Abstract
The aim of this study was to identify genomic regions and genes associated with the fiber diameter (FD), clean fleece weight (CFW), live weight (LW), body condition score (BCS), pregnancy rate (PR) and lambing potential (LP) of Uruguayan Merino sheep. Phenotypic records of approximately 2000 mixed-age ewes were obtained from a Merino nucleus flock. Genome-wide association studies were performed utilizing single-step Bayesian analysis. For wool traits, a total of 35 genomic windows surpassed the significance threshold (PVE ≥ 0.25%). The proportion of the total additive genetic variance explained by those windows was 4.85 and 9.06% for FD and CFW, respectively. There were 42 windows significantly associated with LWM, which collectively explained 43.2% of the additive genetic variance. For BCS, 22 relevant windows accounted for more than 40% of the additive genetic variance, whereas for the reproduction traits, 53 genomic windows (24 and 29 for PR and LP, respectively) reached the suggestive threshold of 0.25% of the PVE. Within the top 10 windows for each trait, we identified several genes showing potential associations with the wool (e.g., IGF-1, TGFB2R, PRKCA), live weight (e.g., CAST, LAP3, MED28, HERC6), body condition score (e.g., CDH10, TMC2, SIRPA, CPXM1) or reproduction traits (e.g., ADCY1, LEPR, GHR, LPAR2) of the mixed-age ewes.
1. Introduction
The genetic improvement of livestock has traditionally been based on phenotypic and pedigree information. Advances in molecular DNA technologies offer the opportunity to increase the rate of genetic gain using genetic markers (e.g., single-nucleotide polymorphisms, SNPs) [1,2]. For several species, including sheep, panels of more than 50,000 SNPs are currently available [3]. This technology enables the identification of genes or chromosomal segments that are associated with the traits of interest (Wenome-Wide Association Studies, GWAS) [4]. A number of statistical methods, including the single-step Bayesian regression approach, which combines all available pedigrees and phenotypic and genomic data, have been employed to conduct GWAS [4,5,6,7] and have been used for a number of livestock species.
In sheep, GWAS analyses have been performed for economically relevant traits such as the fiber diameter (FD), clean fleece weight (CFW), live weight (LW) and reproduction [8,9,10]. These traits are influenced by many genes, each with a small effect, and involve various cell types and tissues [11,12,13]. Nevertheless, candidate genes associated with major wool traits (FD and CFW) have been reported in the Australian and Chinese Merino sheep populations [8,13,14,15]. Genomic regions related to live weight have also been found in Merino sheep in Australia [9] and New Zealand [16]. A French study reported candidate genes associated with ewes’ body condition score (BCS) [17].
In Uruguay, few GWAS of livestock species have been undertaken or published [18,19,20]. There are no published GWAS of the wool, growth, or reproduction traits of Uruguayan sheep. The aim of this study was to detect the genomic regions and genes associated with the FD, CFW, LW, BCS and reproduction traits of Uruguayan ultrafine Merino ewes.
2. Materials and Methods
2.1. Ethical Statement
All animal work was approved by the INIA Animal Ethics Committee (INIA_2018.2).
2.2. Phenotypic and Pedigree Data
The data were derived from a Uruguayan Merino nucleus flock involved in a genetic program, as described by Ramos et al. [21,22]. The selection objectives, nutritional conditions and management of this flock were previously reported by Ramos et al. [22]. Phenotypic records of these six traits were obtained from approximately 2000 mixed-age ewes born between 1999 and 2018. The traits evaluated were the adult fiber diameter and clean fleece weight at late-pregnancy shearing (A_FD and A_CFW, respectively); live weight and body condition score at mating (LWM and BCSM, respectively); pregnancy rate (PR: pregnant or non-pregnant); and lambing potential (LP: the number of ultrasound-scanned fetuses per ewe combined: 0, 1 or ≥2). Details of the trait measurements were described by Ramos et al. [21,22]. The complete pedigree included 7168 animals.
2.3. Genotyping and Quality Control
Genomic DNA extraction from the blood samples was performed as described by Carracelas et al. [23]. The animals were genotyped using the GeneSeek® Genomic Profiler™ Ovine 50K panel. Quality control was performed to remove SNPs with a minor allele frequency (MAF) lower than 1% and call rate below 85%, as well as animals with a call rate lower than 90%. After applying the quality control measures, 1133 animals and 40,036 SNPs were retained and utilized in the analyses.
2.4. Genome-Wide Association Study
Genome-Wide Association Studies were performed utilizing single-step Bayesian regression analyses implemented in the JWAS package [7]. The software tool JWAS is an open-source package for single-trait and multiple-trait genome-enabled prediction and analysis. JWAS is a single-language software that is easy for community members to use. The documentation and examples of JWAS can be found at https://reworkhow.github.io/JWAS.jl/latest/theory/theory accessed on 1 July 2022.
A Bayes C linear mixed model that included the genotyped and non-genotyped ewes was constructed. The model equation for the single-step Bayesian GWAS was as follows for the genotyped animals:
where:
y = Xβ + Zu + Wpe + Mα + e
- y = vector of phenotypes for the genotyped individuals,
- β = vector of fixed effects,
- u = vector of random animal genetic effects not explained by the markers,
- pe = vector of random permanent environmental effects accounting for the covariance between observations of the same individual,
- α = vector of marker effects,
- e = vector of random residual effects,
- X, Z and W = incidence matrices relating records to fixed, animal and permanent environmental effects,
- M = genotype covariate (each coded as 0, 1 or 2).
The model equation for the non-genotyped individuals can be written as:
where:
y = Xβ + Zu + Wpe + Mα + Znϵ + e
- y = vector of phenotypes for the non-genotyped individuals,
- M = genotype covariate matrix for the non-genotyped individuals imputed from the genotyped relatives,
- Zn = incidence matrix corresponding to the imputation residual,
- ϵ = vector of imputation residuals accounting for errors in the genotype imputation.
All the other terms are as described in Equation (1).
A Markov chain Monte Carlo (MCMC) method was utilized to obtain samples from the posterior distributions of all unknown parameters, including the marker effects. A total of 70000 iterations were run after a burn-in of 5000 cycles and a sampling interval each 10 interactions. The probability that the markers would have null effect was set to 99% (parameter π = 0.99), that is, 1% of the 40036 SNPs (approximately 400 SNPs) was assumed to contribute to the genetic variance.
2.5. Detection of Important Windows Associated with the Trait and Candidate Genes
The genome was partitioned into 2015 non-overlapping windows of 20 consecutive SNPs which, on average, represented 1 Mb. Assuming that all the windows explained the same amount of variation, the expected proportion of genetic variance explained (PVE) by each window was 0.05% ([1/2015] ∗ 100). The 1 Mb windows that explained at least 0.25% of the genetic variance, which was 5 times the expected proportion of variance (0.05 × 5 = 0.25%), were considered to be the most important regions associated with the trait [24,25]. The top 10 windows for each trait that explained the largest PVE were examined to identify candidate genes. The annotated genes in those regions were extracted from the OAR v3.1 Ovine (Texel) Genome Assembly, available in the Ensembl database (http://www.ensembl.org/Biomart accessed on 10 August 2022) [26]. The biological functions of the genes were identified using the functional annotation tools in DAVID (https://david.ncifcrf.gov/tools.jsp accessed on 10 August 2022) [27]. Gene ontology (GO) enrichment analysis was conducted using g:Profiler (https://biit.cs.ut.ee/gprofiler/gost accessed on 1 September 2022) [28]. Pathways with a p-value < 0.05 were considered significantly enriched.
