Transcriptome Analysis of Streptococcus mutans Quorum Sensing-Mediated Persisters Reveals an Enrichment in Genes Related to Stress Defense Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Streptococcal Strains and Growth Conditions
2.2. Preparation of Bacterial Inoculum for Persister Cell Collection
2.3. Persister Cell Collection for RNA Sequencing
2.4. RNA Extraction and RNA Sequencing
2.5. RNA Sequencing Data Analysis
2.6. Gene Expression Analysis via RT-qPCR
2.7. Persister Assay
2.8. Toxin Expression in E. coli
3. Results
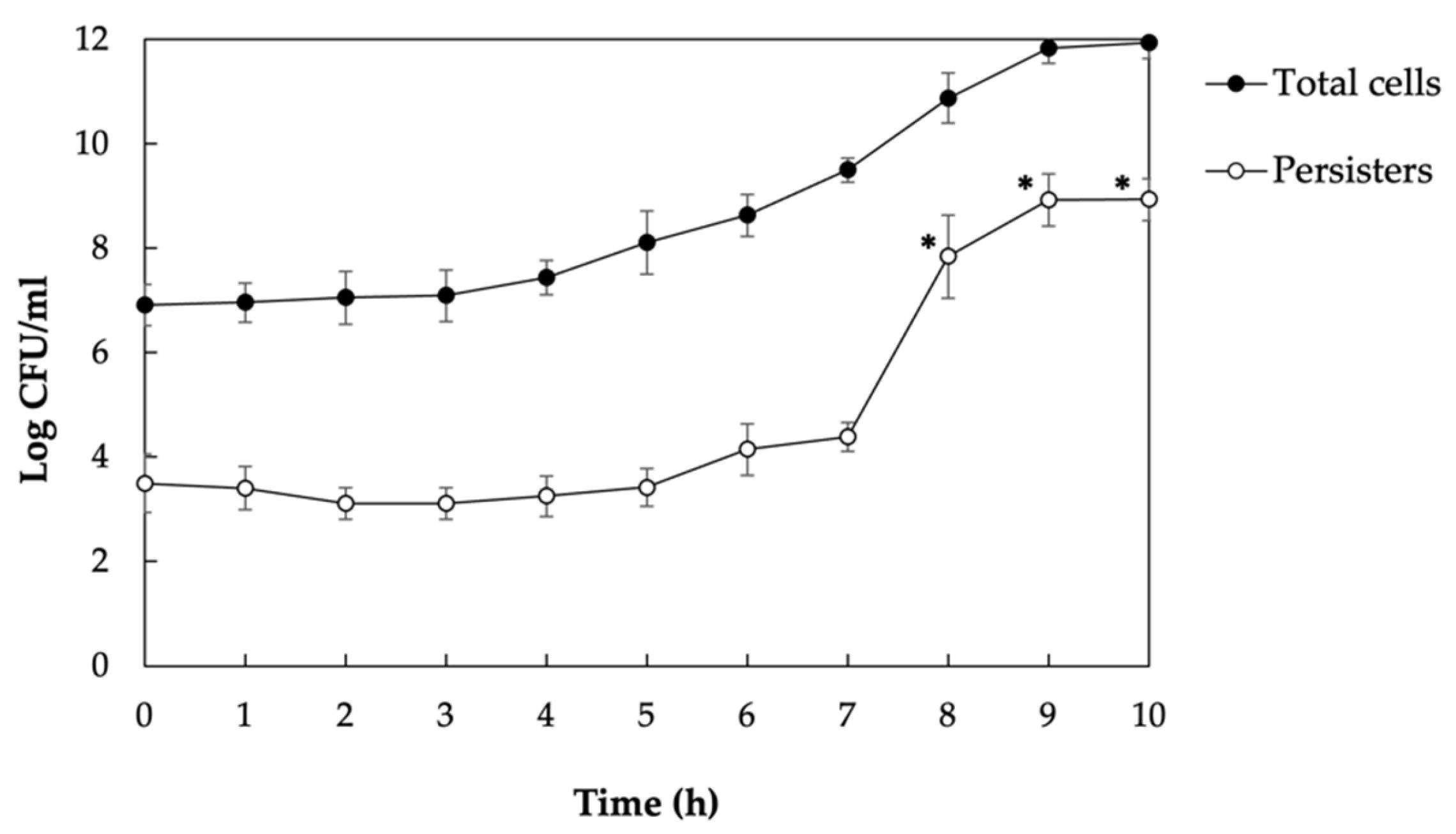
3.1. Persisters Are Metabolically Active Cells, and Their Population Is Enriched at Stationary Phase
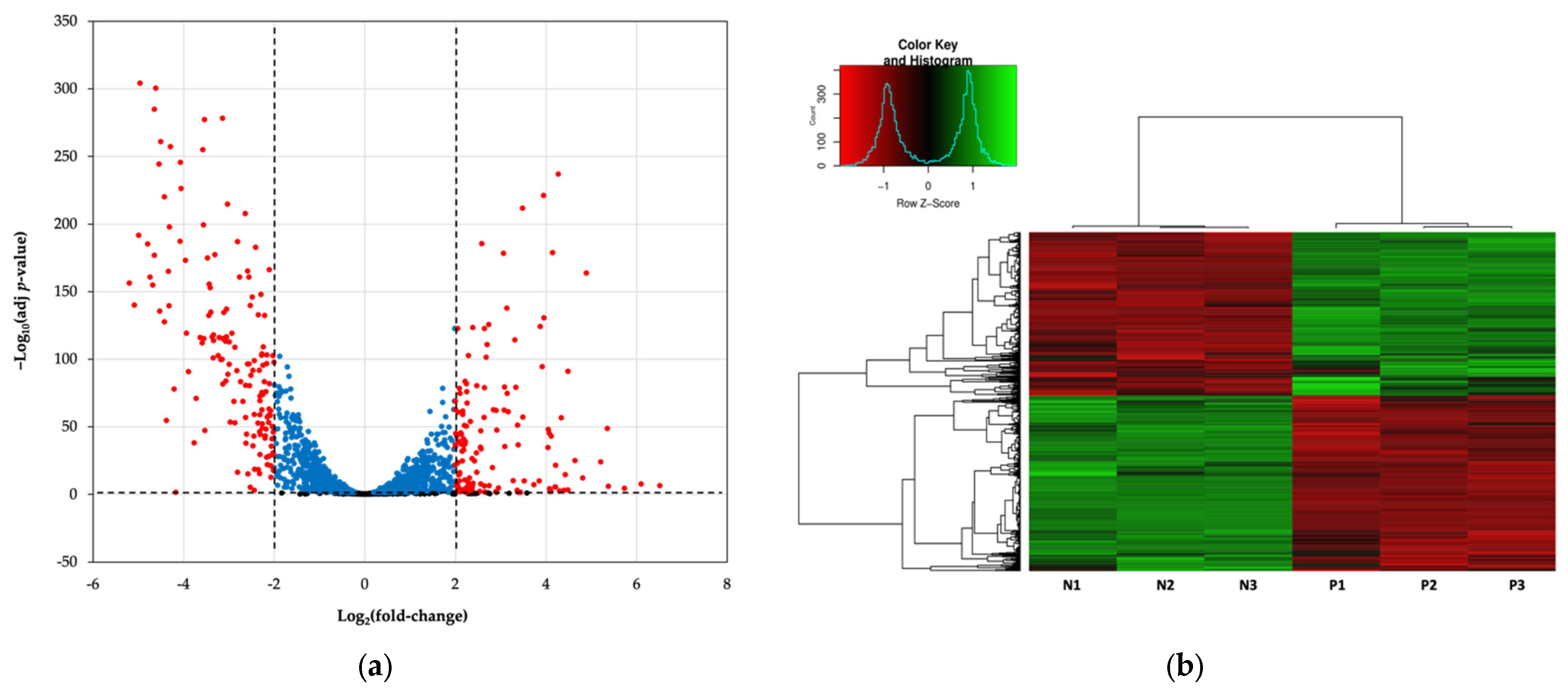
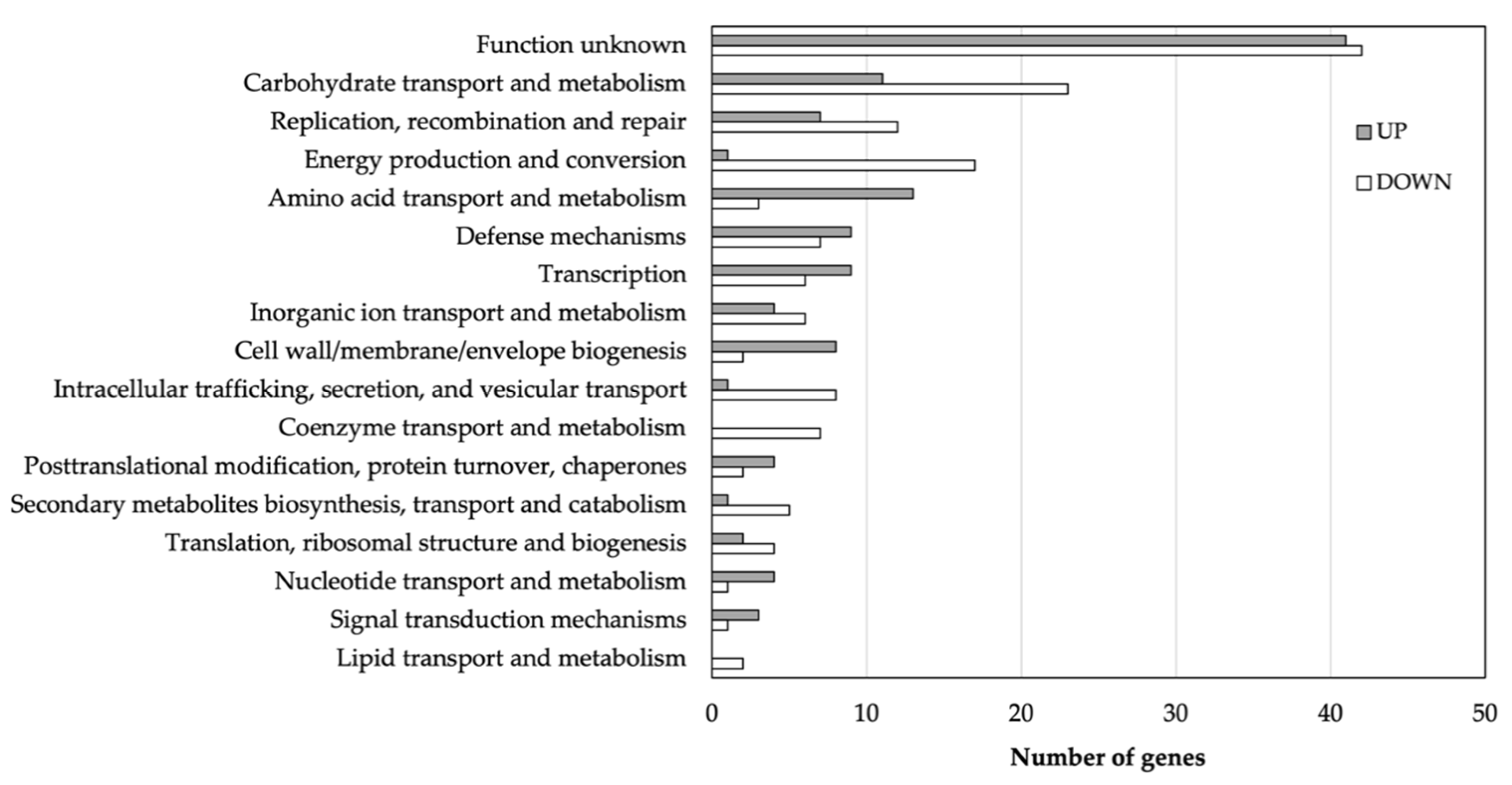
3.2. Isolation and Transcriptome Analysis of Persisters
3.3. Genes Related to Metabolism of Galactose/Lactose
3.4. Genes Related to Stress Defense Mechanisms
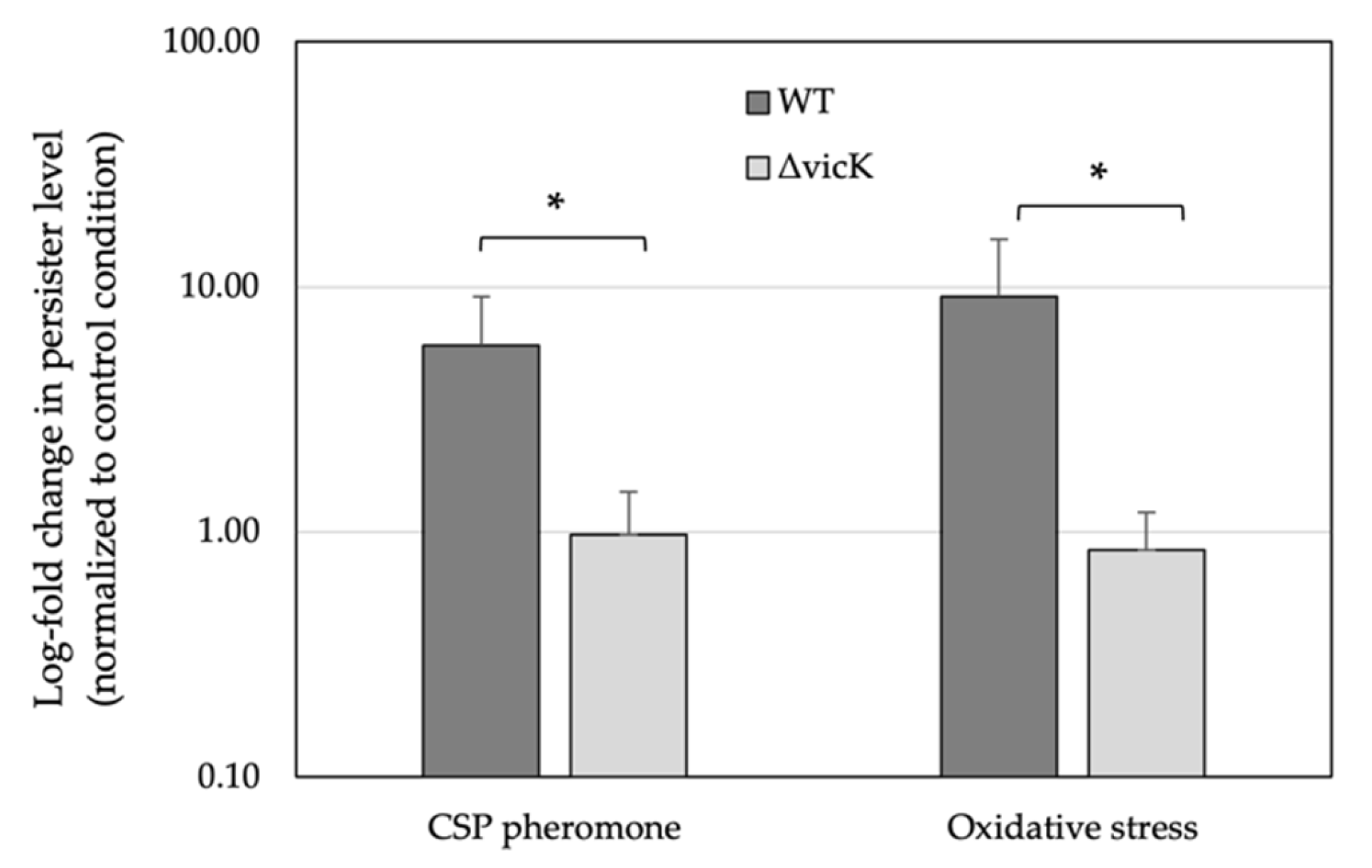
