A Transformation and Genome Editing System for Cassava Cultivar SC8
Abstract
:1. Introduction
2. Materials and Methods
2.1. Production of Friable Embryogenic Calli (FECs) from Cassava SC8
2.2. Vector Constructions
2.3. Effect of Agrobacterium Cell Densities for Transformation Efficiency
2.4. Effect of Acetosyringone Concentrations for Transformation Efficiency
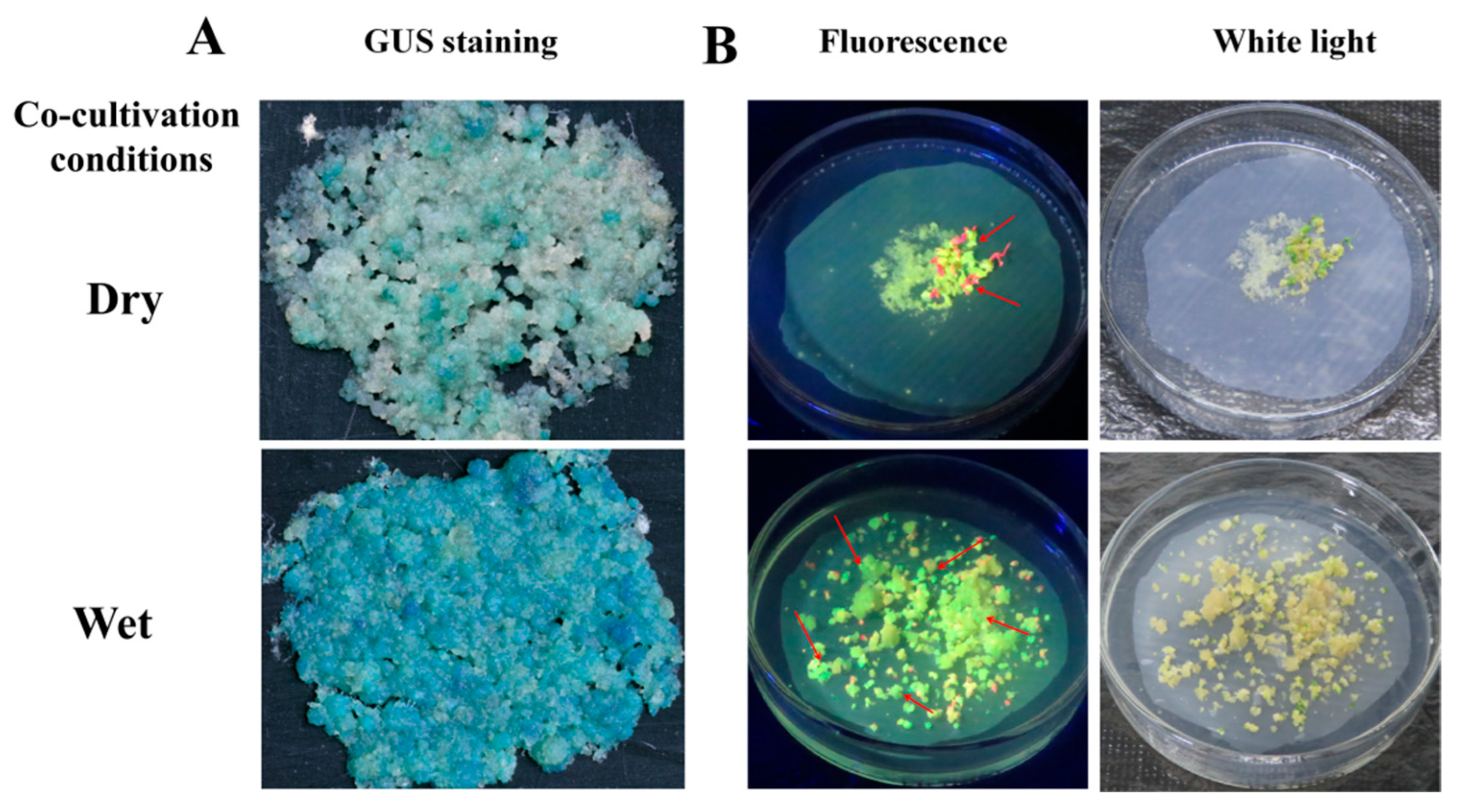
2.5. Effect of Cocultivation Conditions for Transformation Efficiency
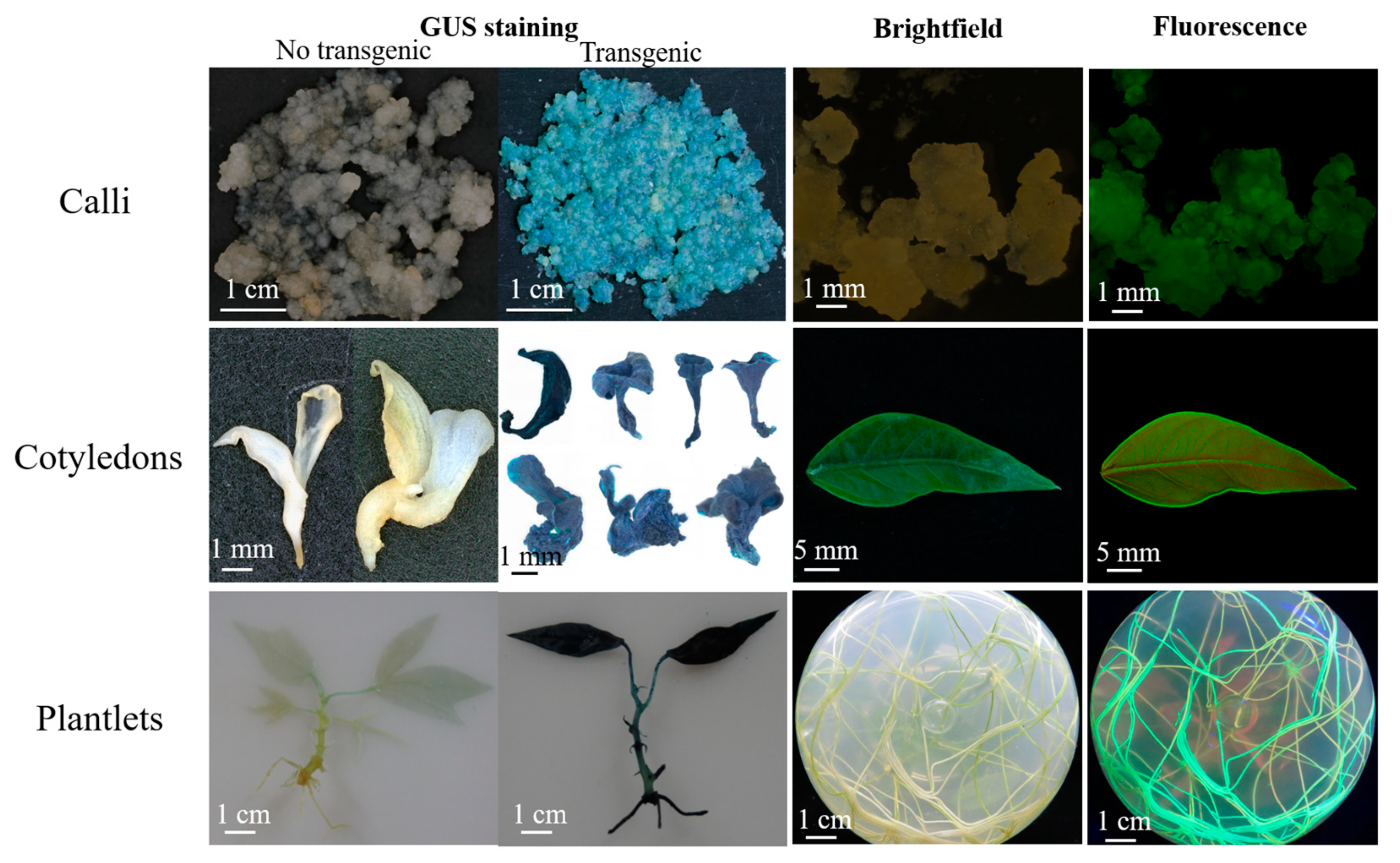
2.6. Selection and Regeneration of Transgenic Plants
2.7. Analysis of GUS and GFP Expression
2.8. PCR Analysis
2.9. Sanger Sequencing and Hi-TOM Sequencing
2.10. Statistical Analysis
3. Results
3.1. Effect of Agrobacterium Cell Concentration on Cassava SC8 Transformation
3.2. Effect of AS Concentration on Cassava SC8 Transformation
3.3. Effect of Cocultivation Conditions on Cassava SC8 Transformation
3.4. Construction of the Optimized Cassava SC8 Transformation Protocol
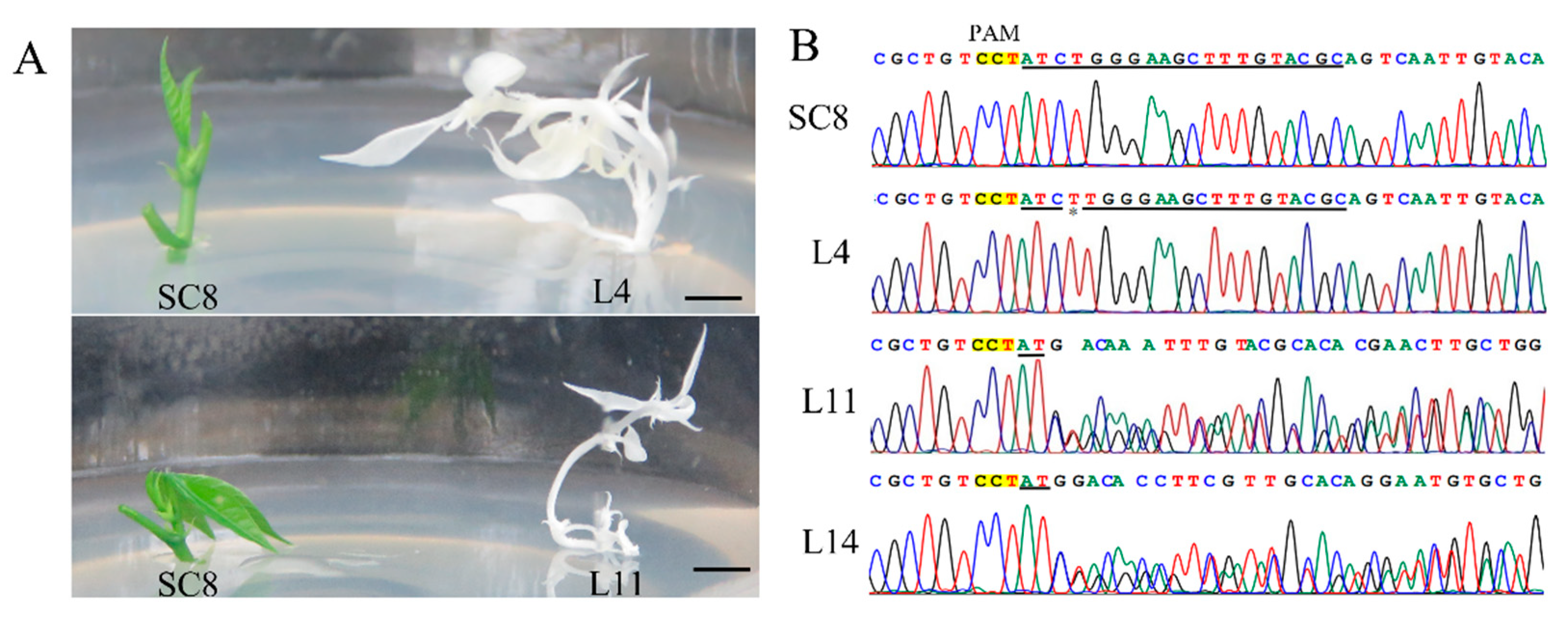
3.5. CRISPR/Cas9-Mediated Mutagenesis in Cassava SC8
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Parmar, A.; Sturm, B.; Hensel, O. Crops that feed the world: Production and improvement of cassava for food, feed, and industrial uses. Food Secur. 2017, 9, 907–927. [Google Scholar] [CrossRef]
- Souza, L.S.; Alves, A.A.C.; de Oliveira, E.J. Phenological diversity of flowering and fruiting in cassava germplasm. Sci. Hortic. 2020, 265, 109253. [Google Scholar] [CrossRef]
