Comparison between the Gametophyte and the Sporophyte Transcriptomes of the Endangered Fern Vandenboschia speciosa
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Sequencing
2.2. In Silico Decontamination of the Gametophyte Reads
2.3. V. speciosa De Novo Transcriptome Assembly
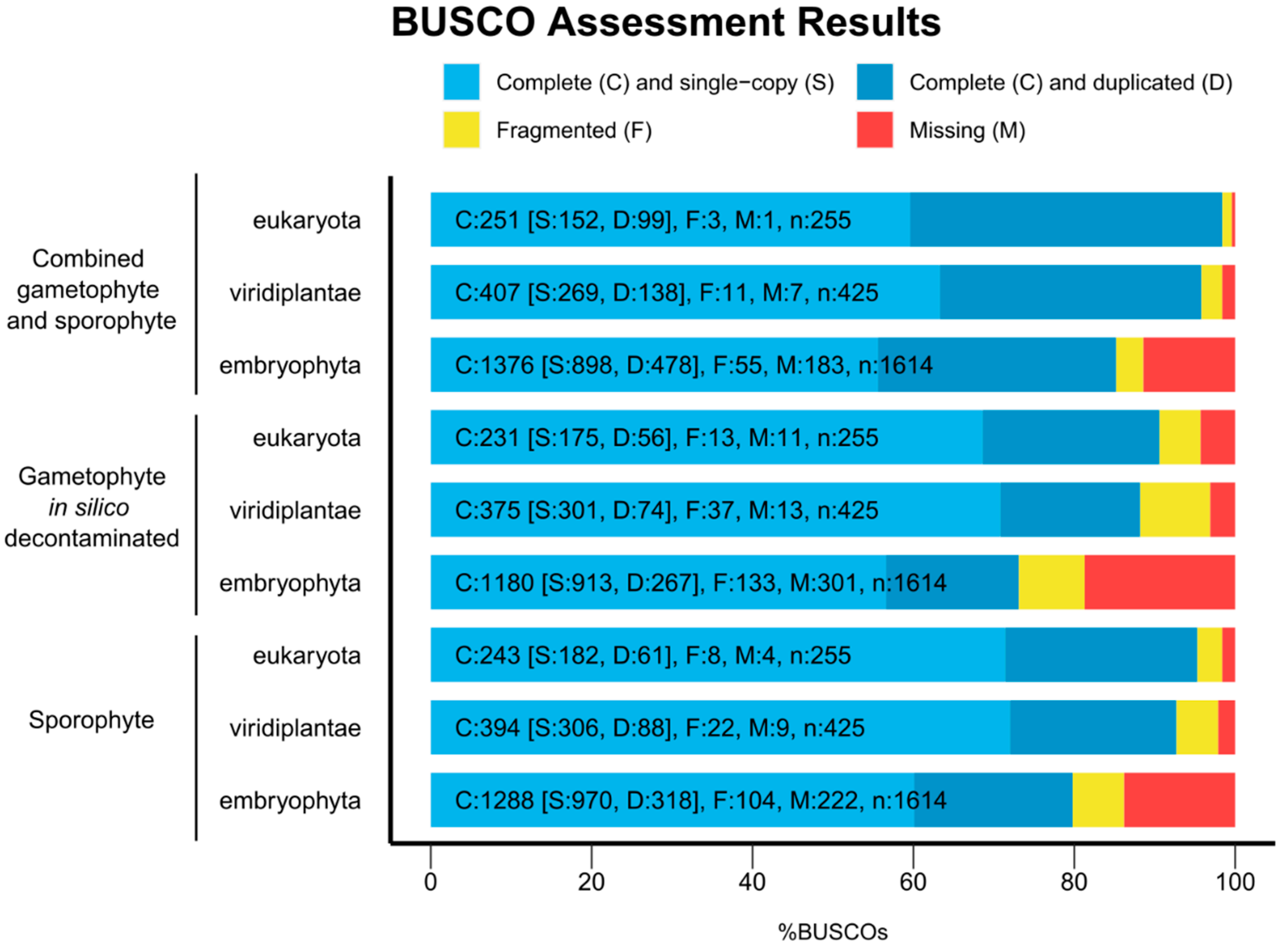
2.4. Assessment of Transcriptome Completion, Coding Sequence Presence, and Functional Annotation
2.5. Transcriptome Expression Profile
3. Results and Discussion
3.1. Cleaning up Cross-Contamination in the Gametophyte Reads
3.2. De Novo Assembly of the V. speciosa Transcriptome
3.3. Differences in Transcript Expression between Tissues
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Matz, M.V. Fantastic Beasts and How To Sequence Them: Ecological Genomics for Obscure Model Organisms. Trends Genet. 2018, 34, 121–132. [Google Scholar] [CrossRef]
- Ellegren, H. Genome sequencing and population genomics in non-model organisms. Trends Ecol. Evol. 2014, 29, 51–63. [Google Scholar] [CrossRef] [PubMed]
- Ekblom, R.; Galindo, J. Applications of next generation sequencing in molecular ecology of non-model organisms. Heredity 2011, 107, 1–15. [Google Scholar] [CrossRef] [Green Version]
- da Fonseca, R.R.; Albrechtsen, A.; Themudo, G.E.; Ramos-Madrigal, J.; Sibbesen, J.A.; Maretty, L.; Zepeda-Mendoza, M.L.; Campos, P.F.; Heller, R.; Pereira, R.J. Next-generation biology: Sequencing and data analysis approaches for non-model organisms. Mar. Genom. 2016, 30, 3–13. [Google Scholar] [CrossRef]
- Yadav, S.; Stow, A.J.; Dudaniec, R.Y. Microgeographical adaptation corresponds to elevational distributions of congeneric montane grasshoppers. Mol. Ecol. 2021, 30, 481–498. [Google Scholar] [CrossRef]
- Hu, S.; Sablok, G.; Wang, B.; Qu, D.; Barbaro, E.; Viola, R.; Li, M.; Varotto, C. Plastome organization and evolution of chloroplast genes in Cardamine species adapted to contrasting habitats. BMC Genom. 2015, 16, 306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Li, L.; Zhang, G. A High-Density SNP Genetic Linkage Map and QTL Analysis of Growth-Related Traits in a Hybrid Family of Oysters (Crassostrea gigas × Crassostrea angulata) Using Genotyping-by-Sequencing. G3 Genes|Genomes|Genetics 2016, 6, 1417–1426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kubota, S.; Iwasaki, T.; Hanada, K.; Nagano, A.J.; Fujiyama, A.; Toyoda, A.; Sugano, S.; Suzuki, Y.; Hikosaka, K.; Ito, M. A genome scan for genes underlying microgeographic-scale local adaptation in a wild Arabidopsis species. PLoS Genet. 2015, 11, e1005361. [Google Scholar]
- Sinha, S.; Schroeder, M.D.; Unnerstall, U.; Gaul, U.; Siggia, E.D. Cross-species comparison significantly improves genome-wide prediction of cis-regulatory modules in Drosophila. BMC Bioinform. 2004, 5, 129. [Google Scholar]