3.5. Inactivation of VicK Abolished the Triggered Persistence Phenotype
3.6. Overexpression of relE40 Promotes the Formation of S. mutans Persisters
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Balaban, N.Q.; Helaine, S.; Lewis, K.; Ackermann, M.; Aldridge, B.; Andersson, D.I.; Brynildsen, M.P.; Bumann, D.; Camilli, A.; Collins, J.J.; et al. Definitions and guidelines for research on antibiotic persistence. Nat. Rev. Microbiol. 2019, 17, 441–448. [Google Scholar] [CrossRef] [PubMed]
- Moldoveanu, A.L.; Rycroft, J.A.; Helaine, S. Impact of bacterial persisters on their host. Curr. Opin. Microbiol. 2021, 59, 65–71. [Google Scholar] [CrossRef] [PubMed]
- Windels, E.M.; Michiels, J.E.; Van den Bergh, B.; Fauvart, M.; Michiels, J. Antibiotics: Combatting tolerance to stop resistance. mBio 2019, 10, e02095-19. [Google Scholar] [CrossRef] [PubMed]
- Gollan, B.; Grabe, G.; Michaux, C.; Helaine, S. Bacterial persisters and infections: Past, present, and progressing. Annu. Rev. Microbiol. 2019, 73, 359–385. [Google Scholar] [CrossRef]
- Van den Bergh, B.; Fauvart, M.; Michiels, J. Formation, physiology, ecology, evolution and clinical importance of bacterial persisters. FEMS Microbiol. Rev. 2017, 41, 219–251. [Google Scholar] [CrossRef]
- Zou, J.; Peng, B.; Qu, J.; Zheng, J. Are bacterial persisters dormant cells only? Front. Microbiol. 2022, 12, 708580. [Google Scholar] [CrossRef]
- Pacios, O.; Blasco, L.; Bleriot, I.; Fernandez-Garcia, L.; Ambroa, A.; López, M.; Bou, G.; Cantón, R.; Garcia-Contreras, R.; Wood, T.K.; et al. (p)ppGpp and its role in bacterial persistence: New challenges. Antimicrob. Agents Chemother. 2020, 64, e01283-20. [Google Scholar] [CrossRef]
- Kaldalu, N.; Hauryliuk, V.; Turnbull, K.J.; La Mensa, A.; Putrinš, M.; Tenson, T. In vitro studies of persister cells. Microbiol. Mol. Biol. Rev. 2020, 84, e00070-20. [Google Scholar] [CrossRef]
