Wide-Range Portrayal of AP2/ERF Transcription Factor Family in Maize (Zea mays L.) Development and Stress Responses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Classification of ZmAP2/ERF Family Members
2.2. Sequence and Phylogenetic Analysis of ZmAP2/ERF Proteins
2.3. Chromosomal Localization, Duplication, and Conserved Motif Analysis of ZmAP2/ERFs
2.4. ZmAP2/ERFs Expression Profiling by RNA-seq Data
2.5. Experimental Material and Treatments
2.6. Expression Quantification by qRT-PCR
3. Results
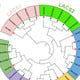
3.1. Phylogenetic Classification of ZmAP2/ERFs Family Members
3.2. Conserved Motif Analysis and Intron-Exon Organization in ZmAP2/ERF TFs
3.3. Chromosomal Arrangement, Paralogous Gene Identification, and Synteny Analysis of the ZmAP2/ERF Transcription Factor Family
3.4. Promoter Region Analysis of Maize AP2/ERFs
3.5. Expression Analysis of ZmAP2/ERFs in Maize Tissues
3.6. Expression Profiling of ZmAP2/ERFs in Response to Biotic and Abiotic Stresses
3.7. Relative Expression of ZmAP2/ERFs by qRT-PCR
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raza, A.; Razzaq, A.; Mehmood, S.S.; Zou, X.; Zhang, X.; Lv, Y.; Xu, J. Impact of Climate Change on Crops Adaptation and Strategies to Tackle Its Outcome: A Review. Plants 2019, 8, 34. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raza, A.; Ashraf, F.; Zou, X.; Zhang, X.; Tosif, H. Plant Adaptation and Tolerance to Environmental Stresses: Mechanisms and Perspectives. In Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives I: General Consequences and Plant Responses; Hasanuzzaman, M., Ed.; Springer: Singapore, 2020; pp. 117–145. [Google Scholar]
- Ramegowda, V.; Senthil-Kumar, M. The interactive effects of simultaneous biotic and abiotic stresses on plants: Mechanistic understanding from drought and pathogen combination. J. Plant Physiol. 2015, 176, 47–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, P.V.V.; Pisipati, S.R.; Momčilović, I.; Ristic, Z. Independent and Combined Effects of High Temperature and Drought Stress During Grain Filling on Plant Yield and Chloroplast EF-Tu Expression in Spring Wheat. J. Agron. Crop. Sci. 2011, 197, 430–441. [Google Scholar] [CrossRef]
- Ciais, P.; Reichstein, M.; Viovy, N.; Granier, A.; Ogée, J.; Allard, V.; Aubinet, M.; Buchmann, N.; Bernhofer, C.; Carrara, A.; et al. Europe-wide reduction in primary productivity caused by the heat and drought in 2003. Nature 2005, 437, 529–533. [Google Scholar] [CrossRef]
- Sullivan, J.H.; Teramura, A.H. Field Study of the Interaction between Solar Ultraviolet-B Radiation and Drought on Photosynthesis and Growth in Soybean. Plant Physiol. 1990, 92, 141–146. [Google Scholar] [CrossRef] [Green Version]
- Gray, S.B.; Brady, S.M. Plant developmental responses to climate change. Dev. Biol. 2016, 419, 64–77. [Google Scholar] [CrossRef] [Green Version]
- Berger, J.; Palta, J.; Vadez, V. Review: An integrated framework for crop adaptation to dry environments: Responses to transient and terminal drought. Plant Sci. 2016, 253, 58–67. [Google Scholar] [CrossRef] [Green Version]
- Bartels. D. Targeting detoxification pathways: An efficient approach to obtain plants with multiple stress tolerance? Trends Plant Sci. 2001, 6, 284–286. [Google Scholar] [CrossRef]
- Chen, J.; Wei, B.; Li, G.; Fan, R.; Zhong, Y.; Wang, X.; Zhang, X. TraeALDH7B1-5A, encoding aldehyde dehydrogenase 7 in wheat, confers improved drought tolerance in Arabidopsis. Planta 2015, 242, 137–151. [Google Scholar] [CrossRef]