- Ceballos, H.; Rojanaridpiched, C.; Phumichai, C.; Becerra, L.A.; Kittipadakul, P.; Iglesias, C.; Gracen, V.E. Excellence in cassava breeding: Perspectives for the future. Crop Breed. Genet. Genom. 2020, 2, e200008. [Google Scholar]
- Zhao, S.S.; Dufour, D.; Sánchez, T.; Ceballos, H.; Zhang, P. Development of waxy cassava with different biological and physico-chemical characteristics of starches for industrial applications. Biotechnol. Bioeng. 2011, 108, 1925–1935. [Google Scholar] [CrossRef]
- Bull, S.E.; Seung, D.; Chanez, C.; Mehta, D.; Kuon, J.E.; Truernit, E.; Hochmuth, A.; Zurkirchen, I.; Zeeman, S.C.; Gruissem, W. Accelerated ex situ breeding of GBSS-and PTST1-edited cassava for modified starch. Sci. Adv. 2018, 4, eaat6086. [Google Scholar] [CrossRef]
- Zhou, W.; Zhao, S.; He, S.; Ma, Q.; Lu, X.; Hao, X.; Wang, H.; Yang, J.; Zhang, P. Production of very-high-amylose cassava by post-transcriptional silencing of branching enzyme genes. J. Integr. Plant Biol. 2020, 62, 832–846. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Hostettler, C.E.; Damberger, F.F.; Kossmann, J.; Zeeman, S.C. Modification of cassava root starch phosphorylation enhances starch functional properties. Front. Plant Sci. 2018, 9, 1562. [Google Scholar] [CrossRef]
- Chen, Q.; Liang, X.; Liu, Y.; Liu, X.; Zhao, H.; Li, K.; Chen, S.; Wang, H.; Han, Z.; Wu, M. Overexpression of leucoanthocyanidin reductase or anthocyanidin reductase elevates tannins content and confers cassava resistance to two-spotted spider mite. Front. Plant Sci. 2022, 13, 994866. [Google Scholar] [CrossRef]
