Genome-Wide Analysis and Expression of the GRAS Transcription Factor Family in Avena sativa
Abstract
1. Introduction
2. Materials & Methods
2.1. AsGRAS Gene Family Identification
2.2. Analysis of A. sativa GRAS Gene Evolution
2.3. Analysis of the Conserved Motif and Gene Structure
2.4. Chromosome Localization and Gene Replication Analysis of GRAS Genes
2.5. Cis-Elements Analysis
2.6. Gene Regulatory Network Analysis of the AsGRAS
2.7. Collinearity Analysis of the A. sativa GRAS Gene Families
2.8. Plant Material and Treatments
2.9. Expression Levels under Stress Treatments
3. Results
3.1. AsGRAS Gene Family Identification and Analysis of Physiochemical Properties
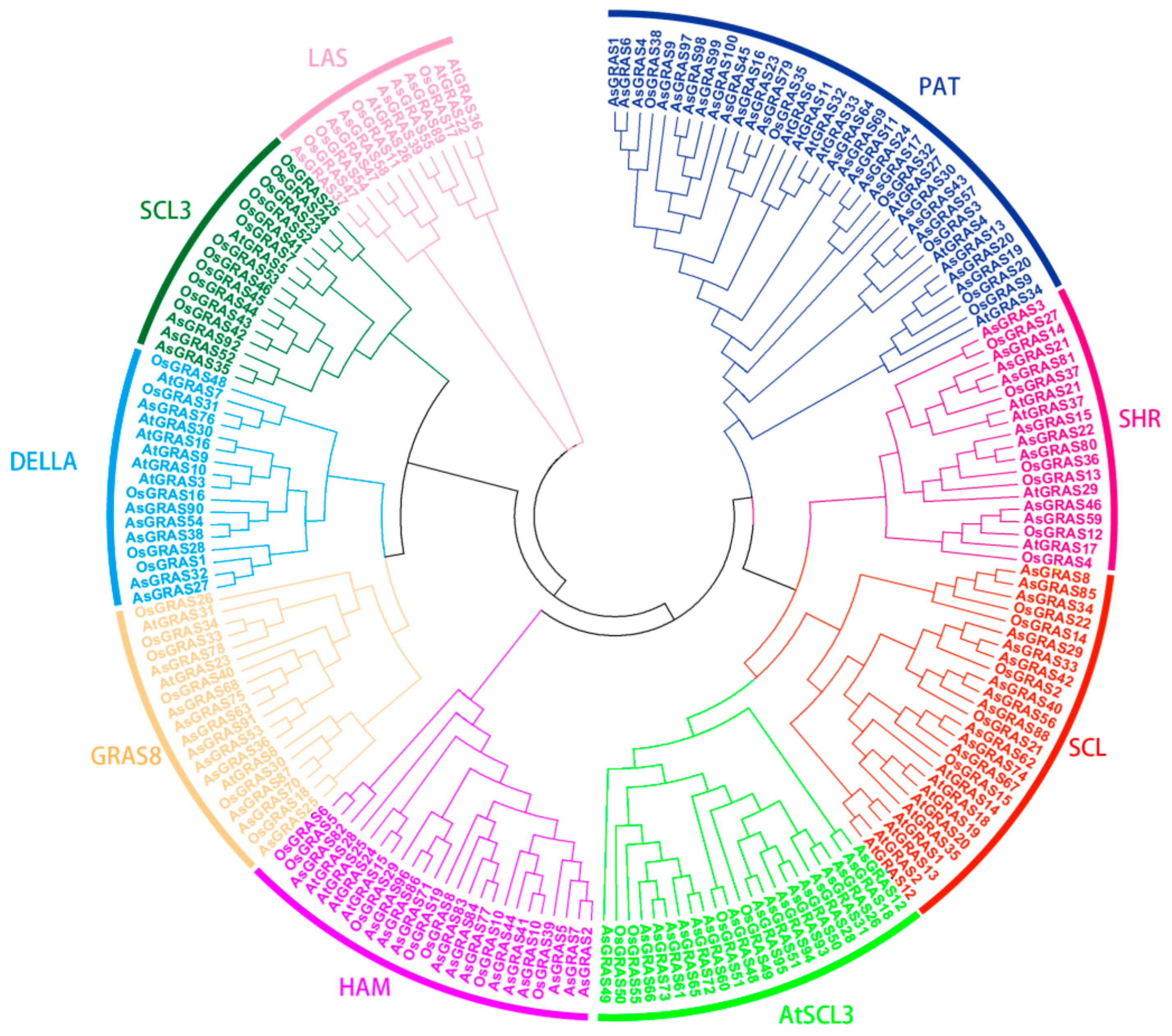
3.2. Analysis of the A. sativa GRAS Gene Family Members and Gene Family Evolution
3.3. A. sativa GRAS Gene Family Motif Analysis and Gene Structure Analysis
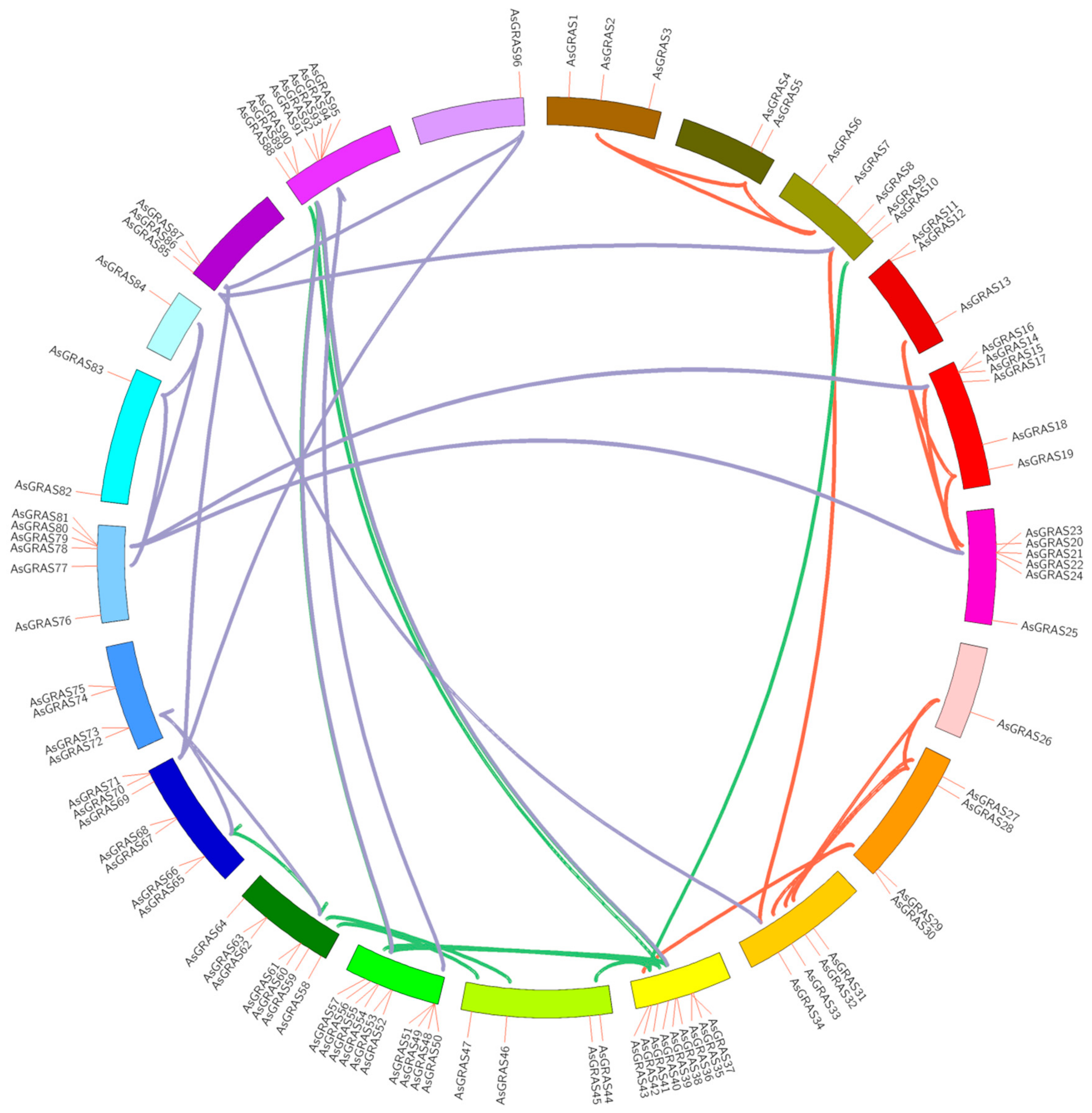
3.4. Location and Gene Duplication of the Chromosomes of the A. sativa GRAS Gene Family
3.5. Analysis of the Promoter Elements of the A. sativa GRAS Gene Family
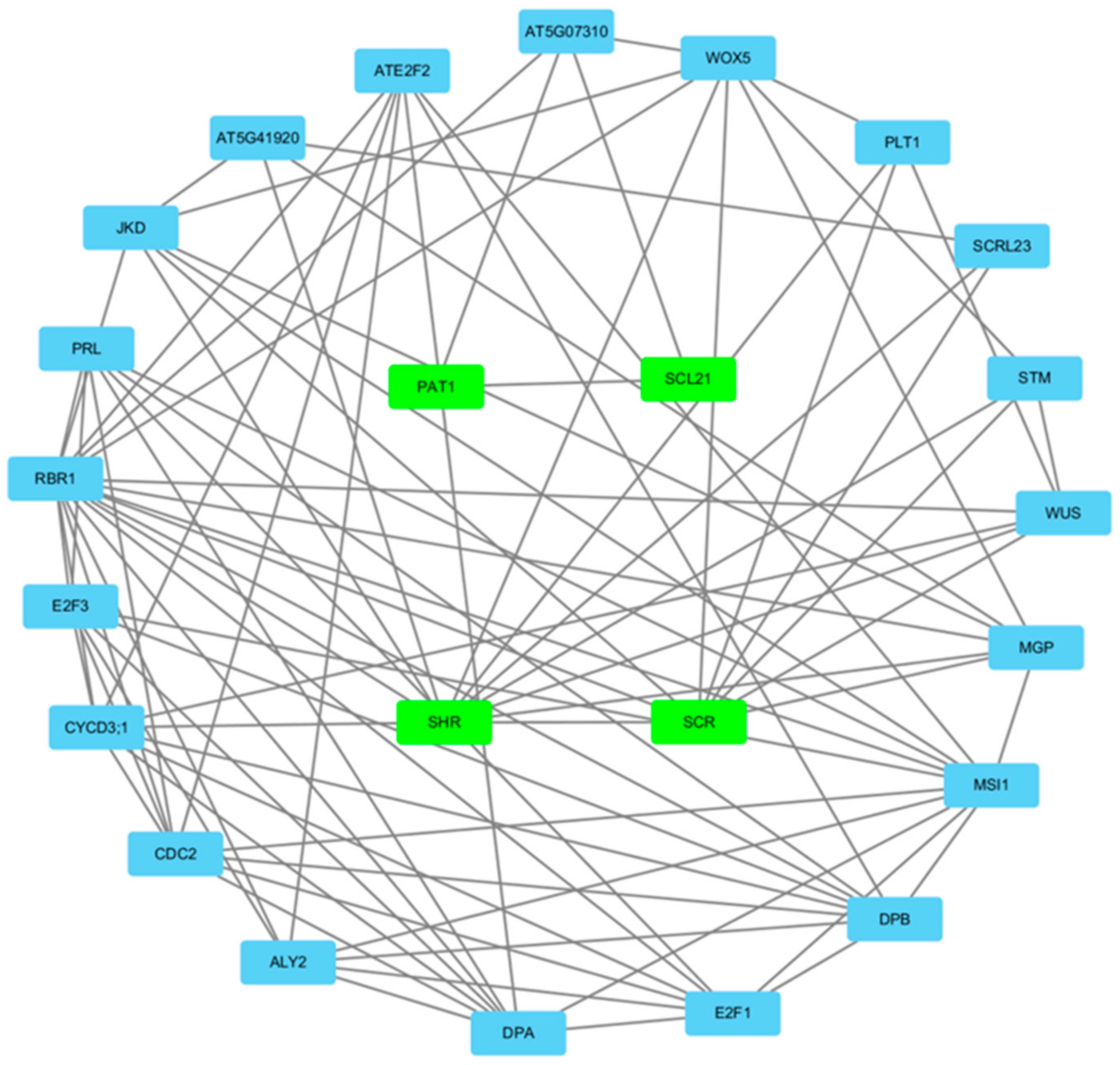
3.6. Gene Regulatory Network Analysis of the A. sativa GRAS Gene Family