- Latchman. D.S. Transcription factors: An overview. Int. J. Exp. Pathol. 1993, 74, 417–422. [Google Scholar]
- Riechmann, J.L.; Heard, J.; Martin, G.; Reuber, L.; Jiang, C.; Keddie, J.; Adam, L.; Pineda, O.; Ratcliffe, O.J.; Samaha, R.R.; et al. Arabidopsis transcription factors: Genome-wide comparative analysis among eukaryotes. Science 2000, 290, 2105–2110. [Google Scholar] [CrossRef] [PubMed]
- Thompson, B.E.; Bartling, L.; Whipple, C.; Hall, D.H.; Sakai, H.; Schmidt, R.; Hake, S. Bearded-ear encodes a MADS box transcription factor critical for maize floral development. Plant Cell 2009, 21, 2578–2590. [Google Scholar] [CrossRef] [Green Version]
- Marzábal, P.; Gas, E.; Fontanet, P.; Vicente-Carbajosa, J.; Torrent, M.; Ludevid, M.D. The maize Dof protein PBF activates transcription of γ-zein during maize seed development. Plant Mol. Biol. 2008, 67, 441–454. [Google Scholar] [CrossRef]
- Sonbol, F.-M.; Fornalé, S.; Capellades, M.; Encina, A.; Touriño, S.; Torres, J.-L.; Rovira, P.; Ruel, K.; Puigdomènech, P.; Rigau, J.; et al. The maize ZmMYB42 represses the phenylpropanoid pathway and affects the cell wall structure, composition and degradability in Arabidopsis thaliana. Plant Mol. Biol. 2009, 70, 283–296. [Google Scholar] [CrossRef] [PubMed]
- Young, T.E.; Ling, J.; Geisler-Lee, C.J.; Tanguay, R.L.; Caldwell, C.; Gallie, D.R. Developmental and thermal regulation of the maize heat shock protein, HSP101. Plant Physiol. 2001, 127, 777–791. [Google Scholar] [CrossRef] [PubMed]
- Nieva, C.; Busk, P.K.; Domínguez-Puigjaner, E.; Lumbreras, V.; Testillano, P.S.; Risueño, M.-C.; Pagès, M. Isolation and Functional Characterisation of Two New bZIP Maize Regulators of the ABA Responsive Gene rab28. Plant Mol. Biol. 2005, 58, 899–914. [Google Scholar] [CrossRef]
- Liu, Z.-J.; Shao, F.-X.; Tang, G.-Y.; Shan, L.; Bi, Y.-P. Cloning and characterization of a transcription factor ZmNAC1 in maize (Zea mays). Yi Chuan 2009, 31, 199–205. [Google Scholar] [CrossRef]
- Licausi, F.; Kosmacz, M.; Weits, D.A.; Giuntoli, B.; Giorgi, F.M.; Voesenek, L.A.C.J.; Perata, P.; van Dongen, J.T. Oxygen sensing in plants is mediated by an N-end rule pathway for protein destabilization. Nature 2011, 479, 419–422. [Google Scholar] [CrossRef]
- Sun, Z.M.; Zhou, M.L.; Tang, Y.X.; Lin, M.; Wu, Y.M. Overexpression of the Lotus corniculatus Soloist Gene LcAP2/ERF107 Enhances Tolerance to Salt Stress. Protein Pept. Lett. 2016, 23, 442–449. [Google Scholar] [CrossRef]
- Nakano, T.; Suzuki, K.; Fujimura, T.; Shinshi, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [Green Version]
- Duan, C.; Argout, X.; Gebelin, V.; Summo, M.; Dufayard, J.F.; Leclercq, J.; Hadi, K.; Piyatrakul, P.; Pirrello, J.; Rio, M.; et al. Identification of the Hevea brasiliensis AP2/ERF superfamily by RNA sequencing. BMC Genom. 2013, 14, 30. [Google Scholar] [CrossRef]
- Xu, W.; Li, F.; Ling, L.; Liu, A. Genome-wide survey and expression profiles of the AP2/ERF family in castor bean (Ricinus communis L.). BMC Genom. 2013, 14, 78. [Google Scholar] [CrossRef] [Green Version]
- Song, X.; Li, Y.; Hou, X. Genome-wide analysis of the AP2/ERF transcription factor superfamily in Chinese cabbage (Brassica rapa ssp. pekinensis). BMC Genom. 2013, 14, 573. [Google Scholar] [CrossRef] [Green Version]
- Lata, C.; Mishra, A.K.; Muthamilarasan, M.; Bonthala, V.S.; Khan, Y.; Parasad, M. Genome-Wide Investigation and Expression Profiling of AP2/ERF Transcription Factor Superfamily in Foxtail Millet (Setaria italica L.). PLoS ONE 2014, 9, e113092. [Google Scholar] [CrossRef] [Green Version]
