Individual Genetic Heterogeneity
Abstract
:1. Introduction
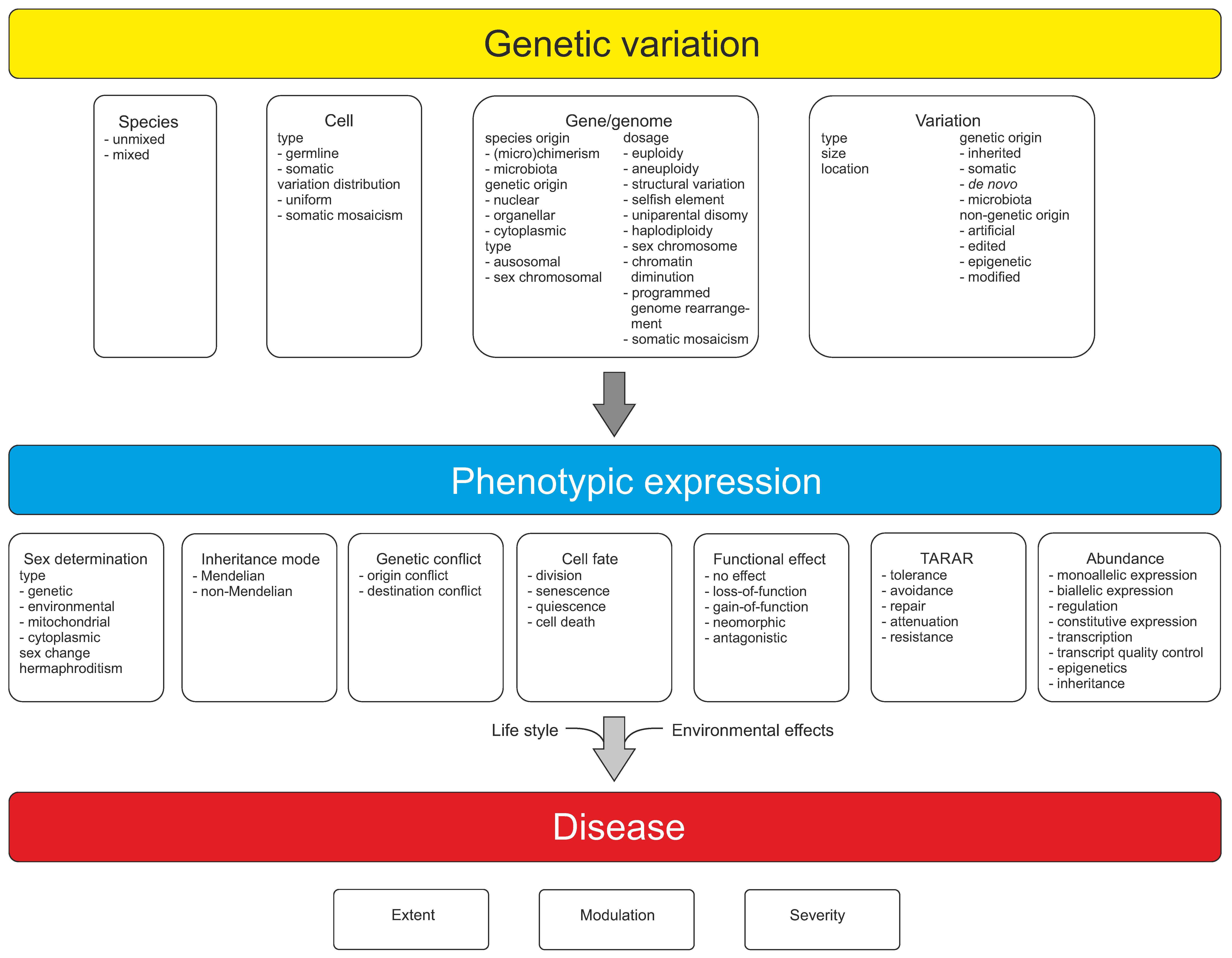
2. Genes and Genome
3. Types of Variations
3.1. Hybrid
3.2. Genome Wide Variations
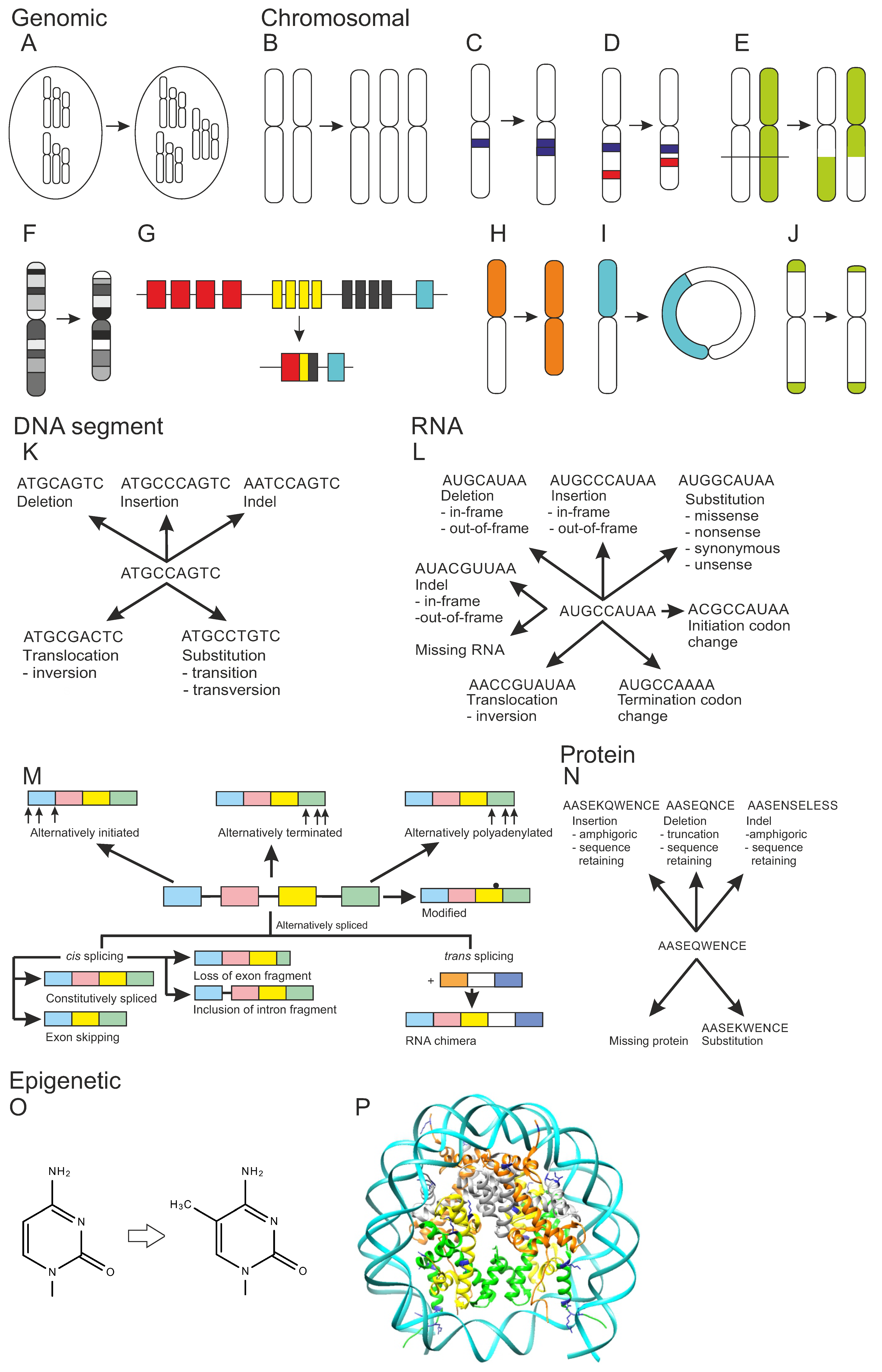
3.3. Chromosomal Variations
3.4. DNA and RNA Chain Variations
3.5. Protein Variations
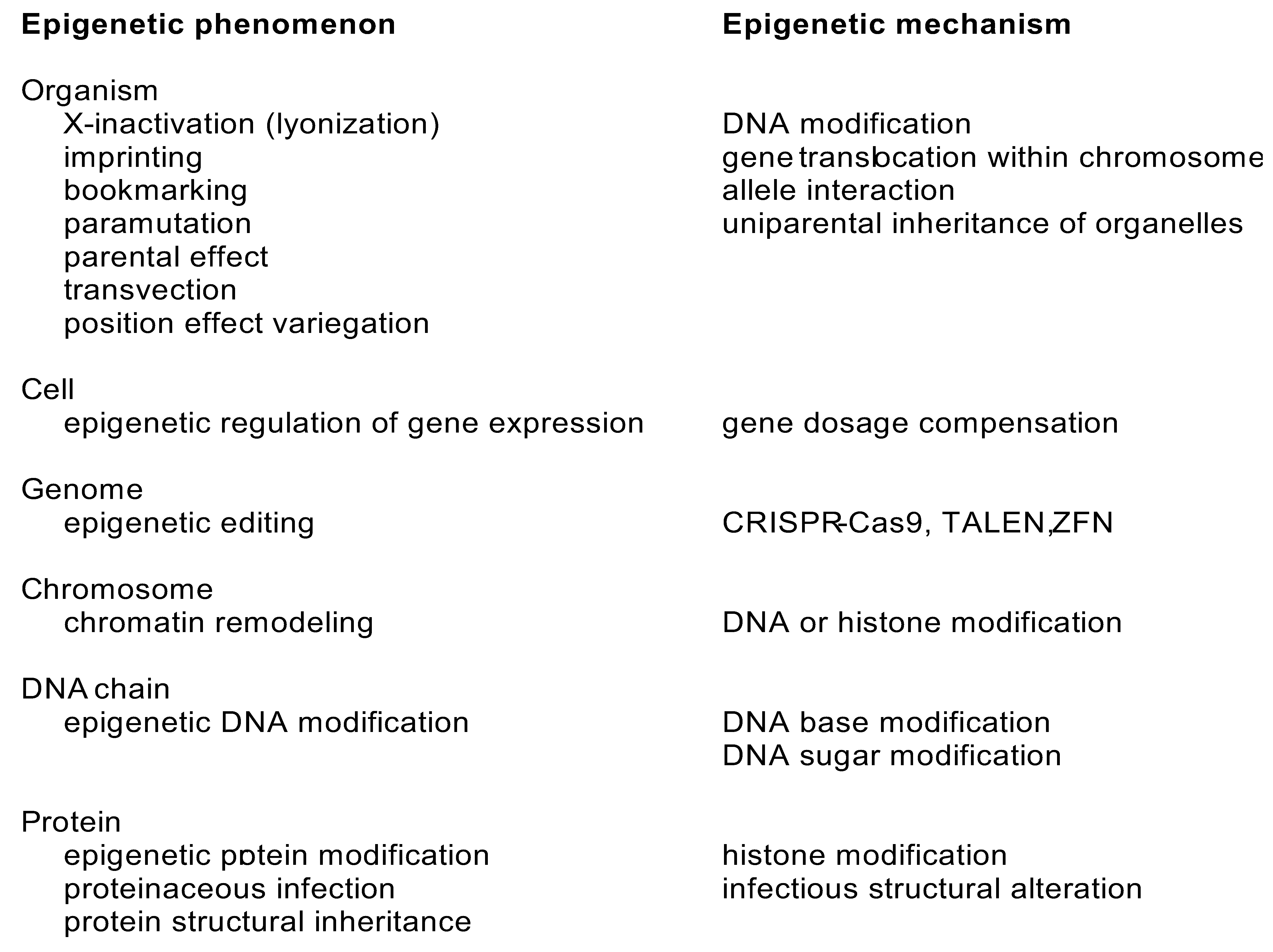
3.6. Epigenetic Variations
3.7. Selfish Genetic Elements
3.8. Genetic Variation in Microbiota
4. Mutation Rate
5. Variation Origin
5.1. Inherited Variants
5.2. Zygosity
5.3. Somatic Genetic Heterogeneity
5.3.1. Somatic Mosaicism
5.3.2. Microchimerism
5.3.3. Ageing-Related Somatic Variation
6. Inheritance
6.1. Monogenic Inheritance
6.2. Non-Mendelian Inheritance
6.2.1. Non-Mendelian Dominance
6.2.2. Multigenic Inheritance
6.2.3. Tropopeissis
6.2.4. Polypleyri Interactions
6.2.5. Gene Dosage Compensation
6.2.6. Genomic Imprinting
6.2.7. Other Forms of Non-Mendelian Inheritance
7. Sexual Reproduction and Sex Determination
8. Functions of DNA
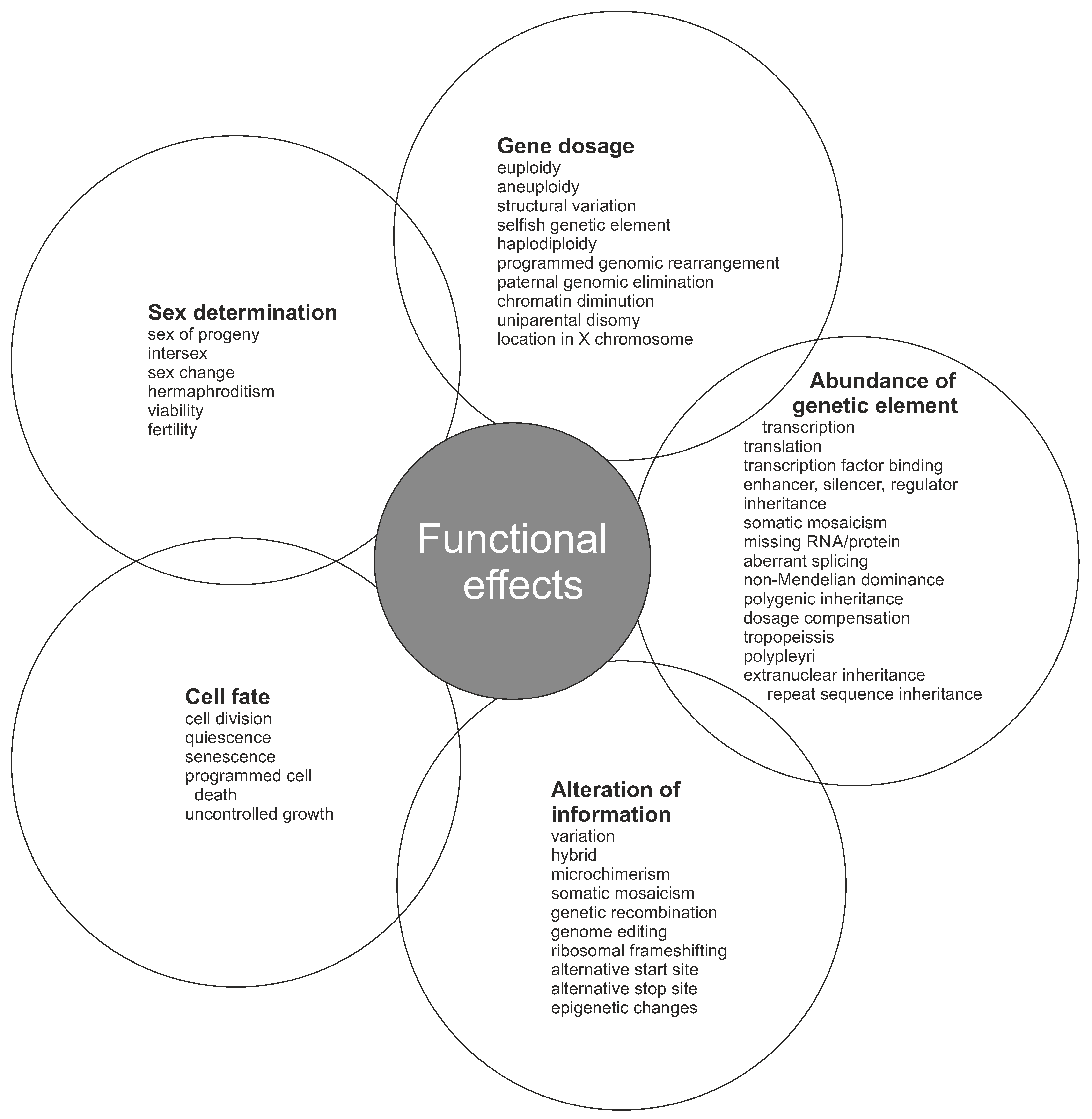
9. Functional Effects of DNA Variations
9.1. Gene Dosage
9.2. Abundance of Gene Products
9.3. Alteration of Information
9.4. Cell Fate
9.5. Sex Determination
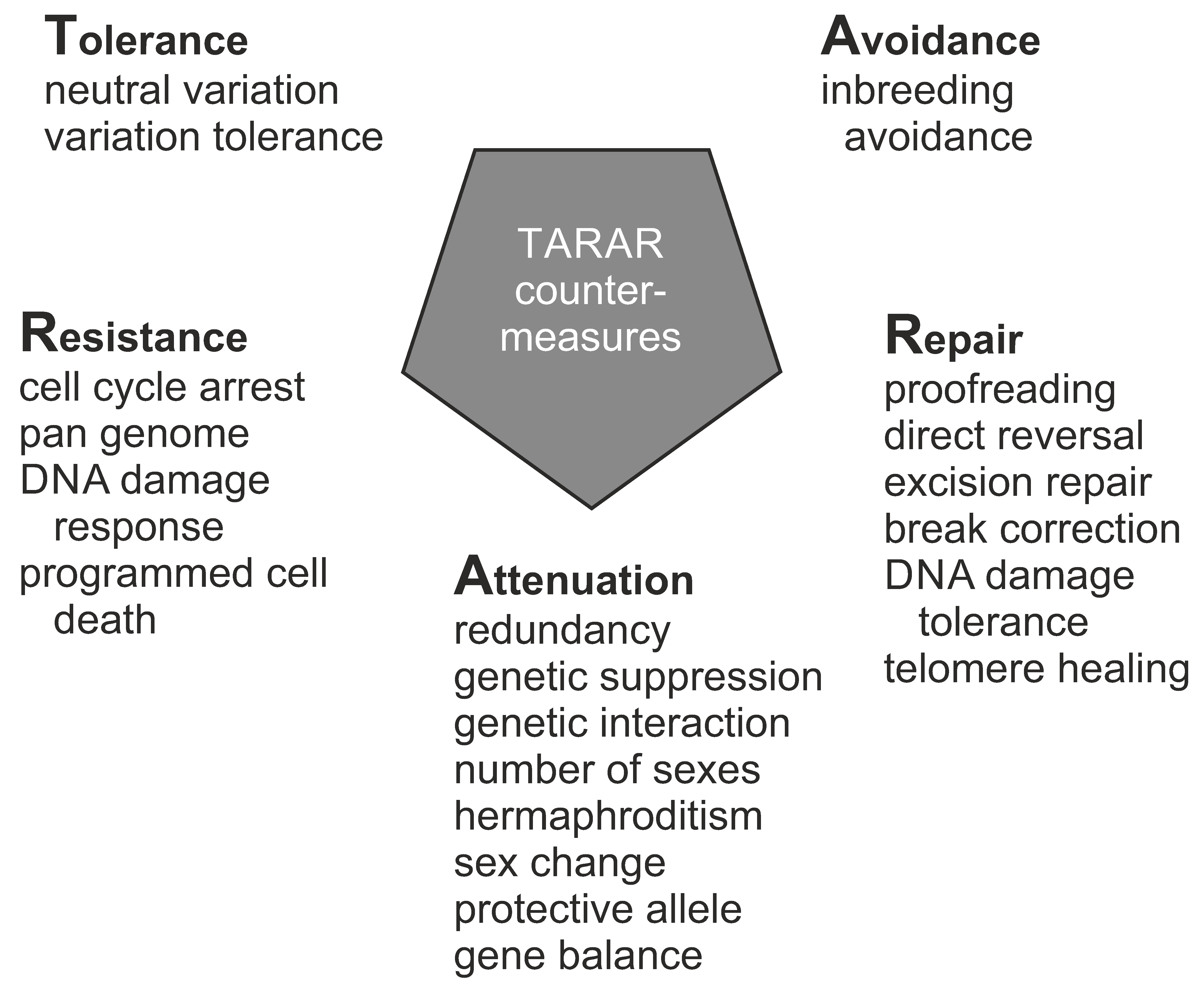
10. TARAR Countermeasures
10.1. Tolerance
10.2. Avoidance
10.3. Repair
10.4. Attenuation
10.5. Resistance
11. Intraindividual Genetic Conflict
12. Genetic Heterogeneity and Diseases
13. Future Perspective
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Shastry, B.S. SNP alleles in human disease and evolution. J. Hum. Genet. 2002, 47, 561–566. [Google Scholar] [CrossRef] [PubMed]
- Brookes, A.J. The essence of SNPs. Gene 1999, 234, 177–186. [Google Scholar] [CrossRef]
- Burrell, R.A.; McGranahan, N.; Bartek, J.; Swanton, C. The causes and consequences of genetic heterogeneity in cancer evolution. Nature 2013, 501, 338–345. [Google Scholar] [CrossRef]