3. Results
3.1. Descriptive Statistics
A summary of the phenotypic records of the traits analyzed is shown in Table 1. The number of records ranged from 6288 to 7079.

Table 1.
Descriptive statistics for the wool, body growth and reproduction traits of mixed-age ewes born between 1999 and 2018.
3.2. Genome-Wide Association Study (GWAS)
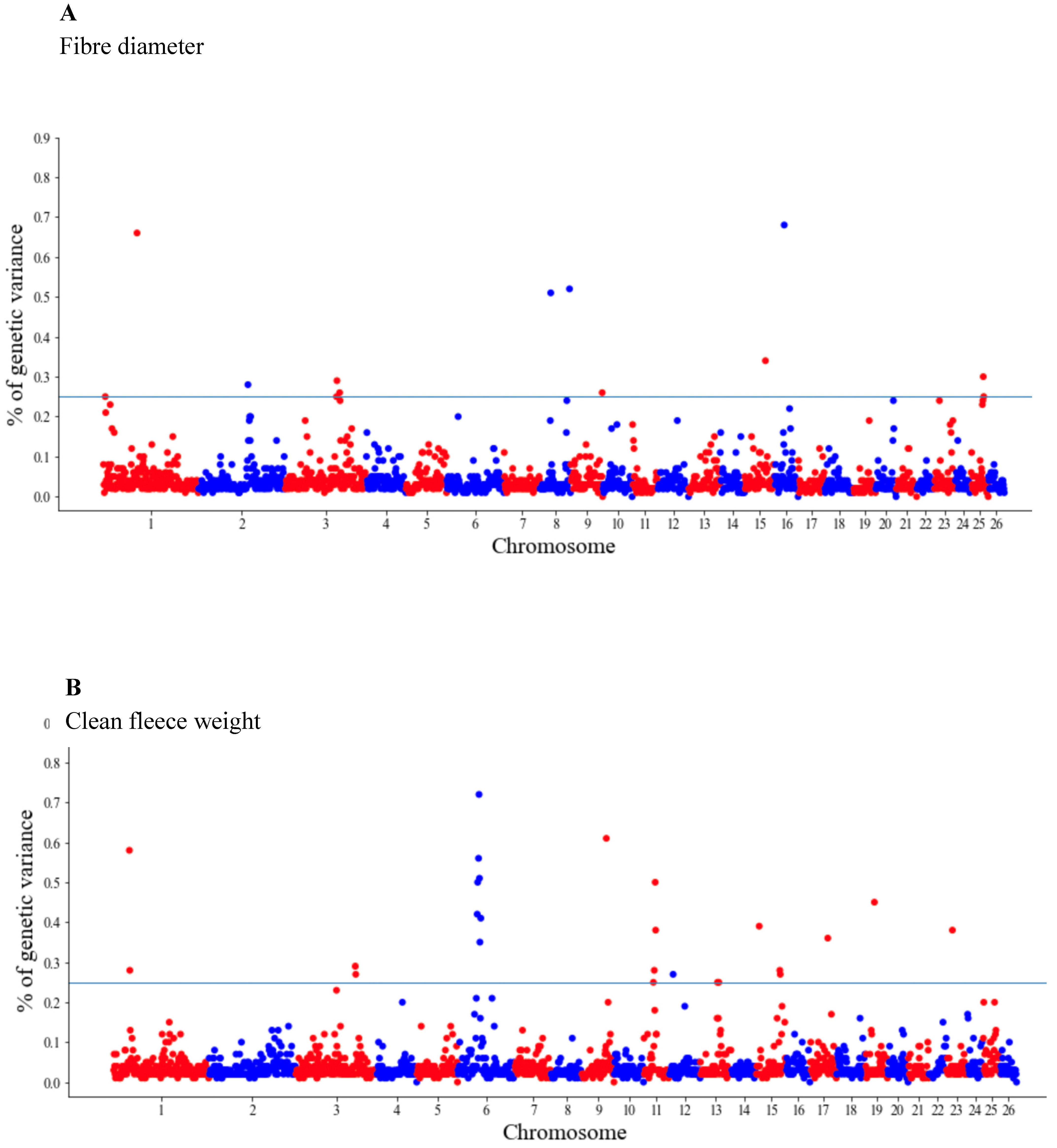
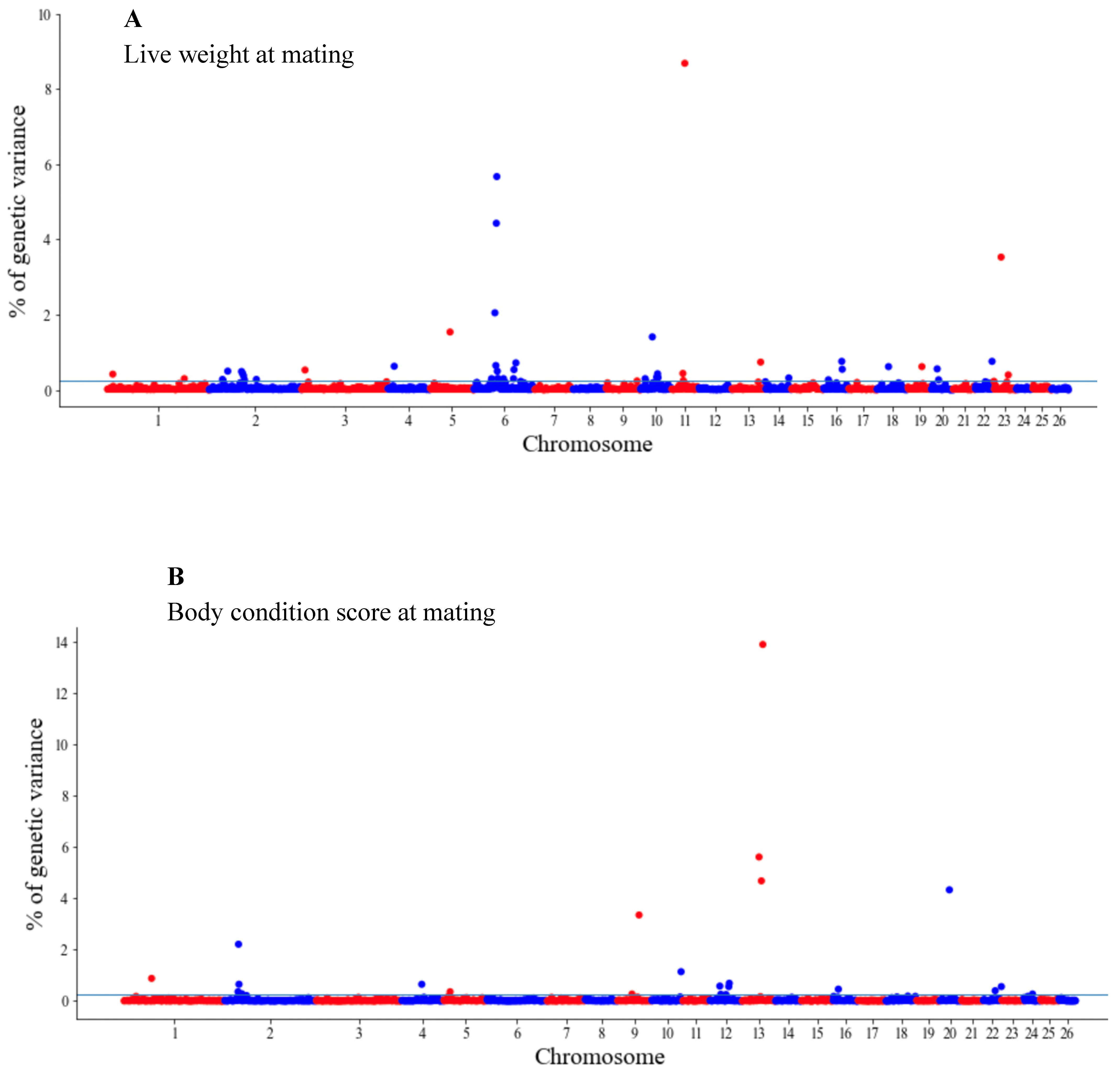
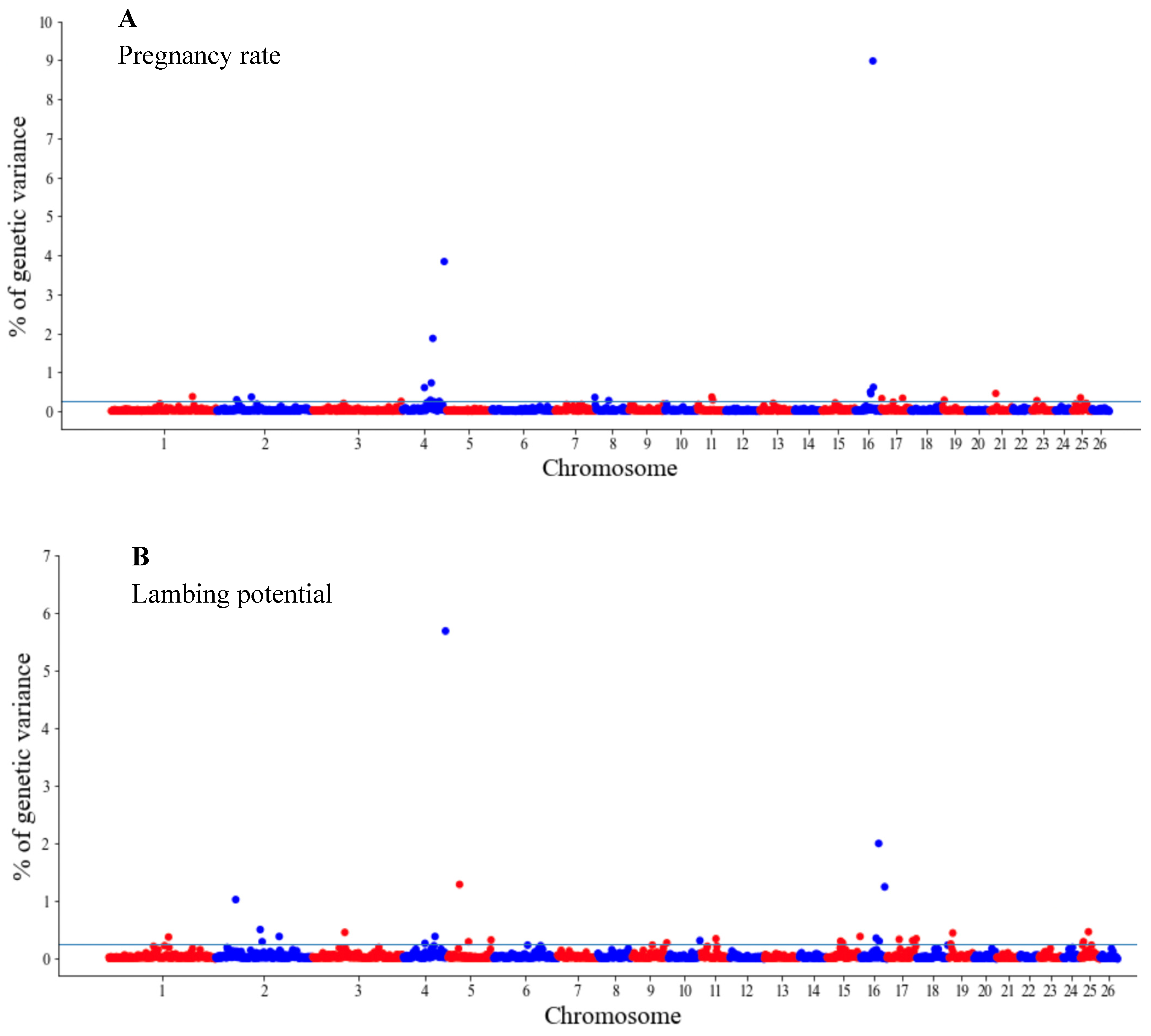