- Lim, Y.W.; Mansfeld, B.N.; Schläpfer, P.; Gilbert, K.B.; Narayanan, N.N.; Qi, W.; Wang, Q.; Zhong, Z.; Boyher, A.; Gehan, J. Mutations in DNA polymerase δ subunit 1 co-segregate with CMD2-type resistance to Cassava Mosaic Geminiviruses. Nat. Commun. 2022, 13, 3933. [Google Scholar] [CrossRef]
- Li, S.; Cheng, Z.; Li, Z.; Dong, S.; Yu, X.; Zhao, P.; Liao, W.; Yu, X.; Peng, M. MeSPL9 attenuates drought resistance by regulating JA signaling and protectant metabolite contents in Cassava. Theor. Appl. Genet. 2022, 135, 817–832. [Google Scholar] [CrossRef]
- Xu, J.; Yang, J.; Duan, X.; Jiang, Y.; Zhang, P. Increased expression of native cytosolic Cu/Zn superoxide dismutase and ascorbate peroxidase improves tolerance to oxidative and chilling stresses in cassava (Manihot esculenta Crantz). BMC Plant Biol. 2014, 14, 208. [Google Scholar] [CrossRef] [PubMed]
- Narayanan, N.; Beyene, G.; Chauhan, R.D.; Gaitán-Solís, E.; Gehan, J.; Butts, P.; Siritunga, D.; Okwuonu, I.; Woll, A.; Jiménez-Aguilar, D.M. Biofortification of field-grown cassava by engineering expression of an iron transporter and ferritin. Nat. Biotechnol. 2019, 37, 144–151. [Google Scholar] [CrossRef] [PubMed]
- Li, K.T.; Moulin, M.; Mangel, N.; Albersen, M.; Verhoeven-Duif, N.M.; Ma, Q.; Zhang, P.; Fitzpatrick, T.B.; Gruissem, W.; Vanderschuren, H. Increased bioavailable vitamin B 6 in field-grown transgenic cassava for dietary sufficiency. Nat. Biotechnol. 2015, 33, 1029–1032. [Google Scholar] [CrossRef] [PubMed]
- Ma, Q.; Xu, J.; Feng, Y.; Wu, X.; Lu, X.; Zhang, P. Knockdown of p-coumaroyl shikimate/quinate 3′-hydroxylase delays the occurrence of post-harvest physiological deterioration in Cassava storage roots. Int. J. Mol. Sci. 2022, 23, 9231. [Google Scholar] [CrossRef]