- Alexandrov, L.B.; Nik-Zainal, S.; Wedge, D.C.; Aparicio, S.A.; Behjati, S.; Biankin, A.V.; Bignell, G.R.; Bolli, N.; Borg, A.; Borresen-Dale, A.L.; et al. Signatures of mutational processes in human cancer. Nature 2013, 500, 415–421. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Poikilosis-pervasive biological variation. F1000Research 2020, 9, 602. [Google Scholar] [CrossRef] [PubMed]
- Strauss, B.S. Mechanisms of mutation. In Genetic Diagnosis of Endocrine Disorders; Weiss, R.E., Refetoff, S., Eds.; Elsevier: Amsterdam, The Netherlands, 2016; pp. 3–18. [Google Scholar]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [CrossRef] [PubMed]
- Payne, S. Introduction to RNA viruses. In Viruses: From Understanding to Investigation; Payne, S., Ed.; Academic Press: Cambridge, MA, USA, 2017; pp. 97–105. [Google Scholar]
- Higgs, P.G.; Lehman, N. The RNA World: Molecular cooperation at the origins of life. Nat. Rev. Genet. 2015, 16, 7–17. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, M.; Stiller, J.; Liu, C. A pan-transcriptome analysis shows that disease resistance genes have undergone more selection pressure during barley domestication. BMC Genom. 2019, 20, 12. [Google Scholar] [CrossRef] [PubMed]
- Sherman, R.M.; Forman, J.; Antonescu, V.; Puiu, D.; Daya, M.; Rafaels, N.; Boorgula, M.P.; Chavan, S.; Vergara, C.; Ortega, V.E.; et al. Assembly of a pan-genome from deep sequencing of 910 humans of African descent. Nat. Genet. 2019, 51, 30–35. [Google Scholar] [CrossRef]
- Sherman, R.M.; Salzberg, S.L. Pan-genomics in the human genome era. Nat. Rev. Genet. 2020, 21, 243–254. [Google Scholar] [CrossRef]
- Waters, K. Molecular genetics. In The Stanford Encyclopedia of Philosophy (Fall 2013 Edition); Zalta, E.N., Ed.; Metaphysics Research Lab, Stanford University: Stanford, CA, USA, 2013; Available online: https://plato.stanford.edu/archives/fall2013/entries/molecular-genetics/ (accessed on 10 August 2022).