The GWAS results are presented as the proportion of additive genetic variation explained by the windows of 20 consecutive SNPs, as reported by other authors [13]. Manhattan plots illustrating the proportion of additive genetic variation explained by each window of 20 adjacent SNPs for the wool, body and reproduction traits are presented in Figure 1, Figure 2 and Figure 3, respectively. The suggestive threshold of 0.25% of the PVE is indicated by the horizonal blue line.
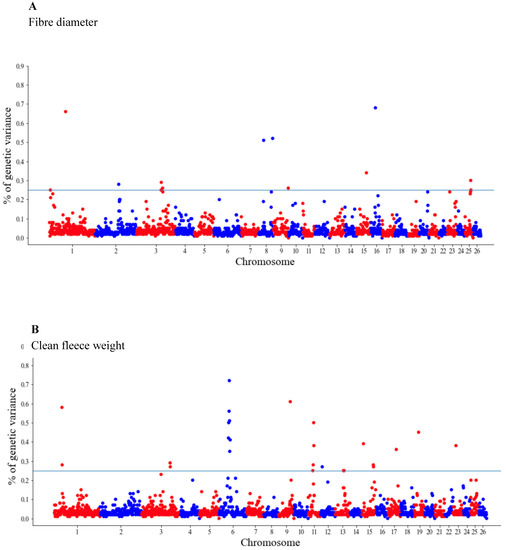
Figure 1.