- Beyene, G.; Chauhan, R.D.; Gehan, J.; Siritunga, D.; Taylor, N. Cassava shrunken-2 homolog MeAPL3 determines storage root starch and dry matter content and modulates storage root postharvest physiological deterioration. Plant Mol. Biol. 2022, 109, 1–17. [Google Scholar] [CrossRef]
- Nyaboga, E.; Njiru, J.; Nguu, E.; Gruissem, W.; Vanderschuren, H.; Tripathi, L. Unlocking the potential of tropical root crop biotechnology in east Africa by establishing a genetic transformation platform for local farmer-preferred cassava cultivars. Front. Plant Sci. 2013, 4, 526. [Google Scholar] [CrossRef]
- Elegba, W.; McCallum, E.; Gruissem, W.; Vanderschuren, H. Efficient genetic transformation and regeneration of a farmer-preferred cassava cultivar from Ghana. Front. Plant Sci. 2021, 12, 668042. [Google Scholar] [CrossRef]
- Chauhan, R.D.; Beyene, G.; Kalyaeva, M.; Fauquet, C.M.; Taylor, N. Improvements in Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) for large-scale production of transgenic plants. Plant Cell Tiss. Org. 2015, 121, 591–603. [Google Scholar] [CrossRef]
- Chetty, C.; Rossin, C.; Gruissem, W.; Vanderschuren, H.; Rey, M. Empowering biotechnology in southern Africa: Establishment of a robust transformation platform for the production of transgenic industry-preferred cassava. New Biotechnol. 2013, 30, 136–143. [Google Scholar] [CrossRef]
- Nyaboga, E.N.; Njiru, J.M.; Tripathi, L. Factors influencing somatic embryogenesis, regeneration, and Agrobacterium-mediated transformation of cassava (Manihot esculenta Crantz) cultivar TME14. Front. Plant Sci. 2015, 6, 411. [Google Scholar] [CrossRef]