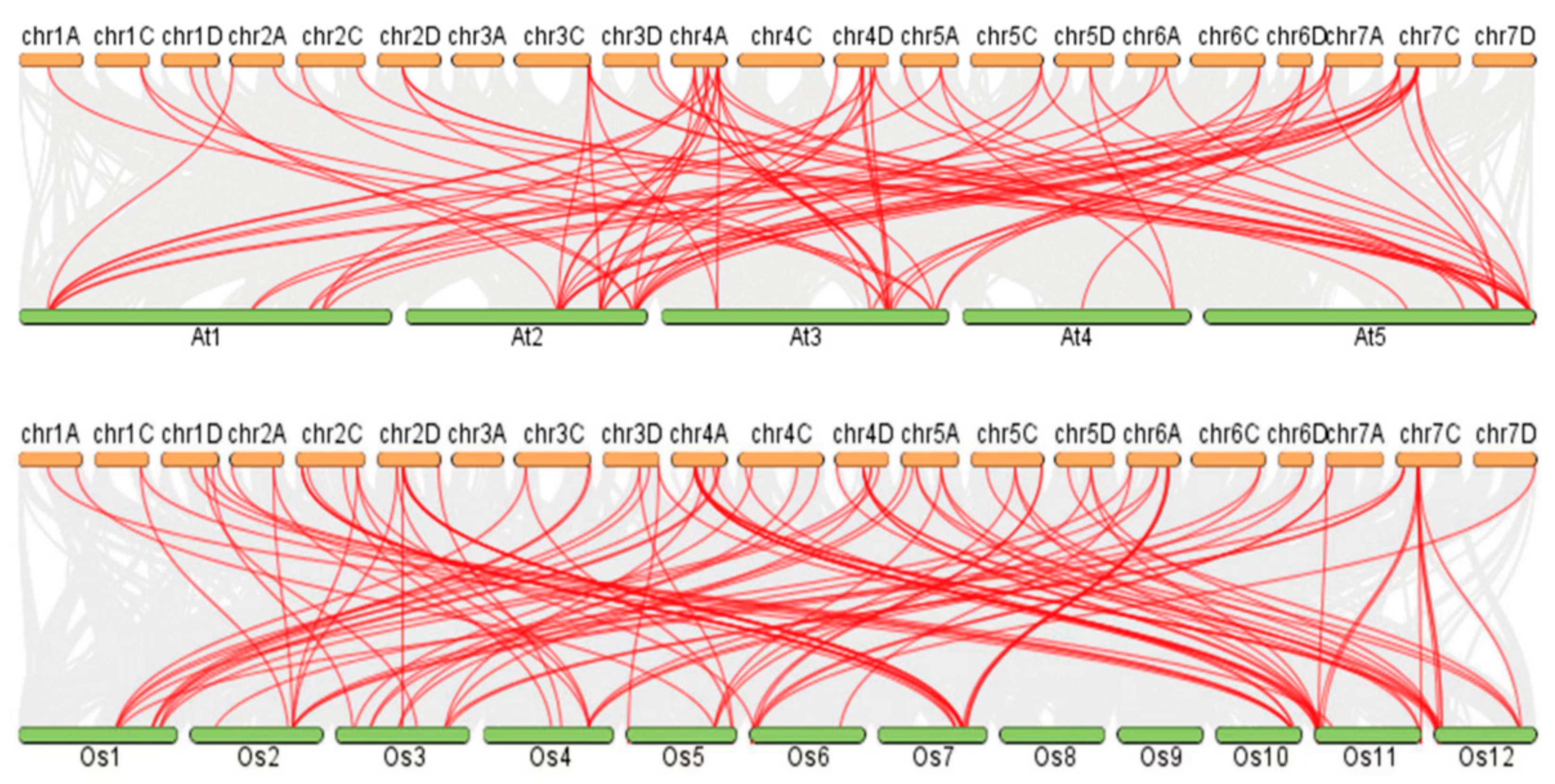
3.7. Collinearity Analysis of the A. sativa GRAS Gene Families
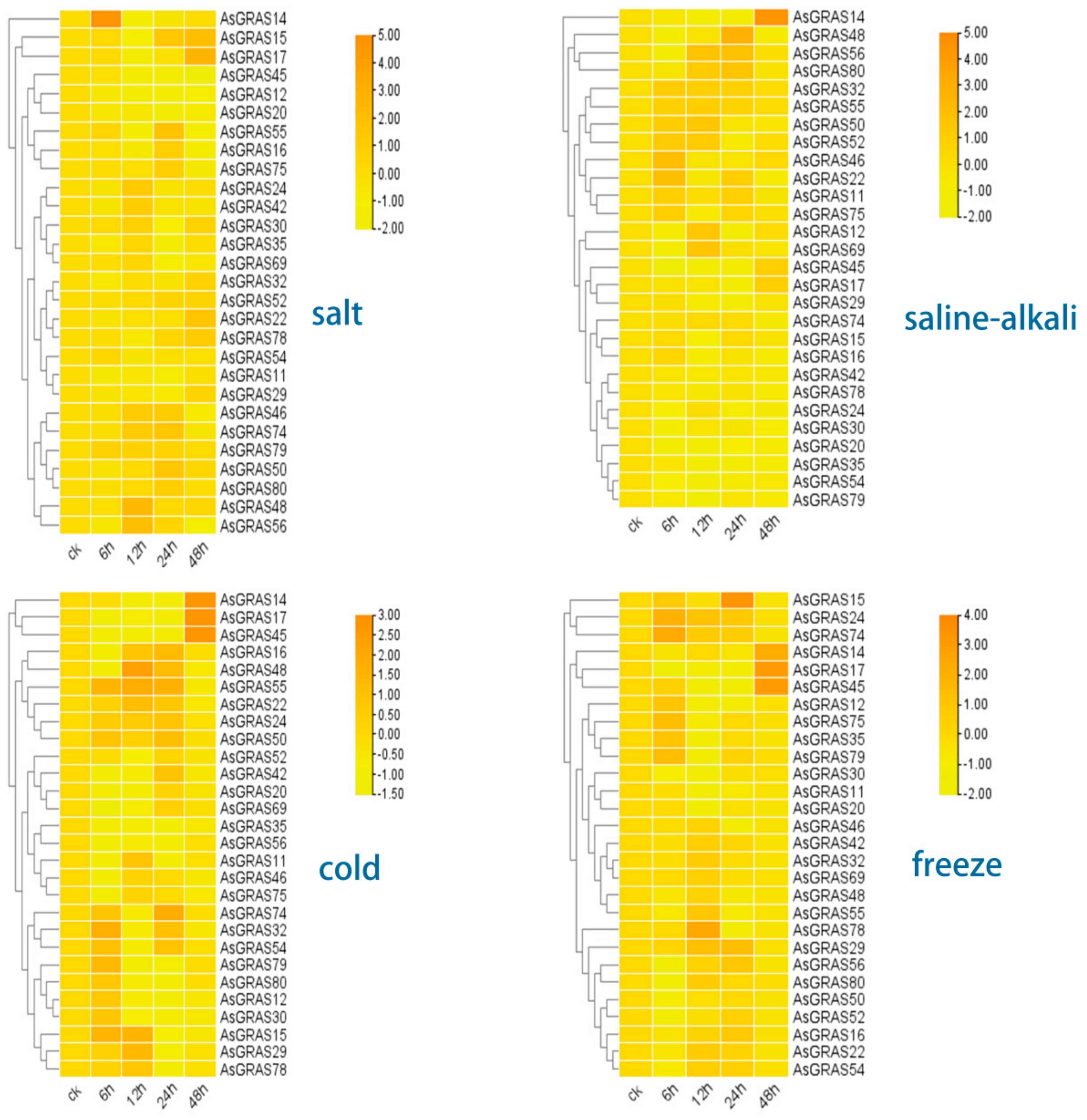
3.8. Response and Expression Pattern of A. sativa GRAS Family Genes under 4 Abiotic Stresses (Salt, Saline–Alkali, Cold, and Freezing Stress Conditions)
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Pysh, L.D.; Wysocka-Diller, J.W.; Camilleri, C.; Bouchez, D.; Benfey, P.N. The GRAS gene family in Arabidopsis sequence characterization and basic expression analysis of the SCARECROW-LIKE genes. Plant J. 1999, 18, 111–119. [Google Scholar] [CrossRef] [PubMed]
- Heery, D.M.; Kalkhoven, E.; Hoare, S.; Parker, M.G. A signature motif in transcriptional co-activators mediates binding to nuclear receptors. Nature 1997, 387, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.; Yang, Q.; Fu, T.; Zhou, Y. Overexpression of the Brassica napus bnlas gene in Arabidopsis affects plant development and increases drought tolerance. Plant Cell Rep. 2011, 30, 373–388. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.S.; Xia, X.L.; Yin, W.L. Cloning and analysis of SCL7 gene from Populus euphratica. J. Beijing For. Univ. 2011, 33, 1–10. [Google Scholar]