- Dossa, K.; Wei, X.; Li, D.; Fonceka, D.; Zhang, Y.; Wang, L.; Yu, J.; Boshou, L.; Diouf, D.; Cisse, N.; et al. Insight into the AP2/ERF transcription factor superfamily in sesame and expression profiling of DREB subfamily under drought stress. BMC Plant Biol. 2016, 16, 171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mathur, S.; Priyadarshini, S.S.; Singh, V.; Vashisht, I.; Jung, K.-H.; Sharma, R.; Sharma, M.K. Comprehensive phylogenomic analysis of ERF genes in sorghum provides clues to the evolution of gene functions and redundancy among gene family members. 3 Biotech 2020, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Guo, Z.H.; Hao, P.P.; Wang, G.-M.; Jin, Z.M.; Zhang, S.L. Multiple regulatory roles of AP2/ERF transcription factor in angiosperm. Bot. Stud. 2017, 58, 6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Licausi, F.; Giorgi, F.M.; Zenoni, S.; Osti, F.; Pezzotti, M.; Perata, P. Genomic and transcriptomic analysis of the AP2/ERF superfamily in Vitis vinifera. BMC Genom. 2010, 11, 719. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kagaya, Y.; Ohmiya, K.; Hattori, T. RAV1, a novel DNA-binding protein, binds to bipartite recognition sequence through two distinct DNA-binding domains uniquely found in higher plants. Nucleic Acids Res. 1999, 27, 470–478. [Google Scholar] [CrossRef]
- Sakumaa, Y.; Liu, Q.; Dubouzet, J.G.; Abea, H.; Shinozaki, K.; Yamaguchi-Shinozaki, K. DNA-binding specificity of the ERF/AP2 domain of Arabidopsis DREBs, transcription factors involved in dehydration- and cold-inducible gene expression. Biochem. Biophys. Res. Commun. 2002, 290, 998–1009. [Google Scholar] [CrossRef]
- Zhuang, J.; Cai, B.; Peng, R.-H.; Zhu, B.; Jin, X.-F.; Xue, Y.; Gao, F.; Fu, X.-Y.; Tian, Y.-S.; Zhao, W.; et al. Genome-wide analysis of the AP2/ERF gene family in Populus trichocarpa. Biochem. Biophys. Res. Commun. 2008, 371, 468–474. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Chen, M.; Chen, X.; Xu, Z.; Guan, S.; Li, L.-C.; Li, A.; Guo, J.; Mao, L.; Ma, Y. Phylogeny, gene structures, and expression patterns of the ERF gene family in soybean (Glycine max L.). J. Exp. Bot. 2008, 59, 4095–4107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharoni, A.M.; Nuruzzaman, M.; Satoh, K.; Shimizu, T.; Kondoh, H.; Sasaya, T.; Choi, I.-R.; Omura, T.; Kikuchi, S. Gene structures, classification and expression models of the AP2/EREBP transcription factor family in rice. Plant Cell Physiol. 2011, 52, 344–360. [Google Scholar] [CrossRef] [PubMed]
- Cao, P.B.; Azar, S.; SanClemente, H.; Mounet, F.; Dunand, C.; Marque, G.; Marque, C.; Teulieres, C. Genome-wide analysis of the AP2/ERF family in Eucalyptus grandis: An intriguing over-representation of stress-responsive DREB1/CBF genes. PLoS ONE 2015, 10, e0121041. [Google Scholar] [CrossRef] [PubMed]
- Hu, L.; Liu, S. Genome-wide identification and phylogenetic analysis of the ERF gene family in cucumbers. Genet. Mol. Biol. 2011, 34, 624–633. [Google Scholar] [CrossRef] [Green Version]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef]
- Schultz, J.; Milpetz, F.; Bork, P.; Ponting, C.P. SMART, a simple modular architecture research tool: Identification of signaling domains. Proc. Natl. Acad. Sci. USA 1998, 95, 5857–5864. [Google Scholar] [CrossRef] [Green Version]