- Lentz, E.M.; Eisner, S.; McCallum, E.J.; Schlegel, K.; Campos, F.A.P.; Gruissem, W.; Vanderschuren, H. Genetic transformation of recalcitrant cassava by embryo selection and increased hormone levels. Methods Protoc. 2018, 1, 42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zainuddin, I.M.; Schlegel, K.; Gruissem, W.; Vanderschuren, H. Robust transformation procedure for the production of transgenic farmer-preferred cassava landraces. Plant Methods 2012, 8, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Gu, B.; Yao, Q.; Li, K.; Chen, S. Change in physicochemical traits of cassava roots and starches associated with genotypes and environmental factors. Starch Starke 2013, 65, 253–263. [Google Scholar] [CrossRef]
- Malik, A.I.; Kongsil, P.; Nguyễn, V.A.; Ou, W.; Sholihin; Srean, P.; Sheela, M.N.; López-Lavalle, L.A.B.; Utsumi, Y.; Lu, C.; et al. Cassava breeding and agronomy in Asia: 50 years of history and future directions. Breed. Sci. 2020, 70, 145–151. [Google Scholar] [CrossRef] [PubMed]
- Fang, J.; Liu, J.; Tang, K. Optimization of the conditions for Agrobacterium-mediated cassava genetic transformation. Chin. J. Trop. Crops 2011, 32, 1697–1703. [Google Scholar]
- Guo, Y.; Yin, Q.; Kong, H.; Huang, Q.; Zuo, J.; He, L.; Guo, A. Effect of silver nitrate on Agrobacterium mediated genetic transformation of cassava. Biotechnol. Bull. 2013, 3, 166–170. [Google Scholar]
- Li, R.M.; Hu, X.W.; Li, K.M.; Fu, S.P.; Guo, J.C. CaCl2 enhanced somatic embryogenesis in Manihot esculenta Crantz. Biosci. Biotechnol. Biochem. 2009, 73, 2513–2515. [Google Scholar] [CrossRef]