- Xu, K.; Chen, S.; Li, T.; Ma, X.; Liang, X.; Ding, X.; Liu, H.; Luo, L. OsGRAS23, a rice gras transcription factor gene, is involved in drought stress response through regulating expression of stress-responsive genes. BMC Plant Biol. 2015, 15, 141. [Google Scholar] [CrossRef]
- Wang, Z.; Wong, D.C.J.; Wang, Y.; Xu, G.; Ren, C.; Liu, Y.; Kuang, Y.; Fan, P.; Li, S.; Xin, H.; et al. GRAS-domain transcription factor PAT1 regulates jasmonic acid biosynthesis in grape cold stress response. Plant Physiol. 2021, 186, 1660–1678. [Google Scholar] [CrossRef] [PubMed]
- Kamal, N.; Tsardakas Renhuldt, N.; Bentzer, J.; Gundlach, H.; Haberer, G.; Juhász, A.; Lux, T.; Bose, U.; Tye-Din, J.A.; Lang, D.; et al. The mosaic oat genome gives insights into a uniquely healthy cereal crop. Nature 2022, 606, 113–119. [Google Scholar] [CrossRef]
- Peng, Y.; Yan, H.; Guo, L.; Deng, C.; Wang, C.; Wang, Y.; Kang, L.; Zhou, P.; Yu, K.; Dong, X.; et al. Reference genome assemblies reveal the origin and evolution of allohexaploid oat. Nat. Genet. 2022, 54, 1248–1258. [Google Scholar] [CrossRef]