- Letunic, I.; Doerks, T.; Bork, P. SMART: Recent updates, new developments and status in 2015. Nucleic Acids Res. 2015, 43, D257–D260. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhang, Y.; Zhang, Z.; Zhu, J.; Yu, J. KaKs_Calculator 2.0: A toolkit incorporating γ-series methods and sliding window strategies. Genom. Proteom. Bioinform. 2010, 8, 77–80. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.-H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Bailey, T.L.; Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994, 2, 28–36. [Google Scholar] [PubMed]
- Walley, J.W.; Sartor, R.C.; Shen, Z.; Schmitz, R.J.; Wu, K.J.; Urich, M.A.; Nery, J.R.; Smith, L.G.; Schnable, J.C.; Ecker, J.R.; et al. Integration of omic networks in a developmental atlas of maize. Science 2016, 353, 814–818. [Google Scholar] [CrossRef] [Green Version]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [Green Version]
- Rashid, M.; Guangyuan, H.; Guangxiao, Y.; Hussain, J.; Xu, Y. AP2/ERF Transcription Factor in Rice: Genome-Wide Canvas and Syntenic Relationships between Monocots and Eudicots. Evol. Bioinform. 2012, 8, 321–355. [Google Scholar] [CrossRef]
- Najafi, S.; Sorkheh, K.; Nasernakhaei, F. Characterization of the APETALA2/Ethylene-responsive factor (AP2/ERF) transcription factor family in sunflower. Sci. Rep. 2018, 8, 11576. [Google Scholar] [CrossRef] [Green Version]
- Jiang, W.; Zhang, X.; Song, X.; Yang, J.; Pang, Y. Genome-Wide Identification and Characterization of APETALA2/Ethylene-Responsive Element Binding Factor Superfamily Genes in Soybean Seed Development. Front. Plant Sci. 2020, 11, 566647. [Google Scholar] [CrossRef]
- Cannon, S.B.; Mitra, A.; Baumgarten, A.; Young, N.D.; May, G. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 2004, 4, 10. [Google Scholar] [CrossRef] [Green Version]
- Bennetzen. J.L. Transposable elements, gene creation and genome rearrangement in flowering plants. Curr. Opin. Genet Dev. 2005, 15, 621–627. [Google Scholar] [CrossRef] [PubMed]
- Elemento, O.; Gascuel, O.; Lefranc, M.P. Reconstructing the Duplication History of Tandemly Repeated Genes. Mol. Biol. Evol. 2002, 19, 278–288. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Holub. E.B. The arms race is ancient history in Arabidopsis, the wildflower. Nat. Rev. Genet. 2001, 2, 516–527. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Cao, W.; Fang, H.; Xu, S.; Yin, S.; Zhang, Y.; Lin, D.; Wang, J.; Chen, Y.; Xu, C.; et al. Transcriptomic Profiling of the Maize (Zea mays L.) Leaf Response to Abiotic Stresses at the Seedling Stage. Front. Plant Sci. 2017, 8, 290. [Google Scholar] [CrossRef] [Green Version]
- Qi, J.; Sun, G.; Wang, L.; Zhao, C.; Hettenhausen, C.; Schuman, M.C.; Baldwin, I.T.; Li, J.; Song, J.; Liu, Z.; et al. Oral secretions from Mythimna separata insects specifically induce defence responses in maize as revealed by high-dimensional biological data. Plant Cell Environ. 2016, 39, 1749–1766. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Li, S.; Teng, S.; Liang, H.; Xin, H.; Gao, H.; Huang, D.; Lang, Z. Transcriptome profiling revealed novel transcriptional regulators in maize responses to Ostrinia furnacalis and jasmonic acid. PLoS ONE 2017, 12, e0177739. [Google Scholar] [CrossRef] [Green Version]
- Van den Broeck, L.; Dubois, M.; Vermeersch, M.; Storme, V.; Matsui, M.; Inzé, D. From network to phenotype: The dynamic wiring of an Arabidopsis transcriptional network induced by osmotic stress. Mol. Syst. Biol. 2017, 13, 961. [Google Scholar] [CrossRef]
- Osnato, M.; Castillejo, C.; Matías-Hernández, L.; Pelaz, S. TEMPRANILLO genes link photoperiod and gibberellin pathways to control flowering in Arabidopsis. Nat. Commun. 2012, 3, 808. [Google Scholar] [CrossRef] [Green Version]