Manhattan plots of the genetic variance explained (%) by 20 adjacent SNP windows for the ewe fiber diameter (A) and clean fleece weight (B). Each dot represents a window. The % of additive genetic variance explained by each window and chromosomes 1–26 are shown on the Y-axis and X-axis, respectively. The horizontal line indicates the suggestive threshold of 0.25% of the PVE.
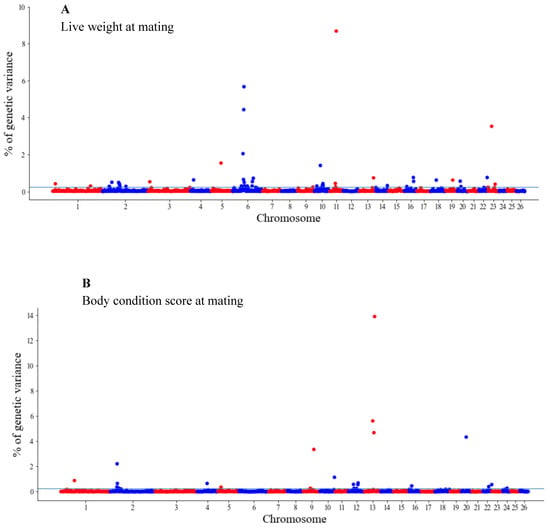
Figure 2.
Manhattan plots of the genetic variance explained (%) by 20 adjacent SNP windows for the ewe live weight (A) and body condition score at mating (B). Each dot represents a window. The % of additive genetic variance explained by each window and chromosomes 1–26 are shown on the Y-axis and X-axis, respectively. The horizontal line indicates the suggestive threshold of 0.25% of the PVE.
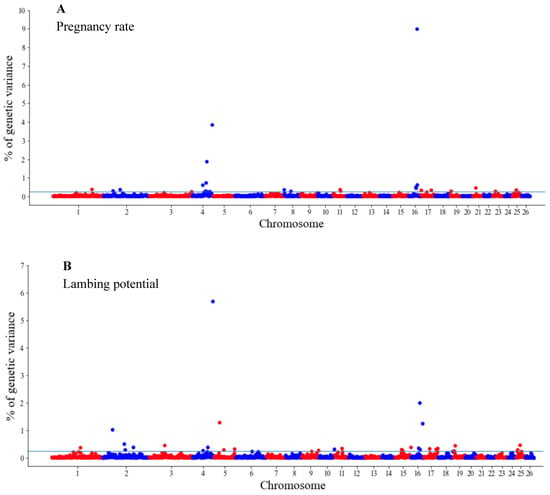
Figure 3.
Manhattan plots of genetic variance explained (%) by 20 adjacent SNP windows for the pregnancy rate (A) and lambing potential (B). Each dot represents a window. The % of additive genetic variance explained by each window and chromosomes 1–26 are shown on the Y-axis and X-axis, respectively. The horizontal line indicates the suggestive threshold of 0.25% of the PVE.
For the wool traits, a total of 35 windows (13 for FD and 22 for CFW) surpassed the significance threshold (PVE ≥ 0.25%, Figure 1). The proportion of the total additive genetic variance explained by these windows was 4.85 and 9.06% for FD and CFW, respectively. There were 42 windows significantly associated with LWM, which collectively explained 43.2% of the additive genetic variance. For BCS, 22 relevant windows accounted for more than 40% of the additive genetic variance (Figure 2). For the reproduction traits, 53 genomic windows (24 and 29 for PR and LP, respectively) reached the suggestive threshold of 0.25% of the PVE (Figure 3).
3.3. Top 10 Genomic Regions and Candidate Genes
The chromosome, location, PVE and candidate genes within the top 10 windows for each trait are shown in Table 2, Table 3 and Table 4. The top 10 windows cumulatively explained 4.1, 5.3, 29.6, 37.4, 18.4 and 13.5% of the additive genetic variance for the A_FD, A_CFW, LWM, BCSM, PR and LP, respectively. Some of these windows were associated with more than one trait. For example, three genomic regions on chromosome 6 were associated with both the CFW and LWM. Similarly, two overlapping regions were associated with the PR and LP. A total of 240 genes were contained within the top 10 genomic regions across the six traits.

Table 2.
Chromosome, location, proportion of additive genetic variance (PVE, %) and candidate genes within the top 10 windows associated with the fiber diameter (A_FD) and clean fleece weight (A_CFW) of Merino ewes.

Table 3.
Chromosome, location, proportion of additive genetic variance (PVE, %) and candidate genes within the top 10 windows associated with the live weight (LWM) and body condition score (BCSM) at mating of Merino ewes.

Table 4.
Chromosome, location, proportion of additive genetic variance (PVE, %) and candidate genes within the top 10 windows associated with the pregnancy rate (PR) and lambing potential (LP) of Merino ewes.
3.4. Enrichment Analysis
A gene ontology (GO) enrichment analysis of the genes within the top 10 windows for each trait was performed. GO terms with a p-value < 0.05 were considered significantly enriched. The enriched terms were associated with molecular functions (MF), biological processes (BP) and/or cellular components (CC). In total, 20 GO terms were enriched (p-value < 0.05). More information from the GO analysis is available in the Supplementary Materials.
4. Discussion
The present study reports on chromosome segments associated with economically relevant traits of Uruguayan Merino sheep. The genomic regions of interest on chromosomes 1, 3, 4, 5, 6, 8, 9, 11, 12, 13, 16, 19 and 22 identified in this study contain known candidate genes related to the wool traits, live weight, body condition score and reproduction traits of sheep and other species (see the following paragraphs). This section focused on some of the genes located within the top 10 windows for each trait that explained the largest proportion of the additive genetic variance.
Wool follicle regulation involves several genes, including IGF and TGFB [29]. The candidate genes for the wool traits identified in this study included IGF-1, TGFBR2, PDE3A, STK3, PRKCA, ZNF704 and EXT1. These genes have previously been associated with hair [30,31,32], cashmere [33,34] and wool [35,36,37]. In our study, the functional analysis indicated that the genes WAPL, ESR1 and IGF-1 were significantly enriched in the regulation of fibroblast proliferation (see Supplementary Materials), which is crucial for hair follicle formation [38]. Overall, the proportion of additive genetic variance explained by each window was relatively small (lower than 0.75%), which reflects the polygenic nature of wool traits.
The identification of genes associated with the live weight is of particular interest for sheep breeding programs [13,39]. Some of the potential genes for the ewe LWM are known to be involved in the LW of young sheep. For example, CAST has been related to the birth weight and growth rate of lambs [40,41]. The genes LAP3, MED28, and HERC6, located on chromosome 6, have previously been identified as candidate genes for the post-weaning LW in Australian Merino sheep [9]. The identification of genes commonly affecting the LW at both early and adult ages is unsurprising, given the moderate to high genetic correlations between these traits [42,43]. The genes HERC6 and MED28 have been also associated with gastrointestinal nematode infection, which is one of the most important health problems in grazing sheep [44]. This is in agreement with earlier studies that suggested that some SNPs associated with gastrointestinal nematode resistance are involved in growth traits [45]. In sheep, the gene GPRIN3 was also reported to be associated with the litter size [46]. Other genes that were found to be associated with the ewe LWM in the present study have been reported as candidate genes for LW-related traits in different species. For example, A1CF, ZNF830, CCT6B and MYO10 were associated with residual feed intake in cattle [47,48,49], while the genes CTBP2 and AP2B1 have been linked to meat quality and lipid metabolism in pigs [50,51].