- Yang, G.; Mozzicafreddo, M.; Ballarini, P.; Pucciarelli, S.; Miceli, C. An in-silico comparative study of lipases from the Antarctic psychrophilic ciliate Euplotes focardii and the mesophilic congeneric species Euplotes crassus: Insight into Molecular Cold-Adaptation. Mar. Drugs 2021, 19, 67. [Google Scholar] [CrossRef]
- Banares Baudet, A.; Blanca, G.; Güemes Heras, J.; Moreno Saiz, J.; Ortiz, S. Red List of Spanish Vascular Flora; Tragsatec: Madrid, Spain, 2008. [Google Scholar]
- Moreno, J. Red List of Spanish Vascular Flora; Ministerio de Medio Ambiente, Rural y Marino: Madrid, Spain, 2008. [Google Scholar]
- Johnson, G.; Rumsey, F.; Headley, A.; Sheffield, E. Adaptations to extreme low light in the fern Trichomanes speciosum. New Phytol. 2000, 148, 423–431. [Google Scholar] [CrossRef] [Green Version]
- Rumsey, F.J.; Vogel, J.C.; Russell, S.J.; Barrett, J.A.; Gibby, M. Population structure and conservation biology of the endangered fern Trichomanes speciosum Willd.(Hymenophyllaceae) at its northern distributional limit. Biol. J. Linn. Soc. 1999, 66, 333–344. [Google Scholar] [CrossRef]
- Makgomol, K.; Sheffield, E. Gametophyte morphology and ultrastructure of the extremely deep shade fern, Trichomanes speciosum. New Phytol. 2001, 151, 243–255. [Google Scholar] [CrossRef]
- Farrar, D.R. Trichomanes intricatum: The independent Trichomanes gametophyte in the eastern United States. Am. Fern J. 1992, 82, 68–74. [Google Scholar] [CrossRef]
- Farrar, D.R.; Mickel, J.T. Vittaria appalachiana: A name for the “Appalachian gametophyte”. Am. Fern J. 1991, 81, 69–75. [Google Scholar] [CrossRef]
- Raine, C.A.; Farrar, D.R.; Sheffield, E. A new Hymenophyllum species in the Appalachians represented by independent gametophyte colonies. Am. Fern J. 1991, 81, 109–118. [Google Scholar] [CrossRef]
- Bakkali, M.; Martín-Blázquez, R.; Ruiz-Estévez, M.; Garrido-Ramos, M.A. De Novo Sporophyte Transcriptome Assembly and Functional Annotation in the Endangered Fern Species Vandenboschia speciosa (Willd.) G. Kunkel. Genes 2021, 12, 1017. [Google Scholar] [CrossRef]
- Ruiz-Estévez, M.; Bakkali, M.; Martín-Blázquez, R.; Garrido-Ramos, M.A. Identification and characterization of TALE homeobox genes in the endangered fern Vandenboschia speciosa. Genes 2017, 8, 275. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Estévez, M.; Bakkali, M.; Martín-Blázquez, R.; Garrido-Ramos, M.A. Differential expression patterns of MIKCC-type MADS-box genes in the endangered fern Vandenboschia speciosa. Plant Gene 2017, 12, 50–56. [Google Scholar] [CrossRef]
- Ruiz-Ruano, F.; Navarro-Domínguez, B.; Camacho, J.; Garrido-Ramos, M. Characterization of the satellitome in lower vascular plants: The case of the endangered fern Vandenboschia speciosa. Ann. Bot. 2019, 123, 587–599. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows–Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bağcı, C.; Patz, S.; Huson, D.H. DIAMOND+ MEGAN: Fast and easy taxonomic and functional analysis of short and long microbiome sequences. Curr. Protoc. 2021, 1, e59. [Google Scholar] [CrossRef] [PubMed]