- Portin, P.; Wilkins, A. The evolving definition of the term “gene”. Genetics 2017, 205, 1353–1364. [Google Scholar] [CrossRef] [PubMed]
- Gerstein, M.B.; Bruce, C.; Rozowsky, J.S.; Zheng, D.; Du, J.; Korbel, J.O.; Emanuelsson, O.; Zhang, Z.D.; Weissman, S.; Snyder, M. What is a gene, post-ENCODE? History and updated definition. Genome Res. 2007, 17, 669–681. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Variation Ontology for annotation of variation effects and mechanisms. Genome Res. 2014, 24, 356–364. [Google Scholar] [CrossRef] [PubMed]
- Famiglietti, M.L.; Estreicher, A.; Gos, A.; Bolleman, J.; Gehant, S.; Breuza, L.; Bridge, A.; Poux, S.; Redaschi, N.; Bougueleret, L.; et al. Genetic variations and diseases in UniProtKB/Swiss-Prot: The ins and outs of expert manual curation. Hum. Mutat. 2014, 35, 927–935. [Google Scholar] [CrossRef] [PubMed]
- Smith, L.M.; Kelleher, N.L. Proteoform: A single term describing protein complexity. Nat. Methods 2013, 10, 186–187. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Functional effects of protein variants. Biochimie 2021, 180, 104–120. [Google Scholar] [CrossRef]
- Chi, X.; Li, Y.; Qiu, X. V(D)J recombination, somatic hypermutation and class switch recombination of immunoglobulins: Mechanism and regulation. Immunology 2020, 160, 233–247. [Google Scholar] [CrossRef]
- Consortium, T.E.P. An integrated encyclopedia of DNA elements in the human genome. Nature 2012, 489, 57–74. [Google Scholar] [CrossRef]
- Djebali, S.; Davis, C.A.; Merkel, A.; Dobin, A.; Lassmann, T.; Mortazavi, A.; Tanzer, A.; Lagarde, J.; Lin, W.; Schlesinger, F.; et al. Landscape of transcription in human cells. Nature 2012, 489, 101–108. [Google Scholar] [CrossRef] [PubMed]
- Moore, J.E.; Purcaro, M.J.; Pratt, H.E.; Epstein, C.B.; Shoresh, N.; Adrian, J.; Kawli, T.; Davis, C.A.; Dobin, A.; Kaul, R.; et al. Expanded encyclopaedias of DNA elements in the human and mouse genomes. Nature 2020, 583, 699–710. [Google Scholar] [CrossRef]
- Cheetham, S.W.; Faulkner, G.J.; Dinger, M.E. Overcoming challenges and dogmas to understand the functions of pseudogenes. Nat. Rev. Genet. 2020, 21, 191–201. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Contribution of pseudogenes to sequence diversity. Methods Mol. Biol. (Clifton N.J.) 2014, 1167, 15–24. [Google Scholar] [CrossRef]
- Ma, Y.; Liu, S.; Gao, J.; Chen, C.; Zhang, X.; Yuan, H.; Chen, Z.; Yin, X.; Sun, C.; Mao, Y.; et al. Genome-wide analysis of pseudogenes reveals HBBP1’s human-specific essentiality in erythropoiesis and implication in β-thalassemia. Dev. Cell 2021, 56, 478–493.e411. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Systematics for types and effects of DNA variations. BMC Genom. 2018, 19, 974. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Systematics for types and effects of RNA variations. RNA Biol. 2021, 18, 481–498. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. Types and effects of protein variations. Hum. Genet. 2015, 134, 405–421. [Google Scholar] [CrossRef]
- Li, J.; Zhou, J.; Zhang, Y.; Yang, Y.; Pu, Q.; Tao, D. New insights into the nature of interspecific hybrid sterility in rice. Front. Plant Sci. 2020, 11, 555572. [Google Scholar] [CrossRef]
- Jolly, C.J.; Woolley-Barker, T.; Beyene, S.; Disotell, T.R.; Phillips-Conroy, J.E. Intergeneric hybrid baboons. Int. J. Primatol. 1997, 18, 597–627. [Google Scholar] [CrossRef]
- Murúa, P.; Edrada-Ebel, R.; Muñoz, L.; Soldatou, S.; Legrave, N.; Müller, D.G.; Patiño, D.J.; van West, P.; Küpper, F.C.; Westermeier, R.; et al. Morphological, genotypic and metabolomic signatures confirm interfamilial hybridization between the ubiquitous kelps Macrocystis (Arthrothamnaceae) and Lessonia (Lessoniaceae). Sci. Rep. 2020, 10, 8279. [Google Scholar] [CrossRef]
- Alvarez, J.B.; Guzmán, C. Interspecific and intergeneric hybridization as a source of variation for wheat grain quality improvement. Appl. Genet. 2018, 131, 225–251. [Google Scholar] [CrossRef]
- Singh, P.P.; Arora, J.; Isambert, H. Identification of ohnolog genes originating from whole genome duplication in early vertebrates, based on synteny comparison across multiple genomes. PLoS Comput. Biol. 2015, 11, e1004394. [Google Scholar] [CrossRef] [PubMed]
- Sender, R.; Fuchs, S.; Milo, R. Revised estimates for the number of human and bacteria cells in the body. PLoS Biol. 2016, 14, e1002533. [Google Scholar] [CrossRef] [PubMed]
- Duncan, A.W. Aneuploidy, polyploidy and ploidy reversal in the liver. Semin. Cell Dev. Biol. 2013, 24, 347–356. [Google Scholar] [CrossRef] [PubMed]
- Park, S.H.; Ayoub, A.; Lee, Y.T.; Xu, J.; Kim, H.; Zheng, W.; Zhang, B.; Sha, L.; An, S.; Zhang, Y.; et al. Cryo-EM structure of the human MLL1 core complex bound to the nucleosome. Nat. Commun. 2019, 10, 5540. [Google Scholar] [CrossRef]
- Barow, M. Endopolyploidy in seed plants. BioEssays 2006, 28, 271–281. [Google Scholar] [CrossRef]
- Ren, D.; Song, J.; Ni, M.; Kang, L.; Guo, W. Regulatory mechanisms of cell polyploidy in insects. Front. Cell Dev. Biol. 2020, 8, 361. [Google Scholar] [CrossRef]
- Wu, C.S.; Lu, W.H.; Hung, M.C.; Huang, Y.S.; Chao, H.W. From polyploidy to polyploidy reversal: Its role in normal and disease states. Trends Genet. 2022. [Google Scholar] [CrossRef]
- Parisod, C.; Holderegger, R.; Brochmann, C. Evolutionary consequences of autopolyploidy. New Phytol. 2010, 186, 5–17. [Google Scholar] [CrossRef]
- Tate, J.A.; Soltis, D.E.; Soltis, P.A. Polyploidy in plants. In The Evolution of the Genome; Gregory, T.R., Ed.; Elsevier: Amsterdam, The Netherlands, 2005; pp. 371–426. [Google Scholar]
- Hassold, T.; Hunt, P. To err (meiotically) is human: The genesis of human aneuploidy. Nat. Rev. Genet. 2001, 2, 280–291. [Google Scholar] [CrossRef]
- Antonarakis, S.E.; Skotko, B.G.; Rafii, M.S.; Strydom, A.; Pape, S.E.; Bianchi, D.W.; Sherman, S.L.; Reeves, R.H. Down syndrome. Nat. Rev. Dis. Prim. 2020, 6, 9. [Google Scholar] [CrossRef]
- Vihinen, M. Muddled genetic terms miss and mess the message. Trends Genet. 2015, 31, 423–425. [Google Scholar] [CrossRef] [PubMed]
- Kurosaki, T.; Popp, M.W.; Maquat, L.E. Quality and quantity control of gene expression by nonsense-mediated mRNA decay. Nat. Rev. Mol. Cell Biol. 2019, 20, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Lindeboom, R.G.H.; Vermeulen, M.; Lehner, B.; Supek, F. The impact of nonsense-mediated mRNA decay on genetic disease, gene editing and cancer immunotherapy. Nat. Genet. 2019, 51, 1645–1651. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. When a synonymous variant is nonsynomous. Genes 2022, 13, 1485. [Google Scholar] [CrossRef]
- Pan, Q.; Shai, O.; Lee, L.J.; Frey, B.J.; Blencowe, B.J. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008, 40, 1413–1415. [Google Scholar] [CrossRef]
- Hu, Z.; Scott, H.S.; Qin, G.; Zheng, G.; Chu, X.; Xie, L.; Adelson, D.L.; Oftedal, B.E.; Venugopal, P.; Babic, M.; et al. Revealing missing human protein isoforms based on ab initio prediction, RNA-seq and proteomics. Sci. Rep. 2015, 5, 10940. [Google Scholar] [CrossRef]
- Tress, M.L.; Abascal, F.; Valencia, A. Alternative splicing may not be the key to proteome complexity. Trends Biochem. Sci. 2017, 42, 98–110. [Google Scholar] [CrossRef]
- Bošković, A.; Rando, O.J. Transgenerational epigenetic inheritance. Annu. Rev. Genet. 2018, 52, 21–41. [Google Scholar] [CrossRef]
- Kumar, S.; Chinnusamy, V.; Mohapatra, T. Epigenetics of modified DNA bases: 5-methylcytosine and beyond. Front. Genet. 2018, 9, 640. [Google Scholar] [CrossRef]
- Tong, T.; Chen, S.; Wang, L.; Tang, Y.; Ryu, J.Y.; Jiang, S.; Wu, X.; Chen, C.; Luo, J.; Deng, Z.; et al. Occurrence, evolution, and functions of DNA phosphorothioate epigenetics in bacteria. Proc. Natl. Acad. Sci. USA 2018, 115, E2988–E2996. [Google Scholar] [CrossRef] [Green Version]
- Zhang, T.; Cooper, S.; Brockdorff, N. The interplay of histone modifications-writers that read. EMBO Rep. 2015, 16, 1467–1481. [Google Scholar] [CrossRef] [PubMed]
- Wilson, P.G. Centriole inheritance. Prion 2008, 2, 9–16. [Google Scholar] [CrossRef] [PubMed]
- Izumi, H.; Kaneko, Y. Evidence of asymmetric cell division and centrosome inheritance in human neuroblastoma cells. Proc. Natl. Acad. Sci. USA 2012, 109, 18048–18053. [Google Scholar] [CrossRef] [PubMed]
- Asher, D.M.; Gregori, L. Human transmissible spongiform encephalopathies: Historic view. Handb. Clin. Neurol. 2018, 153, 1–17. [Google Scholar] [CrossRef]
- Roucou, X.; Gains, M.; LeBlanc, A.C. Neuroprotective functions of prion protein. J. Neurosci. Res. 2004, 75, 153–161. [Google Scholar] [CrossRef]
- Harrison, P.T.; Hart, S. A beginner’s guide to gene editing. Exp. Physiol. 2018, 103, 439–448. [Google Scholar] [CrossRef] [PubMed]
- Court, F.; Martin-Trujillo, A.; Romanelli, V.; Garin, I.; Iglesias-Platas, I.; Salafsky, I.; Guitart, M.; Perez de Nanclares, G.; Lapunzina, P.; Monk, D. Genome-wide allelic methylation analysis reveals disease-specific susceptibility to multiple methylation defects in imprinting syndromes. Hum. Mutat. 2013, 34, 595–602. [Google Scholar] [CrossRef]
- Monk, D.; Mackay, D.J.G.; Eggermann, T.; Maher, E.R.; Riccio, A. Genomic imprinting disorders: Lessons on how genome, epigenome and environment interact. Nat. Rev. Genet. 2019, 20, 235–248. [Google Scholar] [CrossRef]