In sheep, the body condition score is an indicator of the available body reserves (fat and muscle) [52]. Several genes that are known to be involved in fat storage and metabolism were associated with the ewe BCS in the present study. For example, TMC2, SIRPA and CPXM1 have been reported as candidate genes for tail fat deposition in sheep [53]. The gene CPXM1 was also identified as a positive regulator of adipogenesis in mice and humans [54]. In mice, FMO1, a member of the flavin-containing mono-oxygenase (FMO) gene family, was associated with energy homeostasis and metabolic efficiency [55]. TPD52 is a regulator of lipid metabolism and is involved in fatty acid storage [56]. Early studies indicated that the overexpression of ATG3 favors lipid deposition in mice [57]. In humans, LRRC1 is involved in adipocytic differentiation [58], whereas F13A1 is expressed at high levels in the adipose tissue of obese individuals [59].
The present study identified two common regions associated with pregnancy and the lambing potential, suggesting that the same genes may play a role in the regulation of these reproduction traits. These regions are located on chromosomes 4 and 16 and contain genes that have previously been associated with several reproduction traits. For example, ADCY1 has been related to pubertal initiation in sheep [60], fertility in dairy cattle [61] and fecundity in goats [62,63]. The gene CDH10 was associated with several reproduction traits in buffaloes [64]. Furthermore, PDIA4 was found to be involved in the litter size in sheep and pigs [65,66]. In our work, LEPR was identified as a candidate gene for the pregnancy rate. This gene has already been associated with several reproduction traits, including pregnancy [67,68,69,70]. The genes GHR and LPAR2 are associated with sheep reproduction [71,72], while DMXL1 is related to reproduction traits in heifers [73]. Therefore, there is evidence supporting the concept that the genes located on chromosomes 4 and 16 play an important role in the reproduction traits of Merino sheep.
This work was the first to perform a single-step Genome-Wide Association Study of the production and reproduction traits of mixed-age ewes in Uruguay. As mentioned above, several of the candidate genes detected were also reported in other studies, which provides confidence in our results. A limitation of this study was the small sample size, which affected the power of detection. Future analyses based on larger populations would improve the identification of candidate genes for traits of interest in sheep. Our results will contribute to the Uruguayan Merino genetic evaluation, as some of identified genes are good targets for selection. In addition, the genomic regions identified here should be utilized as targets in further studies.
5. Conclusions
This study performed a single-step GWAS of six traits in a Uruguayan Merino sheep population. A total of 13, 22, 42, 22, 24 and 29 genomic regions were significantly associated with the fiber diameter, clean fleece weight, live weight at mating, body condition score at mating, pregnancy rate and lambing potential, respectively. We detected several genes, some of which were novel, showing potential associations with the wool (IGF-1, TGFB2R, PRKCA), live weight (CAST, LAP3, MED28, HERC6), body condition score (CDH10, TMC2, SIRPA, CPXM1) or reproduction traits (ADCY1, LEPR, GHR, LPAR2) of mixed-age ewes. These results require validation using a larger dataset before their implementation in genomic selection among Uruguayan Merino sheep. Overall, our findings will be useful for further genomic studies and genetic improvement programs in Uruguay.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14010167/s1, Table S1: Enrichment analysis.
Author Contributions
Conceptualization, H.T.B., D.J.G., P.R.K., G.C., B.V. and Z.R.; data collection, Z.R.; formal analysis and supervision, H.T.B., D.J.G., G.C. and P.R.K.; analysis, interpretation of results, and preparation of the manuscript, Z.R.; writing—review and editing, H.T.B., D.J.G., P.R.K., G.C. and B.V. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Regional Consortium for Innovation in Ultrafine Wool (CRILU), the European Union’s Horizon 2020 research and innovation program under the grant agreement n°772787 (Smarter) and the National Institute of Agricultural Research of Uruguay (INIA_CL_38: Rumiar). This study was supported by two Ph.D. scholarships (from the National Agency for Investigation and Innovation of Uruguay, ANII, and Massey University, New Zealand), awarded to Zully Ramos.
Institutional Review Board Statement
The study was approved by the INIA Animal Ethics Committee (INIA_2018.2).
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available within the article.
Acknowledgments
The authors would like to thank the staff of the Glencoe Experimental Unit, INIA Tacuarembó, SUL, SCMAU, INIA, and CRILU. We thank Melissa Stephen and Rhiannon Handcock for their assistance in the statistical analysis. We thank the National Institute of Agricultural Research of Uruguay (INIA) for enabling the field data collection, which received funding from the European Union’s Horizon 2020 research and innovation program under the grant agreement n°772787 (Smarter) and from the National Institute of Agricultural Research of Uruguay (INIA_CL_38: Rumiar). The first author acknowledges the financial support for her PhD provided by Massey University, New Zealand, and the National Agency for Investigation and Innovation of Uruguay (ANII).
Conflicts of Interest
The authors declare no conflict of interest.