- Ergashovich, K.A.; Tokhirovna, J.O. Ecophysiological Properties of White Oats. In Proceedings of the International Conference on Innovations in Sciences, Education and Humanities, Rome, Italy, 10 December 2021. [Google Scholar]
- Xu, Z.; Chen, X.; Lu, X.; Zhao, B.; Yang, Y.; Liu, J. Integrative analysis of transcriptome and metabolome reveal mechanism of tolerance to salt stress in oat (Avena sativa L.). Plant Physiol. Biochem. 2021, 160, 315–328. [Google Scholar] [CrossRef] [PubMed]
- Zhao, G.Q.; Ma, B.L.; Ren, C.Z. Salinity Effects on Yield and Yield Components of Contrasting Naked Oat Genotypes. J. Plant Nutr. 2009, 32, 1619–1632. [Google Scholar] [CrossRef]
- Zhang, H.; Liu, X.; Wang, X.; Sun, M.; Song, R.; Mao, P.; Jia, S. Genome-Wide Identification of GRAS Gene Family and Their Responses to Abiotic Stress in Medicago sativa. Int. J. Mol. Sci. 2021, 22, 7729. [Google Scholar] [CrossRef]
- Wang, S.; Duan, Z.; Yan, Q.; Wu, F.; Zhou, P.; Zhang, J. Genome–Wide Identification of the GRAS Family Genes in Melilotus albus and Expression Analysis under Various Tissues and Abiotic Stresses. Int. J. Mol. Sci. 2022, 23, 7403. [Google Scholar] [CrossRef] [PubMed]