- Horstman, A.; Willemsen, V.; Boutilier, K.; Heidstra, R. AINTEGUMENTA-LIKE proteins: Hubs in a plethora of networks. Trends Plant Sci. 2014, 19, 146–157. [Google Scholar] [CrossRef]
- Lata, C.; Prasad, M. Role of DREBs in regulation of abiotic stress responses in plants. J. Exp. Bot. 2011, 62, 4731–4748. [Google Scholar] [CrossRef] [Green Version]
- Mizoi, J.; Shinozaki, K.; Yamaguchi-Shinozaki, K. AP2/ERF family transcription factors in plant abiotic stress responses. Biochim. Et Biophys. Acta 2012, 1819, 86–96. [Google Scholar] [CrossRef] [PubMed]
- Phukan, U.J.; Jeena, G.S.; Tripathi, V.; Shukla, R.K. Regulation of Apetala2/Ethylene Response Factors in Plants. Front. Plant Sci. 2017, 8, 150. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, C.; Zhu, J.-K. The broad roles of CBF genes: From development to abiotic stress. Plant Signal. Behav. 2016, 11, e1215794. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, H.; Cheng, Y.; Sun, Y.; Zhang, X.; He, H.; Liu, J. Genome-Wide Identification of the ARF Gene Family and ARF3 Target Genes Regulating Ovary Initiation in Hazel via ChIP Sequencing. Front. Plant Sci. 2021, 12, 715820. [Google Scholar] [CrossRef]
- Cui, M.; Haider, M.S.; Chai, P.; Guo, J.; Du, P.; Li, H.L.; Dong, W.; Huang, B.; Zheng, Z.; Shi, L.; et al. Genome-Wide Identification and Expression Analysis of AP2/ERF Transcription Factor Related to Drought Stress in Cultivated Peanut (Arachis hypogaea L.). Front. Genet. 2021, 12, 750761. [Google Scholar] [CrossRef]
- Sun, Y.; Liu, Y.; Zhang, J.; Song, L.; Guo, C. Genome-Wide Analysis of the AP2/ERF Superfamily Genes and their Responses to Abiotic Stress in Medicago truncatula. Front. Plant Sci. 2015, 6, 1247. [Google Scholar]
- Liu, J.; Shi, Y.; Yang, S. Insights into the regulation of C-repeat binding factors in plant cold signaling. J. Integr. Plant Biol. 2018, 60, 780–795. [Google Scholar] [CrossRef] [Green Version]
- Dubouzet, J.G.; Sakuma, Y.; Ito, Y.; Kasuga, M.; Dubouzet, E.G.; Miura, S.; Seki, M.; Shinozaki, K.; Shinozaki, K.Y. OsDREB genes in rice, Oryza sativa L. encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003, 33, 751–763. [Google Scholar] [CrossRef]
- Matsukura, S.; Mizoi, J.; Yoshida, T.; Todaka, D.; Ito, Y.; Maruyama, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Comprehensive analysis of rice DREB2-type genes that encode transcription factors involved in the expression of abiotic stress-responsive genes. Mol. Genet. Genom. 2010, 283, 185–196. [Google Scholar] [CrossRef]
- Shen, Y.-G.; Zhang, W.-K.; He, S.-J.; Zhang, J.-S.; Liu, Q.; Chen, S.-Y. An EREBP/AP2-type protein in Triticum aestivum was a DRE-binding transcription factor induced by cold, dehydration and ABA stress. Theor. Appl. Genet. 2003, 106, 923–930. [Google Scholar] [CrossRef]
- Xue, G.P.; Loveridge, C.W. HvDRF1 is involved in abscisic acid-mediated gene regulation in barley and produces two forms of AP2 transcriptional activators, interacting preferably with a CT-rich element. Plant J. 2004, 37, 326–339. [Google Scholar] [CrossRef]
- Kazan, K. Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 2015, 20, 219–229. [Google Scholar] [CrossRef]
- Gibbs, D.J.; Conde, J.V.; Berckhan, S.; Prasad, G.; Mendiondo, G.M.; Holdsworth, M.J. Group VII Ethylene Response Factors Coordinate Oxygen and Nitric Oxide Signal Transduction and Stress Responses in Plants. Plant Physiol. 2015, 169, 23–31. [Google Scholar] [CrossRef] [Green Version]
