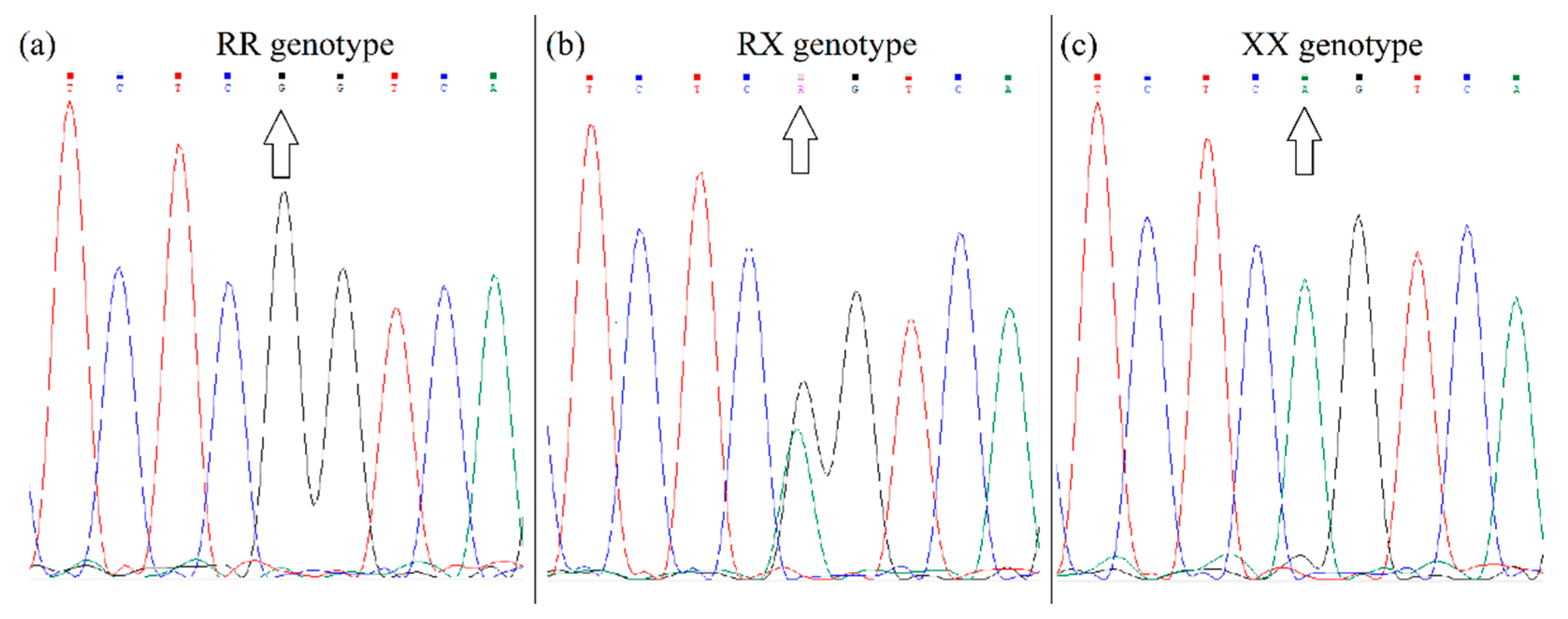

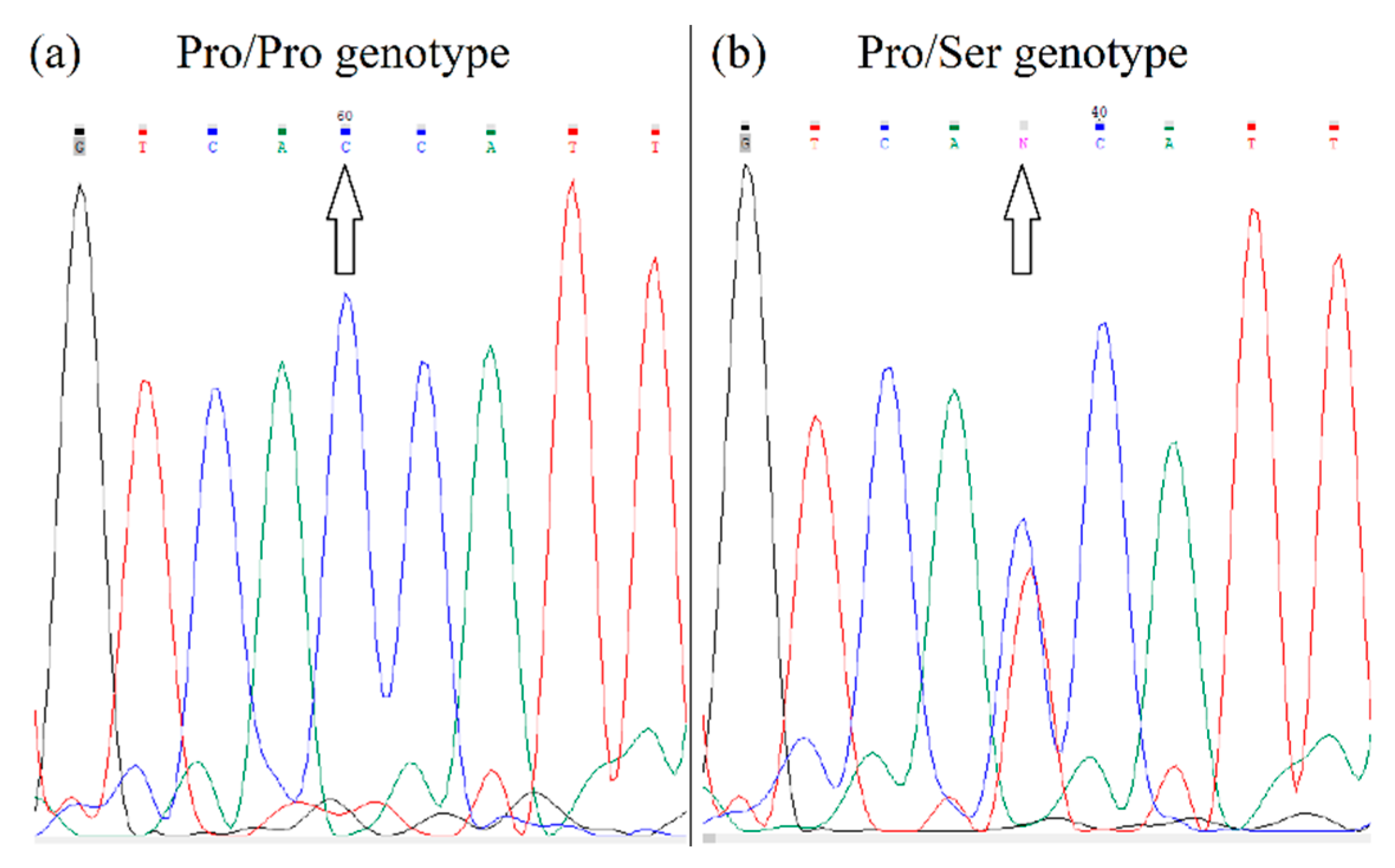
| Gene | Genotype | Experimental Group (n = 64) | Control Group (n = 54) | χ2 | Sig. Level |
|---|---|---|---|---|---|
| Quantity (Frequency) | Quantity (Frequency) | ||||
| ACE | II | 15 (23.44%) | 9 (16.67%) | 9.266 | p < 0.01 |
| ID | 26 (40.63%) | 32 (59.26%) | |||
| DD | 23 (35.94%) | 13 (24.07%) | |||
| ACTN3 | RR | 24 (37.50%) | 20 (37.04%) | 3.979 | p = 0.11 |
| RX | 34 (53.13%) | 24 (44.44%) | |||
| XX | 6 (9.38%) | 10 (18.52%) | |||
| PPARA | GG | 37 (57.81%) | 39 (72.22%) | 67.7 | p < 0.001 |
| GC | 17 (26.56%) | 14 (25.93%) | |||
| CC | 10 (15.63%) | 1 (1.85%) | |||
| HIF1A | Pro/Pro | 53 (82.81%) | 47 (87.04%) | 1.018 | p = 0.25 |
| Pro/Ser | 11 (17.19%) | 7 (12.96%) |
| Experimental Group (n = 64) | Control Group (n = 54) | |||||
|---|---|---|---|---|---|---|
| Alleles | RR | RX | XX | RR | RX | XX |
| n (%) | 24 (37.5) | 34 (51.13) | 6 (9.38) | 20 (37.04) | 24 (44.44) | 10 (18.52) |
| ∆tc [s] | 0.012 (0.023) | 0.010 (0.018) | −0.002 (0.011) | 0.004 (0.015) | 0.000 (0.015) | 0.001 (0.018) |
| ∆h [cm] | −0.09 (3.17) | −0.86 (2.51) | −1.38 (2.40) | 1.30 (3.54) | 0.60 (2.91) | 1.00 (3.97) |
| ∆P [W·kg−1] | −1.55 (4.15) | −2.25 (3.86) | −1.70 (3.82) | 0.98 (2.87) | 0.76 (2.63) | 0.72 (3.23) |
| Experimental Group (n = 64) | Control Group (n = 54) | |||||
|---|---|---|---|---|---|---|
| Alleles | II | ID | DD | II | ID | DD |
| n (%) | 15 (23.44) | 26 (40.63) | 23 (35.94) | 9 (16.67) | 32 (59.26) | 13 (24.07) |