- Mackay, D.J.G.; Temple, I.K. Human imprinting disorders: Principles, practice, problems and progress. Eur. J. Med. Genet. 2017, 60, 618–626. [Google Scholar] [CrossRef]
- Fang, H.; Disteche, C.M.; Berletch, J.B. X Inactivation and escape: Epigenetic and structural features. Front. Cell Dev. Biol. 2019, 7, 219. [Google Scholar] [CrossRef]
- Johnston, C.M.; Lovell, F.L.; Leongamornlert, D.A.; Stranger, B.E.; Dermitzakis, E.T.; Ross, M.T. Large-scale population study of human cell lines indicates that dosage compensation is virtually complete. PLoS Genet. 2008, 4, e9. [Google Scholar] [CrossRef] [PubMed]
- Michieletto, D.; Chiang, M.; Colì, D.; Papantonis, A.; Orlandini, E.; Cook, P.R.; Marenduzzo, D. Shaping epigenetic memory via genomic bookmarking. Nucleic Acids Res. 2018, 46, 83–93. [Google Scholar] [CrossRef] [PubMed]
- Chandler, V.L. Paramutation: From maize to mice. Cell 2007, 128, 641–645. [Google Scholar] [CrossRef] [PubMed]
- Fukaya, T.; Levine, M. Transvection. Curr. Biol. 2017, 27, R1047–R1049. [Google Scholar] [CrossRef] [PubMed]
- Beisson, J. Preformed cell structure and cell heredity. Prion 2008, 2, 1–8. [Google Scholar] [CrossRef]
- Pikaard, C.S. Nucleolar dominance. In Encyclopedia of Life Sciences; John Wiley & Sons: Chischester, UK, 2018. [Google Scholar] [CrossRef]
- Genesio, R.; Melis, D.; Gatto, S.; Izzo, A.; Ronga, V.; Cappuccio, G.; Lanzo, A.; Andria, G.; D’Esposito, M.; Matarazzo, M.R.; et al. Variegated silencing through epigenetic modifications of a large Xq region in a case of balanced X;2 translocation with Incontinentia Pigmenti-like phenotype. Epigenetics 2011, 6, 1242–1247. [Google Scholar] [CrossRef]
- Nurk, S.; Koren, S.; Rhie, A.; Rautiainen, M.; Bzikadze, A.V.; Mikheenko, A.; Vollger, M.R.; Altemose, N.; Uralsky, L.; Gershman, A.; et al. The complete sequence of a human genome. Science 2022, 376, 44–53. [Google Scholar] [CrossRef]
- Stoddard, B.L. Homing endonucleases from mobile group I introns: Discovery to genome engineering. Mob. DNA 2014, 5, 7. [Google Scholar] [CrossRef]
- Lyttle, T.W. Segregation distorters. Annu. Rev. Genet. 1991, 25, 511–557. [Google Scholar] [CrossRef]
- Wedell, N. Selfish genes and sexual selection: The impact of genomic parasites on host reproduction. J. Zool. 2020, 311, 1–12. [Google Scholar] [CrossRef]
- van der Gaag, M.; Debets, A.J.; Oosterhof, J.; Slakhorst, M.; Thijssen, J.A.; Hoekstra, R.F. Spore-killing meiotic drive factors in a natural population of the fungus Podospora anserina. Genetics 2000, 156, 593–605. [Google Scholar] [CrossRef] [PubMed]
- Vogan, A.A.; Martinossi-Allibert, I.; Ament-Velásquez, S.L.; Svedberg, J.; Johannesson, H. The spore killers, fungal meiotic driver elements. Mycologia 2022, 114, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Beeman, R.W.; Friesen, K.S.; Denell, R.E. Maternal-effect selfish genes in flour beetles. Science 1992, 256, 89–92. [Google Scholar] [CrossRef]
- Kobayashi, I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001, 29, 3742–3756. [Google Scholar] [CrossRef] [PubMed]
- Rothschild, D.; Weissbrod, O.; Barkan, E.; Kurilshikov, A.; Korem, T.; Zeevi, D.; Costea, P.I.; Godneva, A.; Kalka, I.N.; Bar, N.; et al. Environment dominates over host genetics in shaping human gut microbiota. Nature 2018, 555, 210–215. [Google Scholar] [CrossRef]
- Valdes, A.M.; Walter, J.; Segal, E.; Spector, T.D. Role of the gut microbiota in nutrition and health. BMJ 2018, 361, k2179. [Google Scholar] [CrossRef]
- Sanna, S.; Kurilshikov, A.; van der Graaf, A.; Fu, J.; Zhernakova, A. Challenges and future directions for studying effects of host genetics on the gut microbiome. Nat. Genet. 2022, 54, 100–106. [Google Scholar] [CrossRef]
- Rafaluk-Mohr, C.; Gerth, M.; Sealey, J.E.; Ekroth, A.K.E.; Aboobaker, A.A.; Kloock, A.; King, K.C. Microbial protection favors parasite tolerance and alters host-parasite coevolutionary dynamics. Curr. Biol. 2022, 32, 1593–1598.e3. [Google Scholar] [CrossRef]
- Ségurel, L.; Wyman, M.J.; Przeworski, M. Determinants of mutation rate variation in the human germline. Annu. Rev. Genom. Hum. Genet. 2014, 15, 47–70. [Google Scholar] [CrossRef]
- Yousefzadeh, M.; Henpita, C.; Vyas, R.; Soto-Palma, C.; Robbins, P.; Niedernhofer, L. DNA damage-how and why we age? eLife 2021, 10, e62852. [Google Scholar] [CrossRef]
- Lindahl, T. DNA glycosylases, endonucleases for apurinic/apyrimidinic sites, and base excision-repair. Prog. Nucleic Acid Res. Mol. Biol. 1979, 22, 135–192. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T.; Barnes, D.E. Repair of endogenous DNA damage. Cold Spring Harb. Symp. Quant. Biol. 2000, 65, 127–133. [Google Scholar] [CrossRef] [PubMed]
- Auton, A.; Brooks, L.D.; Durbin, R.M.; Garrison, E.P.; Kang, H.M.; Korbel, J.O.; Marchini, J.L.; McCarthy, S.; McVean, G.A.; Abecasis, G.R. A global reference for human genetic variation. Nature 2015, 526, 68–74. [Google Scholar] [CrossRef] [PubMed]
- Rahbari, R.; Wuster, A.; Lindsay, S.J.; Hardwick, R.J.; Alexandrov, L.B.; Turki, S.A.; Dominiczak, A.; Morris, A.; Porteous, D.; Smith, B.; et al. Timing, rates and spectra of human germline mutation. Nat. Genet. 2016, 48, 126–133. [Google Scholar] [CrossRef] [PubMed]
- Milholland, B.; Dong, X.; Zhang, L.; Hao, X.; Suh, Y.; Vijg, J. Differences between germline and somatic mutation rates in humans and mice. Nat. Commun. 2017, 8, 15183. [Google Scholar] [CrossRef]
- Kessler, M.D.; Loesch, D.P.; Perry, J.A.; Heard-Costa, N.L.; Taliun, D.; Cade, B.E.; Wang, H.; Daya, M.; Ziniti, J.; Datta, S.; et al. De novo mutations across 1,465 diverse genomes reveal mutational insights and reductions in the Amish founder population. Proc. Natl. Acad. Sci. USA 2020, 117, 2560–2569. [Google Scholar] [CrossRef]
- Lynch, M. Rate, molecular spectrum, and consequences of human mutation. Proc. Natl. Acad. Sci. USA 2010, 107, 961–968. [Google Scholar] [CrossRef]
- Kong, A.; Frigge, M.L.; Masson, G.; Besenbacher, S.; Sulem, P.; Magnusson, G.; Gudjonsson, S.A.; Sigurdsson, A.; Jonasdottir, A.; Jonasdottir, A.; et al. Rate of de novo mutations and the importance of father’s age to disease risk. Nature 2012, 488, 471–475. [Google Scholar] [CrossRef]
- Kloosterman, W.P.; Francioli, L.C.; Hormozdiari, F.; Marschall, T.; Hehir-Kwa, J.Y.; Abdellaoui, A.; Lameijer, E.W.; Moed, M.H.; Koval, V.; Renkens, I.; et al. Characteristics of de novo structural changes in the human genome. Genome Res. 2015, 25, 792–801. [Google Scholar] [CrossRef] [Green Version]
- Sharp, N.P.; Sandell, L.; James, C.G.; Otto, S.P. The genome-wide rate and spectrum of spontaneous mutations differ between haploid and diploid yeast. Proc. Natl. Acad. Sci. USA 2018, 115, E5046–E5055. [Google Scholar] [CrossRef]
- Manders, F.; van Boxtel, R.; Middlekamp, S. The dynamics of somatic mutagenesis during life in humans. Front. Aging 2021, 2, 802407. [Google Scholar] [CrossRef] [PubMed]
- Cagan, A.; Baez-Ortega, A.; Brzozowska, N.; Abascal, F.; Coorens, T.H.H.; Sanders, M.A.; Lawson, A.R.J.; Harvey, L.M.R.; Bhosle, S.; Jones, D.; et al. Somatic mutation rates scale with lifespan across mammals. Nature 2022, 604, 517–524. [Google Scholar] [CrossRef] [PubMed]
- Cabrera, V.M. Human molecular evolutionary rate, time dependency and transient polymorphism effects viewed through ancient and modern mitochondrial DNA genomes. Sci. Rep. 2021, 11, 5036. [Google Scholar] [CrossRef] [PubMed]
- Stewart, J.B.; Chinnery, P.F. Extreme heterogeneity of human mitochondrial DNA from organelles to populations. Nat. Rev. Genet. 2021, 22, 106–118. [Google Scholar] [CrossRef]
- Makova, K.D.; Hardison, R.C. The effects of chromatin organization on variation in mutation rates in the genome. Nat. Rev. Genet. 2015, 16, 213–223. [Google Scholar] [CrossRef]
- Hodgkinson, A.; Eyre-Walker, A. Variation in the mutation rate across mammalian genomes. Nat. Rev. Genet. 2011, 12, 756–766. [Google Scholar] [CrossRef]
- Arnheim, N.; Calabrese, P. Understanding what determines the frequency and pattern of human germline mutations. Nat. Rev. Genet. 2009, 10, 478–488. [Google Scholar] [CrossRef]
- Baer, C.F.; Miyamoto, M.M.; Denver, D.R. Mutation rate variation in multicellular eukaryotes: Causes and consequences. Nat. Rev. Genet. 2007, 8, 619–631. [Google Scholar] [CrossRef]
- Michaelson, J.J.; Shi, Y.; Gujral, M.; Zheng, H.; Malhotra, D.; Jin, X.; Jian, M.; Liu, G.; Greer, D.; Bhandari, A.; et al. Whole-genome sequencing in autism identifies hot spots for de novo germline mutation. Cell 2012, 151, 1431–1442. [Google Scholar] [CrossRef] [Green Version]
- Carlson, J.; Locke, A.E.; Flickinger, M.; Zawistowski, M.; Levy, S.; Myers, R.M.; Boehnke, M.; Kang, H.M.; Scott, L.J.; Li, J.Z.; et al. Extremely rare variants reveal patterns of germline mutation rate heterogeneity in humans. Nat. Commun. 2018, 9, 3753. [Google Scholar] [CrossRef]
- Brinkmann, B.; Klintschar, M.; Neuhuber, F.; Hühne, J.; Rolf, B. Mutation rate in human microsatellites: Influence of the structure and length of the tandem repeat. Am. J. Hum. Genet. 1998, 62, 1408–1415. [Google Scholar] [CrossRef] [PubMed]