- Haas, B.J.; Papanicolaou, A.; Yassour, M.; Grabherr, M.; Blood, P.D.; Bowden, J.; Couger, M.B.; Eccles, D.; Li, B.; Lieber, M. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Madden, T.L.; Schäffer, A.A.; Zhang, J.; Zhang, Z.; Miller, W.; Lipman, D.J. Gapped BLAST and PSI-BLAST: A new generation of protein database search programs. Nucleic Acids Res. 1997, 25, 3389–3402. [Google Scholar] [CrossRef] [Green Version]
- Camacho, C.; Coulouris, G.; Avagyan, V.; Ma, N.; Papadopoulos, J.; Bealer, K.; Madden, T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009, 10, 421. [Google Scholar] [CrossRef] [Green Version]
- Patro, R.; Duggal, G.; Love, M.I.; Irizarry, R.A.; Kingsford, C. Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 2017, 14, 417–419. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; Godzik, A. Cd-hit: A fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics 2006, 22, 1658–1659. [Google Scholar] [CrossRef] [Green Version]
- Simão, F.A.; Waterhouse, R.M.; Ioannidis, P.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 2015, 31, 3210–3212. [Google Scholar] [CrossRef] [Green Version]
- Waterhouse, R.M.; Seppey, M.; Simão, F.A.; Manni, M.; Ioannidis, P.; Klioutchnikov, G.; Kriventseva, E.V.; Zdobnov, E.M. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 2018, 35, 543–548. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The sequence alignment/map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- Mi, H.; Muruganujan, A.; Ebert, D.; Huang, X.; Thomas, P.D. PANTHER version 14: More genomes, a new PANTHER GO-slim and improvements in enrichment analysis tools. Nucleic Acids Res. 2019, 47, D419–D426. [Google Scholar] [CrossRef] [PubMed]
- Thomas, P.D.; Campbell, M.J.; Kejariwal, A.; Mi, H.; Karlak, B.; Daverman, R.; Diemer, K.; Muruganujan, A.; Narechania, A. PANTHER: A library of protein families and subfamilies indexed by function. Genome Res. 2003, 13, 2129–2141. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B (Methodol.) 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Supek, F.; Bošnjak, M.; Škunca, N.; Šmuc, T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011, 6, e21800. [Google Scholar] [CrossRef] [Green Version]
- Ćuković, K.; Dragićević, M.; Bogdanović, M.; Paunović, D.; Giurato, G.; Filipović, B.; Subotić, A.; Todorović, S.; Simonović, A. Plant regeneration in leaf culture of Centaurium erythraea Rafn. Part 3: De novo transcriptome assembly and validation of housekeeping genes for studies of in vitro morphogenesis. Plant Cell Tissue Organ Cult. (PCTOC) 2020, 141, 417–433. [Google Scholar] [CrossRef]