| ∆tc [s] | 0.005 (0.018) | 0.004 (0.017) | 0.017 (0.022) | 0.003 (0.016) | 0.003 (0.015) | 0.001 (0.016) |
| ∆h [cm] | 0.08 (3.38) | −0.99 (2.71) | −0.66 (2.40) | 2.32 (1.80) | 0.23 (3.58) | 1.72 (3.11) |
| ∆P [W·kg−1] | −0.32 (4.53) | −1.85 (3.26) | −3.09 (3.96) | 2.30 (2.25) * | 0.06 (2.77) | 1.73 (2.62) |
| Experimental Group (n = 64) | Control Group (n = 54) | |||
|---|---|---|---|---|
| Alleles | Pro/Pro | Pro/Ser | Pro/Pro | Pro/Ser |
| n (%) | 53 (82.81) | 11 (17.19) | 47 (87.04) | 7 (12.96) |
| ∆tc [s] | 0.007 (0.020) * | 0.020 (0.013) * | 0.001 (0.015) | 0.007 (0.017) |
| ∆h [cm] | −0.62 (2.71) | −0.62 (3.16) | 0.69 (3.20) | 2.59 (3.83) |
| ∆P [W·kg−1] | −1.71 (3.91) | −3.02 (3.96) | 0.50 (2.58) | 3.08 (3.29) * |
| Experimental Group (n = 64) | Control Group (n = 54) | |||||
|---|---|---|---|---|---|---|
| Alleles | GG | GC | CC | GG | GC | CC |
| n (%) | 37 (57.81) | 17 (26.56) | 10 (15.63) | 39 (72.22) | 14 (25.93) | 1 (1.85) |
| ∆tc [s] | 0.011 (0.017) | 0.003 (0.021) | 0.012 (0.026) | 0.001 (0.017) | 0.004 (0.011) | 0.017 |
| ∆h [cm] | −1.13 (2.80) * | 0.09 (2.47) * | 0.06 (2.94) | 0.76 (3.59) | 1.66 (2.34) | −2.41 |
| ∆P [W·kg−1] | −2.91 (3.99) * | −0.16 (2.58) * | −1.36 (4.66) | 0.79 (2.87) | 1.24 (2.44) | −3.34 |
| Parameter | Gene (Genotype) | Effect | Significance Level | ||||
|---|---|---|---|---|---|---|---|
| ∆h | ACTN3 (RR) | + | ACE (ID) | Positive | p < 0.05 | ||
| ACTN3 (RR) | + | PPARA (GC) | Positive | p < 0.05 | |||