- Terekhanova, N.V.; Seplyarskiy, V.B.; Soldatov, R.A.; Bazykin, G.A. Evolution of local mutation rate and its determinants. Mol. Biol. Evol. 2017, 34, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Nesta, A.V.; Tafur, D.; Beck, C.R. Hotspots of human mutation. Trends Genet. 2021, 37, 717–729. [Google Scholar] [CrossRef]
- Ollila, J.; Lappalainen, I.; Vihinen, M. Sequence specificity in CpG mutation hotspots. FEBS Lett. 1996, 396, 119–122. [Google Scholar] [CrossRef]
- Smith, T.; Ho, G.; Christodoulou, J.; Price, E.A.; Onadim, Z.; Gauthier-Villars, M.; Dehainault, C.; Houdayer, C.; Parfait, B.; van Minkelen, R.; et al. Extensive variation in the mutation rate between and within human genes associated with Mendelian disease. Hum. Mutat. 2016, 37, 488–494. [Google Scholar] [CrossRef] [PubMed]
- Alexandrov, L.B.; Jones, P.H.; Wedge, D.C.; Sale, J.E.; Campbell, P.J.; Nik-Zainal, S.; Stratton, M.R. Clock-like mutational processes in human somatic cells. Nat. Genet. 2015, 47, 1402–1407. [Google Scholar] [CrossRef]
- Behjati, S.; Huch, M.; van Boxtel, R.; Karthaus, W.; Wedge, D.C.; Tamuri, A.U.; Martincorena, I.; Petljak, M.; Alexandrov, L.B.; Gundem, G.; et al. Genome sequencing of normal cells reveals developmental lineages and mutational processes. Nature 2014, 513, 422–425. [Google Scholar] [CrossRef]
- Vijg, J.; Dong, X. Pathogenic mechanisms of somatic mutation and genome mosaicism in aging. Cell 2020, 182, 12–23. [Google Scholar] [CrossRef]
- Strich, J.R.; Chertow, D.S. CRISPR-Cas biology and its application to infectious diseases. J. Clin. Microbiol. 2019, 57, e01307-18. [Google Scholar] [CrossRef] [Green Version]
- Weichenhan, D.; Plass, C. The evolving epigenome. Hum. Mol. Genet. 2013, 22, R1–R6. [Google Scholar] [CrossRef]
- Chen, J.M.; Cooper, D.N.; Chuzhanova, N.; Férec, C.; Patrinos, G.P. Gene conversion: Mechanisms, evolution and human disease. Nat. Rev. Genet. 2007, 8, 762–775. [Google Scholar] [CrossRef] [PubMed]
- Camara, P.G.; Rosenbloom, D.I.; Emmett, K.J.; Levine, A.J.; Rabadan, R. Topological data analysis generates high-resolution, genome-wide maps of human recombination. Cell Syst. 2016, 3, 83–94. [Google Scholar] [CrossRef] [PubMed]
- Stapley, J.; Feulner, P.G.D.; Johnston, S.E.; Santure, A.W.; Smadja, C.M. Variation in recombination frequency and distribution across eukaryotes: Patterns and processes. Philos. Trans. R. Soc. B Biol. Sci. 2017, 372, 20160455. [Google Scholar] [CrossRef] [PubMed]
- Hassan, S.; Surakka, I.; Taskinen, M.R.; Salomaa, V.; Palotie, A.; Wessman, M.; Tukiainen, T.; Pirinen, M.; Palta, P.; Ripatti, S. High-resolution population-specific recombination rates and their effect on phasing and genotype imputation. Eur. J. Hum. Genet. 2021, 29, 615–624. [Google Scholar] [CrossRef] [PubMed]
- Baudat, F.; Buard, J.; Grey, C.; Fledel-Alon, A.; Ober, C.; Przeworski, M.; Coop, G.; de Massy, B. PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 2010, 327, 836–840. [Google Scholar] [CrossRef]
- Myers, S.; Bowden, R.; Tumian, A.; Bontrop, R.E.; Freeman, C.; MacFie, T.S.; McVean, G.; Donnelly, P. Drive against hotspot motifs in primates implicates the PRDM9 gene in meiotic recombination. Science 2010, 327, 876–879. [Google Scholar] [CrossRef]
- Bell, A.D.; Mello, C.J.; Nemesh, J.; Brumbaugh, S.A.; Wysoker, A.; McCarroll, S.A. Insights into variation in meiosis from 31,228 human sperm genomes. Nature 2020, 583, 259–264. [Google Scholar] [CrossRef]
- Bianconi, E.; Piovesan, A.; Facchin, F.; Beraudi, A.; Casadei, R.; Frabetti, F.; Vitale, L.; Pelleri, M.C.; Tassani, S.; Piva, F.; et al. An estimation of the number of cells in the human body. Ann. Hum. Biol. 2013, 40, 463–471. [Google Scholar] [CrossRef]
- Iourov, I.Y.; Vorsanova, S.G.; Yurov, Y.B. Somatic genome variations in health and disease. Curr. Genom. 2010, 11, 387–396. [Google Scholar] [CrossRef] [Green Version]
- Diwan, D.; Komazaki, S.; Suzuki, M.; Nemoto, N.; Aita, T.; Satake, A.; Nishigaki, K. Systematic genome sequence differences among leaf cells within individual trees. BMC Genom. 2014, 15, 142. [Google Scholar] [CrossRef]
- García-Nieto, P.E.; Morrison, A.J.; Fraser, H.B. The somatic mutation landscape of the human body. Genome Biol. 2019, 20, 298. [Google Scholar] [CrossRef] [PubMed]
- Sazonova, M.A.; Sinyov, V.V.; Ryzhkova, A.I.; Galitsyna, E.V.; Melnichenko, A.A.; Postnov, A.Y.; Orekhov, A.N.; Sobenin, I.A. Cybrid models of pathological cell processes in different diseases. Oxid. Med. Cell Longev. 2018, 2018, 4647214. [Google Scholar] [CrossRef] [PubMed]
- Rzeszutek, I.; Maurer-Alcalá, X.X.; Nowacki, M. Programmed genome rearrangements in ciliates. Cell. Mol. Life Sci. CMLS 2020, 77, 4615–4629. [Google Scholar] [CrossRef] [PubMed]
- Thompson, D.J.; Genovese, G.; Halvardson, J.; Ulirsch, J.C.; Wright, D.J.; Terao, C.; Davidsson, O.B.; Day, F.R.; Sulem, P.; Jiang, Y.; et al. Genetic predisposition to mosaic Y chromosome loss in blood. Nature 2019, 575, 652–657. [Google Scholar] [CrossRef] [PubMed]
- Sharma, R.; Lewis, S.; Wlodarski, M.W. DNA Repair Syndromes and Cancer: Insights Into Genetics and Phenotype Patterns. Front. Pediatr. 2020, 8, 570084. [Google Scholar] [CrossRef]
- Hasty, P.; Campisi, J.; Hoeijmakers, J.; van Steeg, H.; Vijg, J. Aging and genome maintenance: Lessons from the mouse? Science 2003, 299, 1355–1359. [Google Scholar] [CrossRef]
- Hirschhorn, R. In vivo reversion to normal of inherited mutations in humans. J. Med. Genet. 2003, 40, 721–728. [Google Scholar] [CrossRef]
- Biesecker, L.G.; Spinner, N.B. A genomic view of mosaicism and human disease. Nat. Rev. Genet. 2013, 14, 307–320. [Google Scholar] [CrossRef]
- Forsberg, L.A.; Gisselsson, D.; Dumanski, J.P. Mosaicism in health and disease-clones picking up speed. Nat. Rev. Genet. 2017, 18, 128–142. [Google Scholar] [CrossRef]
- Wu, J.; Greely, H.T.; Jaenisch, R.; Nakauchi, H.; Rossant, J.; Belmonte, J.C. Stem cells and interspecies chimaeras. Nature 2016, 540, 51–59. [Google Scholar] [CrossRef]
- Scandling, J.D.; Busque, S.; Lowsky, R.; Shizuru, J.; Shori, A.; Engleman, E.; Jensen, K.; Strober, S. Macrochimerism and clinical transplant tolerance. Hum. Immunol. 2018, 79, 266–271. [Google Scholar] [CrossRef] [PubMed]
- Loubière, L.S.; Lambert, N.C.; Flinn, L.J.; Erickson, T.D.; Yan, Z.; Guthrie, K.A.; Vickers, K.T.; Nelson, J.L. Maternal microchimerism in healthy adults in lymphocytes, monocyte/macrophages and NK cells. Lab. Investig. 2006, 86, 1185–1192. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, D.W.; Zickwolf, G.K.; Weil, G.J.; Sylvester, S.; DeMaria, M.A. Male fetal progenitor cells persist in maternal blood for as long as 27 years postpartum. Proc. Natl. Acad. Sci. USA 1996, 93, 705–708. [Google Scholar] [CrossRef] [PubMed]
- Narita, S.; Pereira, R.A.S.; Kjellberg, F.; Kageyama, D. Gynandromorphs and intersexes: Potential to understand the mechanism of sex determination in arthropods. Terr. Arthropod Rev. 2010, 3, 63–96. [Google Scholar]
- Frenkel-Morgenstern, M.; Lacroix, V.; Ezkurdia, I.; Levin, Y.; Gabashvili, A.; Prilusky, J.; Del Pozo, A.; Tress, M.; Johnson, R.; Guigo, R.; et al. Chimeras taking shape: Potential functions of proteins encoded by chimeric RNA transcripts. Genome Res. 2012, 22, 1231–1242. [Google Scholar] [CrossRef]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The hallmarks of aging. Cell 2013, 153, 1194–1217. [Google Scholar] [CrossRef]
- Schumacher, B.; Pothof, J.; Vijg, J.; Hoeijmakers, J.H.J. The central role of DNA damage in the ageing process. Nature 2021, 592, 695–703. [Google Scholar] [CrossRef]
- Foo, M.X.R.; Ong, P.F.; Dreesen, O. Premature aging syndromes: From patients to mechanism. J. Derm. Sci. 2019, 96, 58–65. [Google Scholar] [CrossRef]
- Genovese, G.; Kähler, A.K.; Handsaker, R.E.; Lindberg, J.; Rose, S.A.; Bakhoum, S.F.; Chambert, K.; Mick, E.; Neale, B.M.; Fromer, M.; et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N. Engl. J. Med. 2014, 371, 2477–2487. [Google Scholar] [CrossRef]
- Jaiswal, S.; Natarajan, P.; Silver, A.J.; Gibson, C.J.; Bick, A.G.; Shvartz, E.; McConkey, M.; Gupta, N.; Gabriel, S.; Ardissino, D.; et al. Clonal Hematopoiesis and Risk of Atherosclerotic Cardiovascular Disease. N. Engl. J. Med. 2017, 377, 111–121. [Google Scholar] [CrossRef]
- Sano, S.; Horitani, K.; Ogawa, H.; Halvardson, J.; Chavkin, N.W.; Wang, Y.; Sano, M.; Mattisson, J.; Hata, A.; Danielsson, M.; et al. Hematopoietic loss of Y chromosome leads to cardiac fibrosis and heart failure mortality. Science 2022, 377, 292–297. [Google Scholar] [CrossRef] [PubMed]
- Ellison, J.W. Pseudoautosomal inheritance. In Encyclopedia of Life Sicences (ELS); John Wiley Sons: Chichester, UK, 2009. [Google Scholar] [CrossRef]
- McColgan, P.; Tabrizi, S.J. Huntington’s disease: A clinical review. Eur. J. Neurol. 2018, 25, 24–34. [Google Scholar] [CrossRef] [PubMed]
- De Boeck, K. Cystic fibrosis in the year 2020: A disease with a new face. Acta Paediatr. 2020, 109, 893–899. [Google Scholar] [CrossRef] [PubMed]
- Erickson, R.P.; Lai, L.W.; Mustacich, D.J.; Bernas, M.J.; Kuo, P.H.; Witte, M.H. Sex-limited penetrance of lymphedema to females with CELSR1 haploinsufficiency: A second family. Clin. Genet. 2019, 96, 478–482. [Google Scholar] [CrossRef]