- Liu, J.; Chen, X.; Wang, S.; Wang, Y.; Ouyang, Y.; Yao, Y.; Li, R.; Fu, S.; Hu, X.; Guo, J. MeABL5, an ABA insensitive 5-like basic leucine zipper transcription factor, positively regulates MeCWINV3 in cassava (Manihot esculenta Crantz). Front. Plant Sci. 2019, 10, 772. [Google Scholar] [CrossRef]
- Odipio, J.; Alicai, T.; Ingelbrecht, I.; Nusinow, D.A.; Bart, R.; Taylor, N.J. Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 2017, 8, 1780. [Google Scholar] [CrossRef]
- Gomez, M.A.; Lin, Z.D.; Moll, T.; Chauhan, R.D.; Hayden, L.; Renninger, K.; Beyene, G.; Taylor, N.J.; Carrington, J.C.; Staskawicz, B.J. Simultaneous CRISPR/Cas9-mediated editing of cassava eIF 4E isoforms nCBP-1 and nCBP-2 reduces cassava brown streak disease symptom severity and incidence. Plant Biotechnol. J. 2019, 17, 421–434. [Google Scholar] [CrossRef]
- Luo, S.; Ma, Q.; Zhong, Y.; Jing, J.; Wei, Z.; Zhou, W.; Lu, X.; Tian, Y.; Zhang, P. Editing of the starch branching enzyme gene SBE2 generates high-amylose storage roots in cassava. Plant Mol. Biol. 2022, 108, 429–442. [Google Scholar] [CrossRef]
- Gomez, M.A.; Berkoff, K.C.; Gill, B.K.; Iavarone, A.T.; Lieberman, S.E.; Ma, J.M.; Schultink, A.; Wyman, S.K.; Chauhan, R.D.; Taylor, N.J. CRISPR-Cas9-mediated knockout of CYP79D1 and CYP79D2 in cassava attenuates toxic cyanogen production. bioRxiv 2021.
- Wang, Y.; Geng, M.; Pan, R.; Zhang, T.; Zhen, X.; Che, Y.; Li, R.; Liu, J.; Chen, Y.; Guo, J. Engineering bacterial blight-resistant plants through CRISPR/Cas9-targeted editing of the MeSWEET10a promoter in cassava. bioRxiv 2022.