- Fan, Y.; Wei, X.; Lai, D.; Yang, H.; Feng, L.; Li, L.; Niu, K.; Chen, L.; Xiang, D.; Ruan, J.; et al. Genome-wide investigation of the GRAS transcription factor family in foxtail millet (Setaria italica L.). MC Plant Biol. 2021, 21, 508. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Jiao, Z.; Niu, M.X.; Yu, X.; Huang, M.; Liu, C.; Wang, H.L.; Zhou, Y.; Mao, W.; Wang, X.; et al. Genome-Wide Comprehensive Analysis of the GASA Gene Family in Populus. Int. J. Mol. Sci. 2021, 22, 12336. [Google Scholar] [CrossRef]
- Liu, M.; Huang, L.; Ma, Z.; Sun, W.; Wu, Q.; Tang, Z.; Bu, T.; Li, C.; Chen, H. Genome-wide identification, expression analysis and functional study of the GRAS gene family in Tartary buckwheat (Fagopyrum tataricum). BMC Plant Biol. 2019, 19, 342. [Google Scholar] [CrossRef]
- Liu, B.; Sun, Y.; Xue, J.; Jia, X.; Li, R. Genome-wide characterization and expression analysis of GRAS gene family in pepper (Capsicum annuum L.). PeerJ 2018, 6, e4796. [Google Scholar] [CrossRef] [PubMed]
- Ling, L.; Zhang, W.; An, Y.; Du, B.; Wang, D.; Guo, C. Genome-wide analysis of the TCP transcription factor genes in five legume genomes and their response to salt and drought stresses. Funct. Integr. Genom. 2020, 20, 537–550. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef]