References
- Van Eenennaam, A.L.; van der Werf, J.H.; Goddard, M.E. The value of using DNA markers for beef bull selection in the seedstock sector. J. Anim. Sci. 2011, 89, 307–320. [Google Scholar] [CrossRef][Green Version]
- Bouquet, A.; Juga, J. Integrating genomic selection into dairy cattle breeding programmes: A review. Animal. 2013, 7, 705–713. [Google Scholar] [CrossRef]
- Meuwissen, T.; Hayes, B.; Goddard, M. Accelerating improvement of livestock with genomic selection. Annu. Rev. Anim. Biosci. 2013, 1, 221–237. [Google Scholar] [CrossRef]
- Fernando, R.; Toosi, A.; Wolc, A.; Garrick, D.; Dekkers, J. Application of whole-genome prediction methods for genome-wide association studies: A Bayesian approach. J. Agric. Biol. Environ. Stat. 2017, 22, 172–193. [Google Scholar] [CrossRef]
- Fernando, R.L.; Garrick, D. Bayesian methods applied to GWAS. In Genome-Wide Association Studies and Genomic Prediction; Humana Press: Totowa, NJ, USA, 2013; pp. 237–274. [Google Scholar]
- Fernando, R.L.; Dekkers, J.C.; Garrick, D.J. A class of Bayesian methods to combine large numbers of genotyped and non-genotyped animals for whole-genome analyses. Genet. Sel. Evol. 2014, 46, 50. [Google Scholar] [CrossRef]
- Cheng, H.; Fernando, R.; Garrick, D. JWAS: Julia implementation of whole-genome analysis software. In Proceedings of the World Congress on Genetics Applied to Livestock Production, Auckland, New Zealand, 7–11 February 2018. [Google Scholar]
- Wang, Z.; Zhang, H.; Yang, H.; Wang, S.; Rong, E.; Pei, W.; Li, H.; Wang, N. Genome-Wide Association Study for Wool Production Traits in a Chinese Merino Sheep Population. PLoS ONE 2014, 9, e107101. [Google Scholar] [CrossRef]
- Al-Mamun, H.A.; Kwan, P.; Clark, S.A.; Ferdosi, M.H.; Tellam, R.; Gondro, C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. 2015, 47, 66. [Google Scholar] [CrossRef]
- Abdoli, R.; Mirhoseini, S.Z.; Hossein-Zadeh, N.G.; Zamani, P.; Ferdosi, M.H.; Gondro, C. Genome-wide association study of four composite reproductive traits in Iranian fat-tailed sheep. Reprod. Fertil. Dev. 2019, 31, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Parsons, Y.M.; Cooper, D.W.; Piper, L.R. Evidence of linkage between high-glycine-tyrosine keratin gene loci and wool fibre diameter in a Merino half-sib family. Anim. Genet. 1994, 25, 105–108. [Google Scholar] [CrossRef] [PubMed]
- Kominakis, A.; Hager-Theodorides, A.L.; Zoidis, E.; Saridaki, A.; Antonakos, G.; Tsiamis, G. Combined GWAS and ‘guilt by association’-based prioritization analysis identifies functional candidate genes for body size in sheep. Genet. Sel. Evol. 2017, 49, 41. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.; Luo, H.; Huang, X.; Wei, C.; Di, J.; Tian, Y.; Fu, X.; Li, B.; Liu, G.E.; Fang, L.; et al. Integration of a single-step genome-wide association study with a multi-tissue transcriptome analysis provides novel insights into the genetic basis of wool and weight traits in sheep. Genet. Sel. Evol. 2021, 53, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Zhu, S.; Guo, T.; Han, M.; Chen, B.; Qiao, G.; Wu, Y.; Yuan, C.; Liu, J.; Lu, Z.; et al. Whole-genome re-sequencing association study on yearling wool traits in Chinese fine-wool sheep. J. Anim. Sci. 2021, 99, skab210. [Google Scholar] [CrossRef]
- Bolormaa, S.; Swan, A.A.; Stothard, P.; Khansefid, M.; Moghaddar, N.; Duijvesteijn, N.; van der Werf, J.H.; Daetwyler, H.D.; MacLeod, I.M. A conditional multi-trait sequence GWAS discovers pleiotropic candidate genes and variants for sheep wool, skin wrinkle and breech cover traits. Genet. Sel. Evol. 2021, 53, 58. [Google Scholar] [CrossRef] [PubMed]
- Hess, M.K.; Johnson, P.L.; Knowler, K.; Hickey, S.M.; Hess, A.S.; McEwan, J.C.; Rowe, S.J. GWAS for methane yield, residual feed intake and liveweight in New Zealand sheep. In Proceedings of the 23rd Conference of the Association for the Advancement of Animal Breeding and Genetics, Armidale, New South Wales, Australia, 27 October–1 November 2019. [Google Scholar]
- Macé, T.; González-García, E.; Foulquié, D.; Carrière, F.; Pradel, J.; Durand, C.; Douls, S.; Allain, C.; Parisot, S.; Hazard, D. Genome-wide analyses reveal a strong association between LEPR gene variants and body fat reserves in ewes. BMC Genom. 2022, 23, 412. [Google Scholar] [CrossRef]
- Carracelas, B.; Navajas, E.A.; Vera, B.; Ciappesoni, G. Genome-Wide Association Study of Parasite Resistance to Gastrointestinal Nematodes in Corriedale Sheep. Genes 2022, 13, 1548. [Google Scholar] [CrossRef]
- Grasso, N.; Aguilar, I.; Clariget, J.; Lema, M.; Brito, G.; Navajas, E. Genomics of carcass and meat quality traits in Hereford–preliminary results. In Proceedings of the 60th International Congress of Meat Science and Technology, Punta del Este, Uruguay, 17–22 August 2014. [Google Scholar]
- Jara, E.; Peñagaricano, F.; Armstrong, E.; Ciappesoni, G.; Iriarte, A.; Navajas, E.A. Revealing the genetic basis of eyelid pigmentation in Hereford cattle. J. Anim. Sci. 2022, 100, skac110. [Google Scholar] [CrossRef]
- Ramos, Z.; Blair, H.T.; De Barbieri, I.; Ciappesoni, G.; Montossi, F.; Kenyon, P.R. Phenotypic Responses to Selection for Ultrafine Wool in Uruguayan Yearling Lambs. Agriculture 2021, 11, 179. [Google Scholar] [CrossRef]
- Ramos, Z.; Blair, H.T.; De Barbieri, I.; Ciappesoni, G.; Montossi, F.; Kenyon, P.R. Productivity and reproductive performance of mixed-age ewes across 20 years of selection for ultrafine wool in Uruguay. Agriculture 2021, 11, 712. [Google Scholar] [CrossRef]
- Carracelas, B.; Navajas, E.A.; Vera, B.; Ciappesoni, G. SNP arrays evaluation as tools in genetic improvement in Corriedale sheep in Uruguay. Agrociencia 2022, 26, e998. [Google Scholar] [CrossRef]
- Onteru, S.K.; Gorbach, D.M.; Young, J.M.; Garrick, D.J.; Dekkers, J.C.; Rothschild, M.F. Whole genome association studies of residual feed intake and related traits in the pig. PLoS ONE 2013, 8, e61756. [Google Scholar] [CrossRef]