- Veley, K.M.; Okwuonu, I.; Jensen, G.; Yoder, M.; Taylor, N.J.; Meyers, B.C.; Bart, R.S. Gene tagging via CRISPR-mediated homology-directed repair in cassava. G3 2021, 11, jkab028. [Google Scholar] [CrossRef] [PubMed]
- Mehta, D.; Stürchler, A.; Anjanappa, R.B.; Zaidi, S.S.-e.-A.; Hirsch-Hoffmann, M.; Gruissem, W.; Vanderschuren, H. Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 2019, 20, 1–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, Z.; Zhang, Z.; Hua, K.; Gao, X.; Mao, Y.; Botella, J.R.; Zhu, J.-K. A highly efficient cell division-specific CRISPR/Cas9 system generates homozygous mutants for multiple genes in Arabidopsis. Int. J. Mol. Sci. 2018, 19, 3925. [Google Scholar] [CrossRef]
- Gresshoff, P.M.; Doy, C.H. Derivation of a haploid cell line from Vitis vinifera and the importance of the stage of meiotic development of anthers for haploid culture of this and other genera. Z. Pflanzenphysiol. 1974, 73, 132–141. [Google Scholar] [CrossRef]
- Liu, Q.; Wang, C.; Jiao, X.; Zhang, H.; Song, L.; Li, Y.; Gao, C.; Wang, K. Hi-TOM: A platform for high-throughput tracking of mutations induced by CRISPR/Cas systems. Sci. China Life Sci. 2019, 62, 1–7. [Google Scholar] [CrossRef]
- Ruan, M.B.; Guo, X.; Wang, B.; Yang, Y.L.; Li, W.Q.; Yu, X.L.; Zhang, P.; Peng, M. Genome-wide characterization and expression analysis enables identification of abiotic stress-responsive MYB transcription factors in cassava (Manihot esculenta). J. Exp. Bot. 2017, 68, 3657–3672. [Google Scholar] [CrossRef]
- Beyene, G.; Solomon, F.R.; Chauhan, R.D.; Gaitán-Solis, E.; Narayanan, N.; Gehan, J.; Siritunga, D.; Stevens, R.L.; Jifon, J.; Van Eck, J. Provitamin A biofortification of cassava enhances shelf life but reduces dry matter content of storage roots due to altered carbon partitioning into starch. Plant Biotechnol. J. 2018, 16, 1186–1200. [Google Scholar] [CrossRef]
- Ligaba-Osena, A.; Jones, J.; Donkor, E.; Chandrayan, S.; Pole, F.; Wu, C.H.; Vieille, C.; Adams, M.W.; Hankoua, B.B. Novel bioengineered cassava expressing an archaeal starch degradation system and a bacterial ADP-Glucose Pyrophosphorylase for starch self-digestibility and yield increase. Front. Plant Sci. 2018, 9, 192. [Google Scholar] [CrossRef] [PubMed]
- Raemakers, K.; Schreuder, M.; Suurs, L.; Furrer-Verhorst, H.; Vincken, J.P.; de Vetten, N.; Jacobsen, E.; Visser, R.G. Improved cassava starch by antisense inhibition of granule-bound starch synthase I. Mol. Breed. 2005, 16, 163–172. [Google Scholar] [CrossRef]
- Bull, S.E.; Owiti, J.A.; Niklaus, M.; Beeching, J.R.; Gruissem, W.; Vanderschuren, H. Agrobacterium-mediated transformation of friable embryogenic calli and regeneration of transgenic cassava. Nat. Protoc. 2009, 4, 1845–1854. [Google Scholar] [CrossRef] [PubMed]
- Taylor, N.; Gaitán-Solís, E.; Moll, T.; Trauterman, B.; Jones, T.; Pranjal, A.; Trembley, C.; Abernathy, V.; Corbin, D.; Fauquet, C.M. A high-throughput platform for the production and analysis of transgenic cassava (Manihot esculenta) plants. Trop. Plant Biol. 2012, 5, 127–139. [Google Scholar] [CrossRef]