- Vetrie, D.; Vořechovský; Sideras, P.; Holland, J.; Davies, A.; Flinter, F.; Hammarström, L.; Kinnon, C.; Levinsky, R.; Bobrow, M.; et al. The gene involved in X-linked agammaglobulinaemia is a member of the src family of protein-tyrosine kinases. Nature 1993, 361, 226–233. [Google Scholar] [CrossRef] [PubMed]
- White, D.; Rabago-Smith, M. Genotype-phenotype associations and human eye color. J. Hum. Genet. 2011, 56, 5–7. [Google Scholar] [CrossRef]
- Crouch, D.J.M.; Bodmer, W.F. Polygenic inheritance, GWAS, polygenic risk scores, and the search for functional variants. Proc. Natl. Acad. Sci. USA 2020, 117, 18924–18933. [Google Scholar] [CrossRef]
- Cordell, H.J. Epistasis: What it means, what it doesn’t mean, and statistical methods to detect it in humans. Hum. Mol. Genet. 2002, 11, 2463–2468. [Google Scholar] [CrossRef]
- Li, W.; Reich, J. A complete enumeration and classification of two-locus disease models. Hum. Hered 2000, 50, 334–349. [Google Scholar] [CrossRef] [Green Version]
- Hallgrímsdóttir, I.B.; Yuster, D.S. A complete classification of epistatic two-locus models. BMC Genet. 2008, 9, 17. [Google Scholar] [CrossRef]
- Urbanowicz, R.J.; Granizo-Mackenzie, A.L.; Kiralis, J.; Moore, J.H. A classification and characterization of two-locus, pure, strict, epistatic models for simulation and detection. BioData Min. 2014, 7, 8. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Wu, W.; Hong, Z. Genetic interactions of awnness genes in barley. Genes 2021, 12, 606. [Google Scholar] [CrossRef] [PubMed]
- Paaby, A.B.; Rockman, M.V. The many faces of pleiotropy. Trends Genet. 2013, 29, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Hunter, T. Metabolic kinases moonlighting as protein kinases. Trends Biochem. Sci. 2018, 43, 301–310. [Google Scholar] [CrossRef] [PubMed]
- Reissmann, M.; Ludwig, A. Pleiotropic effects of coat colour-associated mutations in humans, mice and other mammals. Semin. Cell Dev. Biol. 2013, 24, 576–586. [Google Scholar] [CrossRef]
- Galupa, R.; Heard, E. X-chromosome inactivation: A crossroads between chromosome architecture and gene regulation. Annu. Rev. Genet. 2018, 52, 535–566. [Google Scholar] [CrossRef]
- Leeb, M.; Wutz, A. Mechanistic concepts in X inactivation underlying dosage compensation in mammals. Heredity 2010, 105, 64–70. [Google Scholar] [CrossRef]
- Birchler, J.A.; Pal-Bhadra, M.; Bhadra, U. Dosage dependent gene regulation and the compensation of the X chromosome in Drosophila males. Genetica 2003, 117, 179–190. [Google Scholar] [CrossRef]
- Nguyen, D.K.; Disteche, C.M. Dosage compensation of the active X chromosome in mammals. Nat. Genet. 2006, 38, 47–53. [Google Scholar] [CrossRef]
- Jans, J.; Gladden, J.M.; Ralston, E.J.; Pickle, C.S.; Michel, A.H.; Pferdehirt, R.R.; Eisen, M.B.; Meyer, B.J. A condensin-like dosage compensation complex acts at a distance to control expression throughout the genome. Genes Dev. 2009, 23, 602–618. [Google Scholar] [CrossRef]
- Kalsner, L.; Chamberlain, S.J. Prader-Willi, Angelman, and 15q11-q13 duplication syndromes. Pediatr. Clin. N. Am. 2015, 62, 587–606. [Google Scholar] [CrossRef] [PubMed]
- Darcy, D.; Atwal, P.S.; Angell, C.; Gadi, I.; Wallerstein, R. Mosaic paternal genome-wide uniparental isodisomy with down syndrome. Am. J. Med. Genet. Part A 2015, 167a, 2463–2469. [Google Scholar] [CrossRef] [PubMed]
- Nissanka, N.; Moraes, C.T. Mitochondrial DNA heteroplasmy in disease and targeted nuclease-based therapeutic approaches. EMBO Rep. 2020, 21, e49612. [Google Scholar] [CrossRef]
- Breton, S.; Stewart, D.T. Atypical mitochondrial inheritance patterns in eukaryotes. Genome 2015, 58, 423–431. [Google Scholar] [CrossRef]
- Breton, S.; Beaupré, H.D.; Stewart, D.T.; Hoeh, W.R.; Blier, P.U. The unusual system of doubly uniparental inheritance of mtDNA: Isn’t one enough? Trends Genet. 2007, 23, 465–474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Sodmergen. Why does biparental plastid inheritance revive in angiosperms? J. Plant Res. 2010, 123, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Bouma, J.E.; Lenski, R.E. Evolution of a bacteria/plasmid association. Nature 1988, 335, 351–352. [Google Scholar] [CrossRef] [PubMed]
- Arbuckle, J.H.; Medveczky, M.M.; Luka, J.; Hadley, S.H.; Luegmayr, A.; Ablashi, D.; Lund, T.C.; Tolar, J.; De Meirleir, K.; Montoya, J.G.; et al. The latent human herpesvirus-6A genome specifically integrates in telomeres of human chromosomes in vivo and in vitro. Proc. Natl. Acad. Sci. USA 2010, 107, 5563–5568. [Google Scholar] [CrossRef] [PubMed]
- Paulson, H. Repeat expansion diseases. Handb. Clin. Neurol. 2018, 147, 105–123. [Google Scholar] [CrossRef]
- Crow, J.F. Advantages of sexual reproduction. Dev. Genet. 1994, 15, 205–213. [Google Scholar] [CrossRef]
- Brown, A.J.; Casselton, L.A. Mating in mushrooms: Increasing the chances but prolonging the affair. Trends Genet. 2001, 17, 393–400. [Google Scholar] [CrossRef]
- MacGillivray, M.H.; Mazur, T. Intersex. Adv. Pediatr. 2005, 52, 295–319. [Google Scholar] [CrossRef] [PubMed]
- Arboleda, V.A.; Sandberg, D.E.; Vilain, E. DSDs: Genetics, underlying pathologies and psychosexual differentiation. Nat. Rev. Endocrinol. 2014, 10, 603–615. [Google Scholar] [CrossRef] [PubMed]
- Weeks, S.C. The role of androdioecy and gynodioecy in mediating evolutionary transitions between dioecy and hermaphroditism in the animalia. Evolution 2012, 66, 3670–3686. [Google Scholar] [CrossRef]
- Renner, S.S. The relative and absolute frequencies of angiosperm sexual systems: Dioecy, monoecy, gynodioecy, and an updated online database. Am. J. Bot. 2014, 101, 1588–1596. [Google Scholar] [CrossRef]
- Chaudhuri, J.; Bose, N.; Tandonnet, S.; Adams, S.; Zuco, G.; Kache, V.; Parihar, M.; von Reuss, S.H.; Schroeder, F.C.; Pires-daSilva, A. Mating dynamics in a nematode with three sexes and its evolutionary implications. Sci. Rep. 2015, 5, 17676. [Google Scholar] [CrossRef]
- Gemmell, N.J.; Todd, E.V.; Goikoetxea, A.; Ortega-Recalde, O.; Hore, T.A. Natural sex change in fish. Curr. Top. Dev. Biol. 2019, 134, 71–117. [Google Scholar] [CrossRef]
- Charnov, E.L.; Bull, J. When is sex environmentally determined? Nature 1977, 266, 828–830. [Google Scholar] [CrossRef]
- Merchant-Larios, H.; Díaz-Hernández, V. Environmental sex determination mechanisms in reptiles. Sex Dev. 2013, 7, 95–103. [Google Scholar] [CrossRef]
- Mancebo Quintana, J.M.; Mancebo Quintana, S. A short-term advantage for syngamy in the origin of eukaryotic sex: Effects of cell fusion on cell cycle duration and other effects related to the duration of the cell cycle-relationship between cell growth curve and the optimal size of the species, and circadian cell cycle in photosynthetic unicellular orrganisms. Int. J. Evol. Biol. 2012, 2012, 746825. [Google Scholar] [CrossRef]
- Bachtrog, D.; Mank, J.E.; Peichel, C.L.; Kirkpatrick, M.; Otto, S.P.; Ashman, T.L.; Hahn, M.W.; Kitano, J.; Mayrose, I.; Ming, R.; et al. Sex determination: Why so many ways of doing it? PLoS Biol. 2014, 12, e1001899. [Google Scholar] [CrossRef] [PubMed]
- Skuse, D.; Printzlau, F.; Wolstencroft, J. Sex chromosome aneuploidies. Handb. Clin. Neurol. 2018, 147, 355–376. [Google Scholar] [CrossRef] [PubMed]
- Moore, E.C.; Roberts, R.B. Polygenic sex determination. Curr. Biol. 2013, 23, R510–R512. [Google Scholar] [CrossRef] [PubMed]
- de la Filia, A.G.; Bain, S.A.; Ross, L. Haplodiploidy and the reproductive ecology of Arthropods. Curr. Opin. Insect. Sci. 2015, 9, 36–43. [Google Scholar] [CrossRef]
- Hodson, C.N.; Hamilton, P.T.; Dilworth, D.; Nelson, C.J.; Curtis, C.I.; Perlman, S.J. Paternal genome elimination in Liposcelis booklice (insecta: Psocodea). Genetics 2017, 206, 1091–1100. [Google Scholar] [CrossRef]
- Ma, W.J.; Vavre, F.; Beukeboom, L.W. Manipulation of arthropod sex determination by endosymbionts: Diversity and molecular mechanisms. Sex Dev. 2014, 8, 59–73. [Google Scholar] [CrossRef]
- Chase, C.D. Cytoplasmic male sterility: A window to the world of plant mitochondrial-nuclear interactions. Trends Genet. 2007, 23, 81–90. [Google Scholar] [CrossRef]
- Horn, S.; Figl, A.; Rachakonda, P.S.; Fischer, C.; Sucker, A.; Gast, A.; Kadel, S.; Moll, I.; Nagore, E.; Hemminki, K.; et al. TERT promoter mutations in familial and sporadic melanoma. Science 2013, 339, 959–961. [Google Scholar] [CrossRef]
- Soukup, A.A.; Zheng, Y.; Mehta, C.; Wu, J.; Liu, P.; Cao, M.; Hofmann, I.; Zhou, Y.; Zhang, J.; Johnson, K.D.; et al. Single-nucleotide human disease mutation inactivates a blood-regenerative GATA2 enhancer. J. Clin. Investig. 2019, 129, 1180–1192. [Google Scholar] [CrossRef] [Green Version]
- Cai, Y.; Patel, D.J.; Broyde, S.; Geacintov, N.E. Base sequence context effects on nucleotide excision repair. J. Nucleic Acids 2010, 2010, 174252. [Google Scholar] [CrossRef]
- Bębenek, A.; Ziuzia-Graczyk, I. Fidelity of DNA replication-a matter of proofreading. Curr. Genet. 2018, 64, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Fresco, J.R.; Amosova, O. Site-specific self-catalyzed DNA depurination: A biological mechanism that leads to mutations and creates sequence diversity. Annu. Rev. Biochem. 2017, 86, 461–484. [Google Scholar] [CrossRef] [PubMed]