| ACTN3 (RR) | + | ACE (ID) | + | HIF1A (Pro/Pro) | Positive | p < 0.05 | |
| ACTN3 (RR) | + | ACE (ID) | + | PPARA (GG) | Negative | p < 0.05 | |
| ∆P | ACTN3 (RR) | + | ACE (ID) | + | HIF1A (Pro/Pro) | Positive | p < 0.05 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Végh, D.; Reichwalderová, K.; Slaninová, M.; Vavák, M. The Effect of Selected Polymorphisms of the ACTN3, ACE, HIF1A and PPARA Genes on the Immediate Supercompensation Training Effect of Elite Slovak Endurance Runners and Football Players. Genes 2022, 13, 1525. https://doi.org/10.3390/genes13091525
Végh D, Reichwalderová K, Slaninová M, Vavák M. The Effect of Selected Polymorphisms of the ACTN3, ACE, HIF1A and PPARA Genes on the Immediate Supercompensation Training Effect of Elite Slovak Endurance Runners and Football Players. Genes. 2022; 13(9):1525. https://doi.org/10.3390/genes13091525
Chicago/Turabian StyleVégh, Dávid, Katarína Reichwalderová, Miroslava Slaninová, and Miroslav Vavák. 2022. "The Effect of Selected Polymorphisms of the ACTN3, ACE, HIF1A and PPARA Genes on the Immediate Supercompensation Training Effect of Elite Slovak Endurance Runners and Football Players" Genes 13, no. 9: 1525. https://doi.org/10.3390/genes13091525
APA StyleVégh, D., Reichwalderová, K., Slaninová, M., & Vavák, M. (2022). The Effect of Selected Polymorphisms of the ACTN3, ACE, HIF1A and PPARA Genes on the Immediate Supercompensation Training Effect of Elite Slovak Endurance Runners and Football Players. Genes, 13(9), 1525. https://doi.org/10.3390/genes13091525