- Muller, M.; Munne, B.S. Ethylene Response Factors: A Key Regulatory Hub in Hormone and Stress Signaling. Plant Physiol. 2015, 169, 32–41. [Google Scholar] [CrossRef] [Green Version]
- Sah, S.K.; Reddy, K.R.; Li, J. Abscisic Acid and Abiotic Stress Tolerance in Crop Plants. Front. Plant Sci. 2016, 7, 571. [Google Scholar] [CrossRef] [Green Version]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [Green Version]
- Gutterson, N.; Reuber, T.L. Regulation of disease resistance pathways by AP2/ERF transcription factors. Curr. Opin. Plant Biol. 2004, 7, 465–471. [Google Scholar] [CrossRef]
- McGrath, K.C.; Dombrecht, B.; Manners, J.M.; Schenk, P.M.; Edgar, C.I.; Maclean, D.J.; Scheible, W.-R.; Udvardi, M.K.; Kazan, K. Repressor- and activator-type ethylene response factors functioning in jasmonate signaling and disease resistance identified via a genome-wide screen of Arabidopsis transcription factor gene expression. Plant Physiol. 2005, 139, 949–959. [Google Scholar] [CrossRef] [Green Version]
- Lorenzo, O.; Piqueras, R.; Sánchez-Serrano, J.J.; Solano, R. ETHYLENE RESPONSE FACTOR1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell 2003, 15, 165–178. [Google Scholar] [CrossRef] [Green Version]
- Van der Fits, L.; Memelink, J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science 2000, 289, 295–297. [Google Scholar] [CrossRef]
- Van der Fits, L.; Memelink, J. The jasmonate-inducible AP2/ERF-domain transcription factor ORCA3 activates gene expression via interaction with a jasmonate-responsive promoter element. Plant J. 2001, 25, 43–53. [Google Scholar]
- Pre, M.; Atallah, M.; Champion, A.; De Vos, M.; Pieterse, C.M.; Memelink, J. The AP2/ERF domain transcription factor ORA59 integrates jasmonic acid and ethylene signals in plant defense. Plant Physiol. 2008, 147, 1347–1357. [Google Scholar] [CrossRef] [Green Version]
- Chen, H.Y.; Hsieh, E.J.; Cheng, M.C.; Chen, C.Y.; Hwang, S.Y.; Lin, T.P. ORA47 (octadecanoid-responsive AP2/ERF-domain transcription factor 47) regulates jasmonic acid and abscisic acid biosynthesis and signaling through binding to a novel cis-element. New Phytol. 2016, 211, 599–613. [Google Scholar] [CrossRef]
- Stelpflug, S.C.; Sekhon, R.S.; Vaillancourt, B.; Hirsch, C.N.; Buell, C.R.; de Leon, N.; Kaeppler, S.M. An Expanded Maize Gene Expression Atlas based on RNA Sequencing and its Use to Explore Root Development. Plant Genome 2016, 9, 25. [Google Scholar] [CrossRef]






Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, C.; An, L.; Li, F.; Ahmad, W.; Aslam, M.; Ul Haq, M.Z.; Yan, Y.; Ahmad, R.M. Wide-Range Portrayal of AP2/ERF Transcription Factor Family in Maize (Zea mays L.) Development and Stress Responses. Genes 2023, 14, 194. https://doi.org/10.3390/genes14010194
Cheng C, An L, Li F, Ahmad W, Aslam M, Ul Haq MZ, Yan Y, Ahmad RM. Wide-Range Portrayal of AP2/ERF Transcription Factor Family in Maize (Zea mays L.) Development and Stress Responses. Genes. 2023; 14(1):194. https://doi.org/10.3390/genes14010194
Chicago/Turabian StyleCheng, Cheng, Likun An, Fangzhe Li, Wahaj Ahmad, Muhammad Aslam, Muhammad Zia Ul Haq, Yuanxin Yan, and Ramala Masood Ahmad. 2023. "Wide-Range Portrayal of AP2/ERF Transcription Factor Family in Maize (Zea mays L.) Development and Stress Responses" Genes 14, no. 1: 194. https://doi.org/10.3390/genes14010194
APA StyleCheng, C., An, L., Li, F., Ahmad, W., Aslam, M., Ul Haq, M. Z., Yan, Y., & Ahmad, R. M. (2023). Wide-Range Portrayal of AP2/ERF Transcription Factor Family in Maize (Zea mays L.) Development and Stress Responses. Genes, 14(1), 194. https://doi.org/10.3390/genes14010194