- Sollero, B.P.; Junqueira, V.S.; Gomes, C.C.; Caetano, A.R.; Cardoso, F.F. Tag SNP selection for prediction of tick resistance in Brazilian Braford and Hereford cattle breeds using Bayesian methods. Genet. Sel. Evol. 2017, 49, 49. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids. Res. 2021, 50, D988–D995. [Google Scholar] [CrossRef] [PubMed]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A web server for functional enrichment analysis and functional annotation of gene lists (2021 update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g: Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Li, H.; Liu, K.; Yu, J.; Cheng, M.; De, W.; Liu, J.; Shi, S.; He, Y.; Zhao, J. Differential expression of genes and proteins associated with wool follicle cycling. Mol. Biol. Rep. 2014, 41, 5343–5349. [Google Scholar] [CrossRef]
- Lin, H.Y.; Yang, L.T. Differential response of epithelial stem cell populations in hair follicles to TGF-β signaling. Dev. Biol. 2013, 373, 394–406. [Google Scholar] [CrossRef] [PubMed]
- Kim, D. The Effects of Phosphodiesterase 3 (PDE3) Inhibitor on Hair Follicle Cell Viability and Hair Growth. Ph.D. Thesis, The Graduate School Seoul National University, Seoul, Republic of Korea, 2020. [Google Scholar]
- Heilmann-Heimbach, S.; Hochfeld, L.M.; Henne, S.K.; Nöthen, M.M. Hormonal regulation in male androgenetic alopecia-Sex hormones and beyond: Evidence from recent genetic studies. Exp. Dermatol. 2020, 29, 814–827. [Google Scholar] [CrossRef]
- Su, R.; Fan, Y.; Qiao, X.; Li, X.; Zhang, L.; Li, C.; Li, J. Transcriptomic analysis reveals critical genes for the hair follicle of Inner Mongolia cashmere goat from catagen to telogen. PLoS ONE 2018, 13, e0204404. [Google Scholar] [CrossRef]
- Han, H.; Yang, M.M.; Dan, J.; Zhang, X.J.; Wei, Q.; Chen, T.; Wang, Q.J.; Yang, C.Y.; Wulan, B.; Zhang, T.T.; et al. Whole-genome sequencing of Chinese native goat offers biological insights into cashmere fiber formation. bioRxiv 2021. [Google Scholar] [CrossRef]
- Damak, S.; Su, H.Y.; Jay, N.P.; Bullock, D.W. Improved wool production in transgenic sheep expressing insulin-like growth factor 1. Nat. Biotechnol. 1996, 14, 185–188. [Google Scholar] [CrossRef]
- Darwish, H.R.; El-Shorbagy, H.M.; Abou-Eisha, A.M.; El-Din, A.E.; Farag, I.M. New polymorphism in the 5′ flanking region of IGF-1 gene and its association with wool traits in Egyptian Barki sheep. J. Genet. Eng. Biotechnol. 2017, 15, 437–441. [Google Scholar] [CrossRef] [PubMed]
- Zhao, H.; Guo, T.; Lu, Z.; Liu, J.; Zhu, S.; Qiao, G.; Han, M.; Yuan, C.; Wang, T.; Li, F.; et al. Genome-wide association studies detects candidate genes for wool traits by re-sequencing in Chinese fine-wool sheep. BMC Genom. 2021, 22, 127. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Jarrell, A.; Guo, C.; Lang, R.; Atit, R. Dermal β-catenin activity in response to epidermal Wnt ligands is required for fibroblast proliferation and hair follicle initiation. Development 2012, 139, 1522–1533. [Google Scholar] [CrossRef] [PubMed]
- Lu, Z.; Yue, Y.; Yuan, C.; Liu, J.; Chen, Z.; Niu, C.; Sun, X.; Zhu, S.; Zhao, H.; Guo, T.; et al. Genome-wide association study of body weight traits in chinese fine-wool sheep. Animals 2020, 10, 170. [Google Scholar] [CrossRef] [PubMed]
- Jawasreh, K.I.; Jadallah, R.; Al-Amareen, A.H.; Abdullah, A.Y.; Al-Qaisi, A.; Alrawashdeh, I.M.; Al-Zghoul, M.B.F.; Ahamed, M.; Obeidat, B. Association between MspI calpastatin gene polymorphisms, growth performance, and meat characteristics of Awassi sheep. Indian J. Anim. Sci. 2017, 87, 635–639. [Google Scholar]
- Armstrong, E.; Ciappesoni, G.; Iriarte, W.; Da Silva, C.; Macedo, F.; Navajas, E.A.; Brito, G.; San Julián, R.; Gimeno, D.; Postiglioni, A. Novel genetic polymorphisms associated with carcass traits in grazing Texel sheep. Meat Sci. 2018, 145, 202–208. [Google Scholar] [CrossRef]
- Safari, E.; Fogarty, N.M.; Gilmour, A.R. A review of genetic parameter estimates for wool, growth, meat and reproduction traits in sheep. Anim. Prod. Sci. 2005, 92, 271–289. [Google Scholar] [CrossRef]
- Huisman, A.E.; Brown, D.J. Genetic parameters for bodyweight, wool, and disease resistance and reproduction traits in Merino sheep. 2. Genetic relationships between bodyweight traits and other traits. Aust. J. Exp. Agric 2008, 48, 1186–1193. [Google Scholar] [CrossRef]
- Al Kalaldeh, M.; Gibson, J.; Lee, S.H.; Gondro, C.; Van Der Werf, J.H. Detection of genomic regions underlying resistance to gastrointestinal parasites in Australian sheep. Genet. Sel. Evol. 2019, 51, 37. [Google Scholar] [CrossRef]
- Álvarez, I.; Fernández, I.; Soudré, A.; Traoré, A.; Pérez-Pardal, L.; Sanou, M.; Tapsoba, S.A.; Menéndez-Arias, N.A.; Goyache, F. Identification of genomic regions and candidate genes of functional importance for gastrointestinal parasite resistance traits in Djallonké sheep of Burkina Faso. Arch. Anim. Breed. 2019, 62, 313–323. [Google Scholar] [CrossRef]
- Tao, L.; He, X.; Jiang, Y.; Liu, Y.; Ouyang, Y.; Shen, Y.; Hong, Q.; Chu, M. Genome-wide analyses reveal genetic convergence of prolificacy between goats and sheep. Genes 2021, 12, 480. [Google Scholar] [CrossRef]
- Karisa, B.K.; Thomson, J.; Wang, Z.; Stothard, P.; Moore, S.S.; Plastow, G.S. Candidate genes and single nucleotide polymorphisms associated with variation in residual feed intake in beef cattle. J. Anim. Sci. 2013, 91, 3502–3513. [Google Scholar] [CrossRef]
- de Las Heras-Saldana, S.; Clark, S.A.; Duijvesteijn, N.; Gondro, C.; van der Werf, J.H.; Chen, Y. Combining information from genome-wide association and multi-tissue gene expression studies to elucidate factors underlying genetic variation for residual feed intake in Australian Angus cattle. BMC Genom. 2019, 20, 939. [Google Scholar] [CrossRef] [PubMed]
- Dlamini, N.M.; Dzomba, E.F.; Magawana, M.; Ngcamu, S.; Muchadeyi, F.C. Linkage Disequilibrium, Haplotype Block Structures, Effective Population Size and Genome-Wide Signatures of Selection of Two Conservation Herds of the South African Nguni Cattle. Animals 2022, 12, 2133. [Google Scholar] [CrossRef]