| Cell Density OD600 | Frequency of GUS Expression (%) | Average No. of Cotyledonary Stage Embryos on Selective Medium |
|---|---|---|
| 0.05 | 11.83 ± 5.56 d | 2.5 ± 2.07 d |
| 0.25 | 12.00 ± 6.06 d | 4.50 ± 2.88 d |
| 0.45 | 22.67 ± 4.27 c | 32.00 ± 8.02 c |
| 0.65 | 40.00 ± 4.47 a | 79.33 ± 10.46 a |
| 0.85 | 31.33 ± 4.89 b | 50.17 ± 8.50 b |
| Treatment | Frequency of GUS Expression (%) | Average No. of Cotyledonary Stage Embryos on Selective Medium |
|---|---|---|
| 50 µM | 11.67 ± 3.61 e | 10.00 ± 5.02 d |
| 100 µM | 12.33 ± 4.63 e | 16.17 ± 6.74 d |
| 150 µM | 21.67 ± 3.98 d | 30.00 ± 6.81 c |
| 200 µM | 38.67 ± 4.92 c | 68.33 ± 8.45 b |
| 250 µM | 69.83 ± 5.95 a | 114.50 ± 7.61 a |
| 300 µM | 50.00 ± 7.21 b | 78.83 ± 11.89 b |
| Treatment | Frequency of GUS Expression (%) | Average No. of Cotyledonary Stage Embryos on Selective Medium |
|---|---|---|
| 1 d | 15.50 ± 5.54 c | 13.17 ± 6.34 d |
| 3 d | 74.33 ± 7.99 a | 122.17 ± 11.69 a |
| 5 d | 46.83 ± 5.60 b | 74.00 ± 9.72 b |
| 7 d | 15.17 ± 4.31 c | 44.50 ± 9.42 c |
| Treatment | Frequency of GUS Expression (%) | Average No. of Cotyledonary Stage Embryos on Selective Medium |
|---|---|---|
| dry | 52.33 ± 8.16 b | 130.33 ± 10.88 b |
| wet | 82.83 ± 6.08 a | 196.00 ± 11.08 a |
| Experiments | Number of Cotyledons | Number of Transgenic Plants |
|---|---|---|
| 1 | 187 | 132 |
| 2 | 170 | 124 |
| 3 | 191 | 143 |
| Variety | Promoter | Plant Lines Analyzed | Mutation Efficiency | Homozygous Mono-Allelic | Homozygous Bi-Allelic | Heterozygous |
|---|---|---|---|---|---|---|
| SC8 a | YAO | 48 | 93.75% (45/48) | 45.83% (22/48) | 29.16% (14/48) | 18.75% (9/14) |
| 60444 b | CaMV35S | 9 | 100.00% (9/9) | 11.11% (1/9) | 11.11% (1/9) | 77.78% (7/9) |
| TME204 b | CaMV35S | 9 | 100.00% (9/9) | 0.00% (0/9) | 33.33% (3/9) | 66.67% (6/9) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Y.-J.; Lu, X.-H.; Zhen, X.-H.; Yang, H.; Che, Y.-N.; Hou, J.-Y.; Geng, M.-T.; Liu, J.; Hu, X.-W.; Li, R.-M.; et al. A Transformation and Genome Editing System for Cassava Cultivar SC8. Genes 2022, 13, 1650. https://doi.org/10.3390/genes13091650
Wang Y-J, Lu X-H, Zhen X-H, Yang H, Che Y-N, Hou J-Y, Geng M-T, Liu J, Hu X-W, Li R-M, et al. A Transformation and Genome Editing System for Cassava Cultivar SC8. Genes. 2022; 13(9):1650. https://doi.org/10.3390/genes13091650
Chicago/Turabian StyleWang, Ya-Jie, Xiao-Hua Lu, Xing-Hou Zhen, Hui Yang, Yan-Nian Che, Jing-Yi Hou, Meng-Ting Geng, Jiao Liu, Xin-Wen Hu, Rui-Mei Li, and et al. 2022. "A Transformation and Genome Editing System for Cassava Cultivar SC8" Genes 13, no. 9: 1650. https://doi.org/10.3390/genes13091650
APA StyleWang, Y.-J., Lu, X.-H., Zhen, X.-H., Yang, H., Che, Y.-N., Hou, J.-Y., Geng, M.-T., Liu, J., Hu, X.-W., Li, R.-M., Guo, J.-C., & Yao, Y. (2022). A Transformation and Genome Editing System for Cassava Cultivar SC8. Genes, 13(9), 1650. https://doi.org/10.3390/genes13091650