- Milligan, M.J.; Lipovich, L. Pseudogene-derived lncRNAs: Emerging regulators of gene expression. Front. Genet. 2014, 5, 476. [Google Scholar] [CrossRef]
- Johnson Pokorná, M.; Reifová, R. Evolution of B Chromosomes: From Dispensable Parasitic Chromosomes to Essential Genomic Players. Front. Genet. 2021, 12, 727570. [Google Scholar] [CrossRef]
- Rancati, G.; Moffat, J.; Typas, A.; Pavelka, N. Emerging and evolving concepts in gene essentiality. Nat. Rev. Genet. 2018, 19, 34–49. [Google Scholar] [CrossRef]
- Eisenberg, E.; Levanon, E.Y. Human housekeeping genes, revisited. Trends Genet. 2013, 29, 569–574. [Google Scholar] [CrossRef]
- Faulkner, G.J. Retrotransposons: Still mobile in humans. Nat. Rev. Genet. 2022, 23, 458–459. [Google Scholar] [CrossRef] [PubMed]
- Chess, A. Monoallelic gene expression in mammals. Annu. Rev. Genet. 2016, 50, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Reinius, B.; Sandberg, R. Random monoallelic expression of autosomal genes: Stochastic transcription and allele-level regulation. Nat. Rev. Genet. 2015, 16, 653–664. [Google Scholar] [CrossRef]
- Eckersley-Maslin, M.A.; Spector, D.L. Random monoallelic expression: Regulating gene expression one allele at a time. Trends Genet. 2014, 30, 237–244. [Google Scholar] [CrossRef]
- Nakamura, J.; Swenberg, J.A. Endogenous apurinic/apyrimidinic sites in genomic DNA of mammalian tissues. Cancer Res. 1999, 59, 2522–2526. [Google Scholar] [PubMed]
- Vihinen, M. Generic model for biological regulation. F1000Research 2022, 11, 419. [Google Scholar] [CrossRef]
- Schaafsma, G.C.P.; Vihinen, M. Large differences in proportions of harmful and benign amino acid substitutions between proteins and diseases. Hum. Mutat. 2017, 38, 839–848. [Google Scholar] [CrossRef] [PubMed]
- Niroula, A.; Vihinen, M. PON-mt-tRNA: A multifactorial probability-based method for classification of mitochondrial tRNA variations. Nucleic Acids Res. 2016, 44, 2020–2027. [Google Scholar] [CrossRef] [PubMed]
- Amarzguioui, M.; Holen, T.; Babaie, E.; Prydz, H. Tolerance for mutations and chemical modifications in a siRNA. Nucleic Acids Res. 2003, 31, 589–595. [Google Scholar] [CrossRef]
- Ribeiro, A.J.M.; Tyzack, J.D.; Borkakoti, N.; Holliday, G.L.; Thornton, J.M. A global analysis of function and conservation of catalytic residues in enzymes. J. Biol. Chem. 2020, 295, 314–324. [Google Scholar] [CrossRef] [PubMed]
- Pusey, A.; Wolf, M. Inbreeding avoidance in animals. Trends Ecol. Evol. 1996, 11, 201–206. [Google Scholar] [CrossRef]
- Pike, V.L.; Cornwallis, C.K.; Griffin, A.S. Why don’t all animals avoid inbreeding? Proc. Biol. Sci. 2021, 288, 20211045. [Google Scholar] [CrossRef]
- Thünken, T.; Bakker, T.C.; Baldauf, S.A.; Kullmann, H. Active inbreeding in a cichlid fish and its adaptive significance. Curr. Biol. 2007, 17, 225–229. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Lu, X. Female ground tits prefer relatives as extra-pair partners: Driven by kin-selection? Mol. Ecol. 2011, 20, 2851–2863. [Google Scholar] [CrossRef]
- Ganai, R.A.; Johansson, E. DNA replication-a matter of fidelity. Mol. Cell 2016, 62, 745–755. [Google Scholar] [CrossRef] [PubMed]
- Yi, C.; He, C. DNA repair by reversal of DNA damage. Cold Spring Harb. Perspect. Biol. 2013, 5, a012575. [Google Scholar] [CrossRef] [PubMed]
- Pilzecker, B.; Buoninfante, O.A.; Jacobs, H. DNA damage tolerance in stem cells, ageing, mutagenesis, disease and cancer therapy. Nucleic Acids Res. 2019, 47, 7163–7181. [Google Scholar] [CrossRef] [PubMed]
- Turner, K.J.; Vasu, V.; Griffin, D.K. Telomere biology and human phenotype. Cells 2019, 8, 73. [Google Scholar] [CrossRef]
- Hodgkin, J. Genetic Suppression; WormBook: Pasadena, CA, USA, 2005; pp. 1–13. [Google Scholar] [CrossRef]
- Hartman, J.L.t.; Garvik, B.; Hartwell, L. Principles for the buffering of genetic variation. Science 2001, 291, 1001–1004. [Google Scholar] [CrossRef]
- Rossi, A.; Kontarakis, Z. Beyond Mendelian inheritance: Genetic buffering and phenotype variability. Phenomics 2021, 2, 79–87. [Google Scholar] [CrossRef]
- Dehal, P.; Boore, J.L. Two rounds of whole genome duplication in the ancestral vertebrate. PLoS Biol. 2005, 3, e314. [Google Scholar] [CrossRef]
- Singh, P.P.; Isambert, H. OHNOLOGS v2: A comprehensive resource for the genes retained from whole genome duplication in vertebrates. Nucleic Acids Res. 2020, 48, D724–D730. [Google Scholar] [CrossRef]
- Hannay, K.; Marcotte, E.M.; Vogel, C. Buffering by gene duplicates: An analysis of molecular correlates and evolutionary conservation. BMC Genom. 2008, 9, 609. [Google Scholar] [CrossRef]
- Dean, E.J.; Davis, J.C.; Davis, R.W.; Petrov, D.A. Pervasive and persistent redundancy among duplicated genes in yeast. PLoS Genet. 2008, 4, e1000113. [Google Scholar] [CrossRef]
- Ihmels, J.; Collins, S.R.; Schuldiner, M.; Krogan, N.J.; Weissman, J.S. Backup without redundancy: Genetic interactions reveal the cost of duplicate gene loss. Mol. Syst. Biol. 2007, 3, 86. [Google Scholar] [CrossRef] [PubMed]
- Kafri, R.; Bar-Even, A.; Pilpel, Y. Transcription control reprogramming in genetic backup circuits. Nat. Genet. 2005, 37, 295–299. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.H.; Zhao, X.M.; van Noort, V.; Bork, P. Human monogenic disease genes have frequently functionally redundant paralogs. PLoS Comput. Biol. 2013, 9, e1003073. [Google Scholar] [CrossRef]
- Blomen, V.A.; Majek, P.; Jae, L.T.; Bigenzahn, J.W.; Nieuwenhuis, J.; Staring, J.; Sacco, R.; van Diemen, F.R.; Olk, N.; Stukalov, A.; et al. Gene essentiality and synthetic lethality in haploid human cells. Science 2015, 350, 1092–1096. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Birsoy, K.; Hughes, N.W.; Krupczak, K.M.; Post, Y.; Wei, J.J.; Lander, E.S.; Sabatini, D.M. Identification and characterization of essential genes in the human genome. Science 2015, 350, 1096–1101. [Google Scholar] [CrossRef]
- Hart, T.; Chandrashekhar, M.; Aregger, M.; Steinhart, Z.; Brown, K.R.; MacLeod, G.; Mis, M.; Zimmermann, M.; Fradet-Turcotte, A.; Sun, S.; et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell 2015, 163, 1515–1526. [Google Scholar] [CrossRef]
- Lv, W.; Zheng, J.; Luan, M.; Shi, M.; Zhu, H.; Zhang, M.; Lv, H.; Shang, Z.; Duan, L.; Zhang, R.; et al. Comparing the evolutionary conservation between human essential genes, human orthologs of mouse essential genes and human housekeeping genes. Brief. Bioinform. 2015, 16, 922–931. [Google Scholar] [CrossRef] [PubMed]
- Harper, A.R.; Nayee, S.; Topol, E.J. Protective alleles and modifier variants in human health and disease. Nat. Rev. Genet. 2015, 16, 689–701. [Google Scholar] [CrossRef]
- Zeberg, H.; Pääbo, S. A genomic region associated with protection against severe COVID-19 is inherited from Neandertals. Proc. Natl. Acad. Sci. USA 2021, 118, e2026309118. [Google Scholar] [CrossRef]
- Zeberg, H.; Pääbo, S. The major genetic risk factor for severe COVID-19 is inherited from Neanderthals. Nature 2020, 587, 610–612. [Google Scholar] [CrossRef]
- Ciccia, A.; Elledge, S.J. The DNA damage response: Making it safe to play with knives. Mol. Cell 2010, 40, 179–204. [Google Scholar] [CrossRef]
- Blackford, A.N.; Jackson, S.P. ATM, ATR, and DNA-PK: The Trinity at the Heart of the DNA Damage Response. Mol. Cell 2017, 66, 801–817. [Google Scholar] [CrossRef]
- Pack, L.R.; Daigh, L.H.; Meyer, T. Putting the brakes on the cell cycle: Mechanisms of cellular growth arrest. Curr. Opin Cell Biol. 2019, 60, 106–113. [Google Scholar] [CrossRef]
- Roos, W.P.; Kaina, B. DNA damage-induced cell death: From specific DNA lesions to the DNA damage response and apoptosis. Cancer Lett. 2013, 332, 237–248. [Google Scholar] [CrossRef]
- Kreuzer, K.N. DNA damage responses in prokaryotes: Regulating gene expression, modulating growth patterns, and manipulating replication forks. Cold Spring Harb. Perspect. Biol. 2013, 5, a012674. [Google Scholar] [CrossRef]
- Tian, D.; Traw, M.B.; Chen, J.Q.; Kreitman, M.; Bergelson, J. Fitness costs of R-gene-mediated resistance in Arabidopsis thaliana. Nature 2003, 423, 74–77. [Google Scholar] [CrossRef]
- McLaughlin, R.N., Jr.; Malik, H.S. Genetic conflicts: The usual suspects and beyond. J. Exp. Biol. 2017, 220, 6–17. [Google Scholar] [CrossRef]
- Gardner, A.; Úbeda, F. The meaning of intragenomic conflict. Nat. Ecol. Evol. 2017, 1, 1807–1815. [Google Scholar] [CrossRef] [Green Version]
- Werren, J.H. Selfish genetic elements, genetic conflict, and evolutionary innovation. Proc. Natl. Acad. Sci. USA 2011, 108 (Suppl. 2), 10863–10870. [Google Scholar] [CrossRef]
- Luo, S.; Valencia, C.A.; Zhang, J.; Lee, N.C.; Slone, J.; Gui, B.; Wang, X.; Li, Z.; Dell, S.; Brown, J.; et al. Biparental inheritance of mitochondrial DNA in humans. Proc. Natl. Acad. Sci. USA 2018, 115, 13039–13044. [Google Scholar] [CrossRef]
- Hoffmann, A.A.; Turelli, M.; Harshman, L.G. Factors affecting the distribution of cytoplasmic incompatibility in Drosophila simulans. Genetics 1990, 126, 933–948. [Google Scholar] [CrossRef]
- Kageyama, D.; Nishimura, G.; Hoshizaki, S.; Ishikawa, Y. Feminizing Wolbachia in an insect, Ostrinia furnacalis (Lepidoptera: Crambidae). Heredity 2002, 88, 444–449. [Google Scholar] [CrossRef]