- Niu, S.-C.; Xu, Q.; Zhang, G.-Q.; Zhang, Y.-Q.; Tsai, W.-C.; Hsu, J.-L.; Liang, C.-K.; Luo, Y.-B.; Liu, Z.-J. De novo transcriptome assembly databases for the butterfly orchid Phalaenopsis equestris. Sci. Data 2016, 3, 160083. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Li, X.; Zhou, W.; Li, T.; Tian, C. De novo assembly and transcriptome characterization of spruce dwarf mistletoe Arceuthobium sichuanense uncovers gene expression profiling associated with plant development. BMC Genom. 2016, 17, 771. [Google Scholar] [CrossRef] [Green Version]
- Hu, R.; Yu, C.; Wang, X.; Jia, C.; Pei, S.; He, K.; He, G.; Kong, Y.; Zhou, G. De novo transcriptome analysis of Miscanthus lutarioriparius identifies candidate genes in rhizome development. Front. Plant Sci. 2017, 8, 492. [Google Scholar] [CrossRef] [Green Version]
- Sigel, E.M.; Schuettpelz, E.; Pryer, K.M.; Der, J.P. Overlapping patterns of gene expression between gametophyte and sporophyte phases in the fern Polypodium amorphum (Polypodiales). Front. Plant Sci. 2018, 9, 1450. [Google Scholar] [CrossRef]
- Geng, Y.; Cai, C.; McAdam, S.A.; Banks, J.A.; Wisecaver, J.H.; Zhou, Y. A de novo transcriptome assembly of Ceratopteris richardii provides insights into the evolutionary dynamics of complex gene families in land plants. Genome Biol. Evol. 2021, 13, evab042. [Google Scholar] [CrossRef]
- Jo, Y.; Choi, H.; Kim, S.-M.; Kim, S.-L.; Lee, B.C.; Cho, W.K. Integrated analyses using RNA-Seq data reveal viral genomes, single nucleotide variations, the phylogenetic relationship, and recombination for Apple stem grooving virus. BMC Genom. 2016, 17, 579. [Google Scholar] [CrossRef] [Green Version]
- Torrens-Spence, M.; Fallon, T.; Weng, J. A workflow for studying specialized metabolism in nonmodel eukaryotic organisms. In Methods in Enzymology; Elsevier: Amsterdam, The Netherlands, 2016; Volume 576, pp. 69–97. [Google Scholar]
- Aya, K.; Kobayashi, M.; Tanaka, J.; Ohyanagi, H.; Suzuki, T.; Yano, K.; Takano, T.; Yano, K.; Matsuoka, M. De novo transcriptome assembly of a fern, Lygodium japonicum, and a web resource database, Ljtrans DB. Plant Cell Physiol. 2015, 56, e5. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; You, S.; Taylor-Teeples, M.; Li, W.L.; Schuetz, M.; Brady, S.M.; Douglas, C.J. BEL1-LIKE HOMEODOMAIN6 and KNOTTED ARABIDOPSIS THALIANA7 interact and regulate secondary cell wall formation via repression of REVOLUTA. Plant Cell 2014, 26, 4843–4861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, J.; Yang, X.; Zhao, W.; Lang, T.; Samuelsson, T. Evolution, diversification, and expression of KNOX proteins in plants. Front. Plant Sci. 2015, 6, 882. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horst, N.A.; Katz, A.; Pereman, I.; Decker, E.L.; Ohad, N.; Reski, R. A single homeobox gene triggers phase transition, embryogenesis and asexual reproduction. Nat. Plants 2016, 2, 15209. [Google Scholar] [CrossRef]
- Sakakibara, K.; Ando, S.; Yip, H.K.; Tamada, Y.; Hiwatashi, Y.; Murata, T.; Deguchi, H.; Hasebe, M.; Bowman, J.L. KNOX2 genes regulate the haploid-to-diploid morphological transition in land plants. Science 2013, 339, 1067–1070. [Google Scholar] [CrossRef]