- Ponsuksili, S.; Murani, E.; Phatsara, C.; Schwerin, M.; Schellander, K.; Wimmers, K. Porcine muscle sensory attributes associate with major changes in gene networks involving CAPZB, ANKRD1, and CTBP2. Funct. Integr. Genom. 2009, 9, 455–471. [Google Scholar] [CrossRef] [PubMed]
- Piórkowska, K.; Żukowski, K.; Ropka-Molik, K.; Tyra, M.; Gurgul, A. A comprehensive transcriptome analysis of skeletal muscles in two Polish pig breeds differing in fat and meat quality traits. Genet. Mol. Biol. 2018, 41, 125–136. [Google Scholar] [CrossRef] [PubMed]
- Kenyon, P.R.; Maloney, S.K.; Blache, D. Review of sheep body condition score in relation to production characteristics. N. Z. J. Agric. Res. 2014, 57, 38–64. [Google Scholar] [CrossRef]
- Yuan, Z.; Liu, E.; Liu, Z.; Kijas, J.W.; Zhu, C.; Hu, S.; Ma, X.; Zhang, L.; Du, L.; Wang, H.; et al. Selection signature analysis reveals genes associated with tail type in Chinese indigenous sheep. Anim. Genet. 2017, 48, 55–66. [Google Scholar] [CrossRef]
- Kim, Y.H.; Barclay, J.L.; He, J.; Luo, X.; O’Neill, H.M.; Keshvari, S.; Webster, J.A.; Ng, C.; Hutley, L.J.; Prins, J.B.; et al. Identification of carboxypeptidase X (CPX)-1 as a positive regulator of adipogenesis. FASEB J. 2016, 30, 2528–2540. [Google Scholar] [CrossRef]
- Veeravalli, S.; Omar, B.A.; Houseman, L.; Hancock, M.; Malagon, S.G.G.; Scott, F.; Janmohamed, A.; Phillips, I.R.; Shephard, E.A. The phenotype of a flavin-containing monooyxgenase knockout mouse implicates the drug-metabolizing enzyme FMO1 as a novel regulator of energy balance. Biochem. Pharmacol. 2014, 90, 88–95. [Google Scholar] [CrossRef]
- Ha, M.; Han, M.E.; Kim, J.Y.; Jeong, D.C.; Oh, S.O.; Kim, Y.H. Prognostic role of TPD52 in acute myeloid leukemia: A retrospective multicohort analysis. J. Cell. Biochem. 2019, 120, 3672–3678. [Google Scholar] [CrossRef] [PubMed]
- da Silva Lima, N.; Fondevila, M.F.; Nóvoa, E.; Buqué, X.; Mercado-Gómez, M.; Gallet, S.; González-Rellan, M.J.; Fernandez, U.; Loyens, A.; Garcia-Vence, M.; et al. Inhibition of ATG3 ameliorates liver steatosis by increasing mitochondrial function. J. Hepatol. 2022, 76, 11–24. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, J.; Wang, T.; Ma, B.; Wu, P.; Xu, X.; Xiong, J. The downstream PPARγ target LRRC1 participates in early-stage adipocytic differentiation. Mol. Cell. Bioachem. 2022, 1–9. [Google Scholar] [CrossRef]
- Dull, K.; Fazekas, F.; Törőcsik, D. Factor XIII-A in Diseases: Role Beyond Blood Coagulation. Int. J. Mol. Sci. 2021, 22, 1459. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Sui, Z.; Zhang, J.; Li, Q.; Zhang, Y.; Wang, C.; Li, X.; Xing, F. Identification of Signatures of Selection for Litter Size and Pubertal Initiation in Two Sheep Populations. Animals 2022, 12, 2520. [Google Scholar] [CrossRef]
- Höglund, J.K.; Sahana, G.; Guldbrandtsen, B.; Lund, M.S. Validation of associations for female fertility traits in Nordic Holstein, Nordic Red and Jersey dairy cattle. BMC Genet. 2014, 15, 8. [Google Scholar] [CrossRef]
- Lai, F.N.; Zhai, H.L.; Cheng, M.; Ma, J.Y.; Cheng, S.F.; Ge, W.; Zhang, G.L.; Wang, J.J.; Zhang, R.Q.; Wang, X.; et al. Whole-genome scanning for the litter size trait associated genes and SNPs under selection in dairy goat (Capra hircus). Sci. Rep. 2016, 6, 38096. [Google Scholar] [CrossRef] [PubMed]
- Islam, R.; Li, Y.; Liu, X.; Berihulay, H.; Abied, A.; Gebreselassie, G.; Ma, Q.; Ma, Y. Genome-wide runs of homozygosity, effective population size, and detection of positive selection signatures in six Chinese goat breeds. Genes 2019, 10, 938. [Google Scholar] [CrossRef]
- Li, J.; Liu, J.; Liu, S.; Plastow, G.; Zhang, C.; Wang, Z.; Campanile, G.; Salzano, A.; Gasparrini, B.; Hua, G.; et al. Integrating RNA-seq and GWAS reveals novel genetic mutations for buffalo reproductive traits. Anim. Reprod. Sci. 2018, 197, 290–295. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Niu, Z.; Zeng, Z.; Jiang, Y.; Jiang, Y.; Ding, Y.; Tang, S.; Shi, H.; Ding, X. Using High-Density SNP Array to Reveal Selection Signatures Related to Prolificacy in Chinese and Kazakhstan Sheep Breeds. Animals 2020, 10, 1633. [Google Scholar] [CrossRef]
- Liu, C.; Ran, X.; Niu, X.; Li, S.; Wang, J.; Zhang, Q. Insertion of 275-bp SINE into first intron of PDIA4 gene is associated with litter size in Xiang pigs. Anim. Reprod. Sci. 2018, 195, 16–23. [Google Scholar] [CrossRef] [PubMed]
- Moschos, S.; Chan, J.L.; Mantzoros, C.S. Leptin and reproduction: A review. Fertil. Steril. 2002, 77, 433–444. [Google Scholar] [CrossRef] [PubMed]
- Taheri, S.J.; Parham, A. Sheep oocyte expresses leptin and functional leptin receptor mRNA. Asian Pac. J. Reprod. 2016, 5, 395–399. [Google Scholar] [CrossRef]
- Juengel, J.L.; French, M.C.; O’Connell, A.R.; Edwards, S.J.; Haldar, A.; Brauning, R.; Farquhar, P.A.; Dodds, K.G.; Galloway, S.M.; Johnstone, P.D.; et al. Mutations in the leptin receptor gene associated with delayed onset of puberty are also associated with decreased ovulation and lambing rates in prolific Davisdale sheep. Reprod. Fertil. Dev. 2016, 28, 1318–1325. [Google Scholar] [CrossRef]
- Lakhssassi, K.; Serrano, M.; Lahoz, B.; Sarto, M.P.; Iguácel, L.P.; Folch, J.; Alabart, J.L.; Calvo, J.H. The LEPR gene is associated with reproductive seasonality traits in Rasa Aragonesa sheep. Animals 2020, 10, 2448. [Google Scholar] [CrossRef] [PubMed]
- Akhatayeva, Z.; Mao, C.; Jiang, F.; Pan, C.; Lin, C.; Hao, K.; Lan, T.; Chen, H.; Zhang, Q.; Lan, X. Indel variants within the PRL and GHR genes associated with sheep litter size. Reprod. Domest. Anim. 2020, 55, 1470–1478. [Google Scholar] [CrossRef]
- Li, Z.; He, X.; Zhang, X.; Zhang, J.; Guo, X.; Sun, W.; Chu, M. Transcriptome Profile of Key CircRNAs and MiRNAs in Oviduct that Affect Sheep Reproduction. Res. Sq. 2020. [Google Scholar] [CrossRef]
- Mohammadi, A.; Alijani, S.; Rafat, S.A.; Abdollahi-Arpanahi, R. Single-step genome-wide association study and candidate genes networks affecting reproductive traits in Iranian Holstein cattle. Livest. Sci. 2022, 262, 104971. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).