- Touzet, P.; Budar, F. Unveiling the molecular arms race between two conflicting genomes in cytoplasmic male sterility? Trends Plant Sci. 2004, 9, 568–570. [Google Scholar] [CrossRef]
- Madgwick, P.G.; Belcher, L.J.; Wolf, J.B. Greenbeard genes: Theory and reality. Trends Ecol. Evol. 2019, 34, 1092–1103. [Google Scholar] [CrossRef]
- James, P.A.; Fortuno, C.; Li, N.; Lim, B.W.X.; Campbell, I.G.; Spurdle, A.B. Estimating the proportion of pathogenic variants from breast cancer case-control data: Application to calibration of ACMG/AMP variant classification criteria. Hum. Mutat. 2022, 43, 882–888. [Google Scholar] [CrossRef]
- Väliaho, J.; Faisal, I.; Ortutay, C.; Smith CI, E.; Vihinen, M. Characterization of all possible single nucleotide change –caused amino acid substitutions in the kinase domain of Bruton tyrosine kinase. Hum. Mutat. 2015, 36, 638–647. [Google Scholar] [CrossRef]
- Sondka, Z.; Bamford, S.; Cole, C.G.; Ward, S.A.; Dunham, I.; Forbes, S.A. The COSMIC Cancer Gene Census: Describing genetic dysfunction across all human cancers. Nat. Rev. Cancer 2018, 18, 696–705. [Google Scholar] [CrossRef]
- Niroula, A.; Vihinen, M. Harmful somatic amino acid substitutions affect key pathways in cancers. BMC Med. Genom. 2015, 8, 53. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Need, A.C.; Petrovski, S.; Goldstein, D.B. One gene, many neuropsychiatric disorders: Lessons from Mendelian diseases. Nat. Neurosci. 2014, 17, 773–781. [Google Scholar] [CrossRef]
- Zhu, A.Y.; Costain, G.; Cytrynbaum, C.; Weksberg, R.; Cohn, R.D.; Ali, A. Novel heterozygous variants in PXDN cause different anterior segment dysgenesis phenotypes in monozygotic twins. Ophthalmic Genet. 2021, 42, 624–630. [Google Scholar] [CrossRef]
- Dulovic-Mahlow, M.; König, I.R.; Trinh, J.; Diaw, S.H.; Urban, P.P.; Knappe, E.; Kuhnke, N.; Ingwersen, L.C.; Hinrichs, F.; Weber, J.; et al. Discordant monozygotic Parkinson disease twins: Role of mitochondrial integrity. Ann. Neurol. 2021, 89, 158–164. [Google Scholar] [CrossRef] [PubMed]
- Ketelaar, M.E.; Hofstra, E.M.; Hayden, M.R. What monozygotic twins discordant for phenotype illustrate about mechanisms influencing genetic forms of neurodegeneration. Clin. Genet. 2012, 81, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Manchia, M.; Cullis, J.; Turecki, G.; Rouleau, G.A.; Uher, R.; Alda, M. The impact of phenotypic and genetic heterogeneity on results of genome wide association studies of complex diseases. PLoS ONE 2013, 8, e76295. [Google Scholar] [CrossRef]
- Zlotogora, J. Penetrance and expressivity in the molecular age. Genet. Med. 2003, 5, 347–352. [Google Scholar] [CrossRef]
- Gruber, C.; Bogunovic, D. Incomplete penetrance in primary immunodeficiency: A skeleton in the closet. Hum. Genet. 2020, 139, 745–757. [Google Scholar] [CrossRef] [PubMed]
- Vihinen, M. How to define pathogenicity, health, and disease? Hum. Mutat. 2017, 38, 129–136. [Google Scholar] [CrossRef]
- Vihinen, M. Strategy for disease diagnosis, progression prediction, risk group stratification and teatment-Case of COVID-19. Front. Med. 2020, 7, 294. [Google Scholar] [CrossRef]
- Hanahan, D.; Weinberg, R.A. The hallmarks of cancer. Cell 2000, 100, 57–70. [Google Scholar] [CrossRef] [Green Version]
- Mizuguchi, T.; Nakashima, M.; Kato, M.; Okamoto, N.; Kurahashi, H.; Ekhilevitch, N.; Shiina, M.; Nishimura, G.; Shibata, T.; Matsuo, M.; et al. Loss-of-function and gain-of-function mutations in PPP3CA cause two distinct disorders. Hum. Mol. Genet. 2018, 27, 1421–1433. [Google Scholar] [CrossRef]
- Juric, M.K.; Ghimire, S.; Ogonek, J.; Weissinger, E.M.; Holler, E.; van Rood, J.J.; Oudshoorn, M.; Dickinson, A.; Greinix, H.T. Milestones of hematopoietic stem cell transplantation-from first human studies to current developments. Front. Immunol. 2016, 7, 470. [Google Scholar] [CrossRef]
- Lukashev, A.N.; Zamyatnin, A.A., Jr. Viral vectors for gene therapy: Current state and clinical perspectives. Biochemistry (Moscow) 2016, 81, 700–708. [Google Scholar] [CrossRef] [PubMed]
- Zu, H.; Gao, D. Non-viral vectors in gene therapy: Recent development, challenges, and prospects. AAPS J. 2021, 23, 78. [Google Scholar] [CrossRef] [PubMed]
- Doudna, J.A. The promise and challenge of therapeutic genome editing. Nature 2020, 578, 229–236. [Google Scholar] [CrossRef] [PubMed]
- Patch, C.; Middleton, A. Genetic counselling in the era of genomic medicine. Br. Med. Bull 2018, 126, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Niroula, A.; Vihinen, M. Variation interpretation predictors: Principles, types, performance, and choice. Hum. Mutat. 2016, 37, 579–597. [Google Scholar] [CrossRef]
- Vihinen, M. Problems in variation interpretation guidelines and in their implementation in computational tools. Mol. Genet. Genom. Med. 2020, 8, e1206. [Google Scholar] [CrossRef]
- Niroula, A.; Urolagin, S.; Vihinen, M. PON-P2: Prediction method for fast and reliable identification of harmful variants. PLoS ONE 2015, 10, e0117380. [Google Scholar] [CrossRef]
- Yang, Y.; Shao, A.; Vihinen, M. PON-All, amino acid substitution tolerance predictor for all organisms. Front. Mol. Biosci 2022, 9, 867572. [Google Scholar] [CrossRef]
- Niroula, A.; Vihinen, M. Predicting severity of disease-causing variants. Hum. Mutat. 2017, 38, 357–364. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–423. [Google Scholar] [CrossRef]
- Thompson, B.A.; Spurdle, A.B.; Plazzer, J.P.; Greenblatt, M.S.; Akagi, K.; Al-Mulla, F.; Bapat, B.; Bernstein, I.; Capella, G.; den Dunnen, J.T.; et al. Application of a 5-tiered scheme for standardized classification of 2,360 unique mismatch repair gene variants in the InSiGHT locus-specific database. Nat. Genet. 2014, 46, 107–115. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Hoover, J.; et al. ClinVar: Public archive of interpretations of clinically relevant variants. Nucleic Acids Res. 2016, 44, D862–D868. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Lappalainen, I.; Lopez, J.; Skipper, L.; Hefferon, T.; Spalding, J.D.; Garner, J.; Chen, C.; Maguire, M.; Corbett, M.; Zhou, G.; et al. DbVar and DGVa: Public archives for genomic structural variation. Nucleic Acids Res. 2013, 41, D936–D941. [Google Scholar] [CrossRef]
- Sirugo, G.; Williams, S.M.; Tishkoff, S.A. The missing diversity in human genetic studies. Cell 2019, 177, 26–31. [Google Scholar] [CrossRef]
- Alfredsson, L.; Olsson, T. Lifestyle and environmental factors in multiple sclerosis. Cold Spring Harb. Perspect. Med. 2019, 9, a028944. [Google Scholar] [CrossRef]
- Zhang, Y.B.; Pan, X.F.; Chen, J.; Cao, A.; Xia, L.; Zhang, Y.; Wang, J.; Li, H.; Liu, G.; Pan, A. Combined lifestyle factors, all-cause mortality and cardiovascular disease: A systematic review and meta-analysis of prospective cohort studies. J. Epidemiol. Community Health 2021, 75, 92–99. [Google Scholar] [CrossRef]
- Marras, C.; Canning, C.G.; Goldman, S.M. Environment, lifestyle, and Parkinson’s disease: Implications for prevention in the next decade. Mov. Disord. 2019, 34, 801–811. [Google Scholar] [CrossRef]
- Raraigh, K.S.; Han, S.T.; Davis, E.; Evans, T.A.; Pellicore, M.J.; McCague, A.F.; Joynt, A.T.; Lu, Z.; Atalar, M.; Sharma, N.; et al. Functional assays are essential for interpretation of missense variants associated with variable expressivity. Am. J. Hum. Genet. 2018, 102, 1062–1077. [Google Scholar] [CrossRef]
- Goetz, L.H.; Schork, N.J. Personalized medicine: Motivation, challenges, and progress. Fertil. Steril. 2018, 109, 952–963. [Google Scholar] [CrossRef]
- Carrasco-Ramiro, F.; Peiró-Pastor, R.; Aguado, B. Human genomics projects and precision medicine. Gene 2017, 24, 551–561. [Google Scholar] [CrossRef]

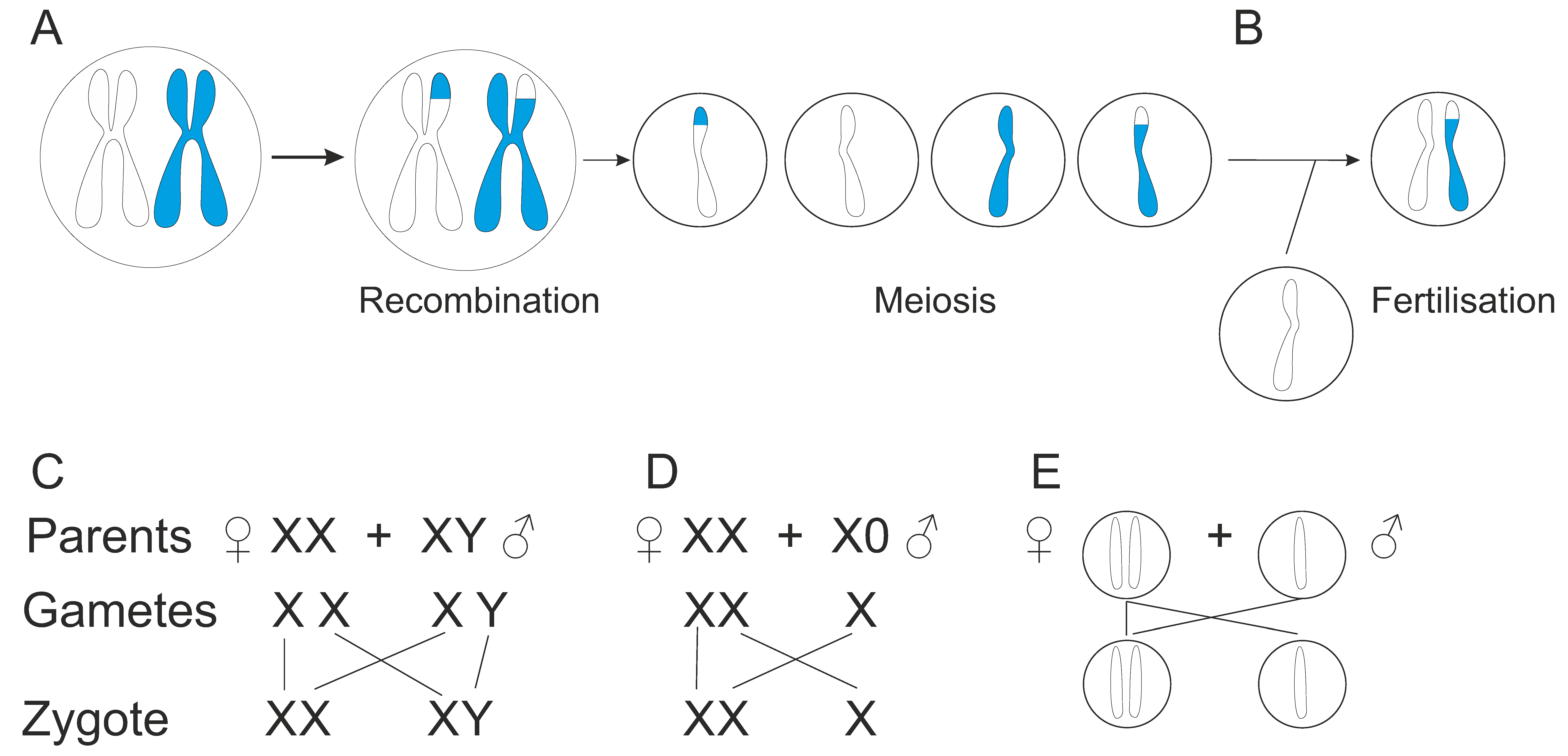
| DNA Lesion | Frequency per Cell per Day |
|---|---|
| Endogeneous | |
| Cytosine deamination | 100 |
| Cyclopurine adduct | 100 |
| Depyrimidation | 100 |
| Depurination | 10,000 |
| 8-oxoG | 1000 |
| Monodialdehyde adducts | 1000 |
| Alkylation adducts | 1000 |
| Single strand breaks | 10,000 |
| Double strand breaks | 10 |
| Environmental | |
| Damaged bases | 10 |
| Photodimers | 100 |
| Single strand breaks | 2–5 |
| Double strand breaks | 0.25 |
| Variation | Mean Value, Range |
|---|---|
| Variation type | |
| SNV | 3.53–4.31 × 106 |
| Insertion or deletion | 546,000–625,000 |
| Large deletion | 939–1100 |
| CNV | 153–170 |
| Inversion | 9–12 |
| Mobile genetic element | 1012–1222 |
| Variation effect or site | |
| Non-synonymous | 10,200–12,200 |
| Synonymous | 11,200–13,800 |
| Untranslated region | 30,000–37,200 |
| Intron | 1.68–2.06 × 106 |
| Transcription factor binding site | 748–927 |
| Promoter | 81,600–102,000 |
| Insulator | 57,700–70,900 |
| Enhancer | 288,000–354,000 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vihinen, M. Individual Genetic Heterogeneity. Genes 2022, 13, 1626. https://doi.org/10.3390/genes13091626
Vihinen M. Individual Genetic Heterogeneity. Genes. 2022; 13(9):1626. https://doi.org/10.3390/genes13091626
Chicago/Turabian StyleVihinen, Mauno. 2022. "Individual Genetic Heterogeneity" Genes 13, no. 9: 1626. https://doi.org/10.3390/genes13091626
APA StyleVihinen, M. (2022). Individual Genetic Heterogeneity. Genes, 13(9), 1626. https://doi.org/10.3390/genes13091626