- Furumizu, C.; Alvarez, J.P.; Sakakibara, K.; Bowman, J.L. Antagonistic roles for KNOX1 and KNOX2 genes in patterning the land plant body plan following an ancient gene duplication. PLoS Genet. 2015, 11, e1004980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hu, J.-Y.; Zhou, Y.; He, F.; Dong, X.; Liu, L.-Y.; Coupland, G.; Turck, F.; de Meaux, J. miR824-regulated AGAMOUS-LIKE16 contributes to flowering time repression in Arabidopsis. Plant Cell 2014, 26, 2024–2037. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hirsch, S.; Oldroyd, G.E. GRAS-domain transcription factors that regulate plant development. Plant Signal. Behav. 2009, 4, 698–700. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, G.B.; Ionashiro, M.; Carrara, T.B.; Crivellari, A.C.; Tiné, M.A.; Prado, J.; Carpita, N.C.; Buckeridge, M.S. Cell wall polysaccharides from fern leaves: Evidence for a mannan-rich Type III cell wall in Adiantum raddianum. Phytochemistry 2011, 72, 2352–2360. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Singh, G.; Arya, S.K. Mannans: An overview of properties and application in food products. Int. J. Biol. Macromol. 2018, 119, 79–95. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Ruano, F.J.; Navarro-Domínguez, B.; Camacho, J.P.M.; Garrido-Ramos, M.A. Transposable element landscapes illuminate past evolutionary events in the endangered fern Vandenboschia speciosa. Genome 2022, 65, 95–103. [Google Scholar] [CrossRef]




| Tissue | Raw Data | After QC | |
|---|---|---|---|
| Number of paired-end reads | Gametophyte | 48.7 million | 24.7 million |
| Combined tissues | 115 million | 89.9 million | |
| Sporophyte | 66.3 million | 65.2 million | |
| Number of bases | Gametophyte | 4900 million | 2500 million |
| Combined tissues | 10,800 million | 9090 million | |
| Sporophyte | 6700 million | 6590 million |
| Percentage of BLAST Hits | Taxon | Raw Gametophyte | In Silico Decontaminated Gametophyte | Combined Tissues | Sporophyte |
|---|---|---|---|---|---|
| 50 most represented species | Plant | 30 | 60 | 60 | 68 |
| Animal | 40 | 30 | 30 | 26 | |
| Fungi | 24 | 10 | 10 | 6 | |
| Protozoa | 6 | 0 | 0 | 0 | |
| All species | A. thaliana | 30.29 | 55.88 | 55.82 | 70.00 |
| Gametophyte | Combined Tissues | Sporophyte | ||||
|---|---|---|---|---|---|---|
| Before Filtering | After Filtering | Before Filtering | After Filtering | Before Filtering | After Filtering | |
| Total transcripts | 44,455 | 43,139 | 88,383 | 42,918 | 84,759 | 36,430 |
| Percent GC | 45.48 | 45.47 | 45.23 | 45.18 | 45.18 | 45.18 |
| Contig N50 (bp) | 2101 | 2102 | 2264 | 2243 | 1955 | 2085 |
| Contig N70 (bp) | 1509 | 1400 | 1632 | 1640 | 1332 | 1511 |
| Contig N90 (bp) | 786 | 659 | 807 | 855 | 479 | 729 |
| Ex90N50 (bp) | 2236 | 2235 | 2511 | 2615 | 2039 | 2299 |