- Bailey, T.L.; Williams, N.; Misleh, C.; Li, W.W. MEME: Discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006, 34, W369–W373. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Ye, J.; McGinnis, S.; Madden, T.L. BLAST: Improvements for better sequence analysis. Nucleic Acids Res. 2006, 34, W6–W9. [Google Scholar] [CrossRef] [PubMed]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Hoagland, D.R.; Arnon, D.I. The water culture method for growing plants without soil. Circ. Calif. Agric. Exp. Stn. 1950, 347, 1–38. [Google Scholar]
- Huang, W.; Xian, Z.; Kang, X.; Tang, N.; Li, Z. Genome-wide identification, phylogeny and expression analysis of GRAS gene family in tomato. MC Plant Biol. 2015, 15, 209. [Google Scholar] [CrossRef]
- Zhang, H.; Cao, Y.; Shang, C.; Li, J.; Wang, J.; Wu, Z.; Ma, L.; Qi, T.; Fu, C.; Bai, Z.; et al. Genome-wide characterization of GRAS family genes in Medicago truncatula reveals their evolutionary dynamics and functional diversification. PLoS ONE 2017, 12, e0185439. [Google Scholar] [CrossRef]
- Zhang, C.; Liu, S.; Liu, D.; Guo, F.; Yang, Y.; Dong, T.; Zhang, Y.; Ma, C.; Tang, Z.; Li, F.; et al. Genome-wide survey and expression analysis of GRAS transcription factor family in sweetpotato provides insights into their potential roles in stress response. BMC Plant Biol. 2022, 22, 232. [Google Scholar] [CrossRef]
- To, V.T.; Shi, Q.; Zhang, Y.; Shi, J.; Shen, C.; Zhang, D.; Cai, W. Genome-Wide Analysis of the GRAS Gene Family in Barley (Hordeum vulgare L.). Genes 2020, 11, 553. [Google Scholar] [CrossRef]
- Tian, C.; Wan, P.; Sun, S.; Li, J.; Chen, M. Genome-wide analysis of the GRAS gene family in rice and Arabidopsis. Plant Mol. Biol. 2004, 54, 519–532. [Google Scholar] [CrossRef]
- Zou, M.; Guo, B.; He, S. The Roles and Evolutionary Patterns of Intronless Genes in Deuterostomes. Comp. Funct. Genom. 2011, 2011, 680673. [Google Scholar] [CrossRef]
- Zhang, B.; Liu, J.; Yang, Z.E.; Chen, E.Y.; Zhang, C.J.; Zhang, X.Y.; Li, F.G. Genome-wide analysis of GRAS transcription factor gene family in Gossypium hirsutum L. BMC Genom. 2018, 19, 348. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Fang, L.; Karungo, S.K.; Zhang, L.; Gao, Y.; Li, S.; Xin, H. Overexpression of VaPAT1, a GRAS transcription factor from Vitis amurensis, confers abiotic stress tolerance in Arabidopsis. Plant Cell Rep. 2016, 35, 655–666. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ling, L.; Li, M.; Chen, N.; Ren, G.; Qu, L.; Yue, H.; Wu, X.; Zhao, J. Genome-Wide Analysis and Expression of the GRAS Transcription Factor Family in Avena sativa. Genes 2023, 14, 164. https://doi.org/10.3390/genes14010164
Ling L, Li M, Chen N, Ren G, Qu L, Yue H, Wu X, Zhao J. Genome-Wide Analysis and Expression of the GRAS Transcription Factor Family in Avena sativa. Genes. 2023; 14(1):164. https://doi.org/10.3390/genes14010164
Chicago/Turabian StyleLing, Lei, Mingjing Li, Naiyu Chen, Guoling Ren, Lina Qu, Hua Yue, Xinyu Wu, and Jing Zhao. 2023. "Genome-Wide Analysis and Expression of the GRAS Transcription Factor Family in Avena sativa" Genes 14, no. 1: 164. https://doi.org/10.3390/genes14010164
APA StyleLing, L., Li, M., Chen, N., Ren, G., Qu, L., Yue, H., Wu, X., & Zhao, J. (2023). Genome-Wide Analysis and Expression of the GRAS Transcription Factor Family in Avena sativa. Genes, 14(1), 164. https://doi.org/10.3390/genes14010164