| Number transcripts corresponding to the Ex90 peak | 11,445 | 11,519 | 13,665 | 13,743 | 14,645 | 21,543 |
| Size of the smallest contig (bp) | 201 | 201 | 194 | 196 | 201 | 201 |
| Size of the largest contig (bp) | 9541 | 9541 | 16715 | 16715 | 13,225 | 13,224 |
| Number of contigs greater than 1 Kb long | 25,786 | 16,605 | 50,272 | 25,454 | 35,801 | 20,532 |
| Number of contigs greater than 10 Kb long | 0 | 0 | 36 | 20 | 18 | 12 |
| Median contig length (bp) | 1223 | 1227 | 1234 | 1299 | 722 | 1197 |
| Average contig (bp) | 1462.28 | 1465.00 | 1504.37 | 1534.61 | 1144.86 | 1437.37 |
| Total number of assembled bases | 65,005,800 | 63,198,512 | 132,960,361 | 65,862,246 | 97,037,551 | 52,363,571 |
| Gametophyte | Combined Tissues | |||||||
|---|---|---|---|---|---|---|---|---|
| Before Filtering | After Filtering | Before Filtering | After Filtering | |||||
| Percentage of Covered Length Intervals | Number of Proteins | Accumulated Number of Proteins | Number of Proteins | Accumulated Number of Proteins | Number of Proteins | Accumulated Number of Proteins | Number of Proteins | Accumulated Number of Proteins |
| 91–100 | 3097 | 3097 | 3097 | 3097 | 3739 | 3739 | 3585 | 3585 |
| 81–90 | 1328 | 4425 | 1325 | 4422 | 1564 | 5303 | 1493 | 5078 |
| 71–80 | 938 | 5363 | 937 | 5359 | 1070 | 6373 | 997 | 6075 |
| 61–70 | 649 | 6012 | 646 | 6005 | 769 | 7142 | 690 | 6765 |
| 51–60 | 589 | 6601 | 587 | 6592 | 713 | 7855 | 619 | 7384 |
| 41–50 | 653 | 7254 | 648 | 7240 | 753 | 8608 | 635 | 8019 |
| 31–40 | 634 | 7888 | 623 | 7863 | 777 | 9385 | 623 | 8642 |
| 21–30 | 769 | 8657 | 762 | 8625 | 866 | 10,251 | 653 | 9295 |
| 11–20 | 849 | 9506 | 844 | 9469 | 1036 | 11,287 | 722 | 10,017 |
| 1–10 | 427 | 9933 | 426 | 9895 | 584 | 11,871 | 395 | 10,412 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Martín-Blázquez, R.; Bakkali, M.; Ruiz-Estévez, M.; Garrido-Ramos, M.A. Comparison between the Gametophyte and the Sporophyte Transcriptomes of the Endangered Fern Vandenboschia speciosa. Genes 2023, 14, 166. https://doi.org/10.3390/genes14010166
Martín-Blázquez R, Bakkali M, Ruiz-Estévez M, Garrido-Ramos MA. Comparison between the Gametophyte and the Sporophyte Transcriptomes of the Endangered Fern Vandenboschia speciosa. Genes. 2023; 14(1):166. https://doi.org/10.3390/genes14010166
Chicago/Turabian StyleMartín-Blázquez, Rubén, Mohammed Bakkali, Mercedes Ruiz-Estévez, and Manuel A. Garrido-Ramos. 2023. "Comparison between the Gametophyte and the Sporophyte Transcriptomes of the Endangered Fern Vandenboschia speciosa" Genes 14, no. 1: 166. https://doi.org/10.3390/genes14010166
APA StyleMartín-Blázquez, R., Bakkali, M., Ruiz-Estévez, M., & Garrido-Ramos, M. A. (2023). Comparison between the Gametophyte and the Sporophyte Transcriptomes of the Endangered Fern Vandenboschia speciosa. Genes, 14(1), 166. https://doi.org/10.3390/genes14010166