- Cohen, N.R.; Lobritz, M.A.; Collins, J.J. Microbial persistence and the road to drug resistance. Cell Host Microbe 2013, 13, 632–642. [Google Scholar] [CrossRef]
- Papadimitriou, K.; Alegría, Á.; Bron, P.A.; de Angelis, M.; Gobbetti, M.; Kleerebezem, M.; Lemos, J.A.; Linares, D.M.; Ross, P.; Stanton, C.; et al. Stress physiology of lactic acid bacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 837–890. [Google Scholar] [CrossRef]
- Claverys, J.P.; Prudhomme, M.; Martin, B. Induction of competence regulons as a general response to stress in gram-positive bacteria. Annu. Rev. Microbiol. 2006, 60, 451–475. [Google Scholar] [CrossRef]
- Charpentier, X.; Polard, P.; Claverys, J.P. Induction of competence for genetic transformation by antibiotics: Convergent evolution of stress responses in distant bacterial species lacking SOS? Curr. Opin. Microbiol. 2012, 15, 570–576. [Google Scholar] [CrossRef]
- Dufour, D.; Lévesque, C.M. Bacterial behaviors associated with the quorum-sensing peptide pheromone (‘alarmone’) in streptococci. Future Microbiol. 2013, 8, 593–605. [Google Scholar] [CrossRef]
- Li, Y.H.; Lau, P.C.; Lee, J.H.; Ellen, R.P.; Cvitkovitch, D.G. Natural genetic transformation of Streptococcus mutans growing in biofilms. J. Bacteriol. 2001, 183, 897–908. [Google Scholar] [CrossRef] [PubMed]
- Shanker, E.; Federle, M.J. Quorum sensing regulation of competence and bacteriocins in Streptococcus pneumoniae and mutans. Genes 2017, 8, 15. [Google Scholar] [CrossRef] [PubMed]
- Perry, J.A.; Jones, M.B.; Peterson, S.N.; Cvitkovitch, D.G.; Lévesque, C.M. Peptide alarmone signalling triggers an auto-active bacteriocin necessary for genetic competence. Mol. Microbiol. 2009, 72, 905–917. [Google Scholar] [CrossRef] [PubMed]
- Leung, V.; Lévesque, C.M. A stress-inducible quorum-sensing peptide mediates the formation of persister cells with noninherited multidrug tolerance. J. Bacteriol. 2012, 194, 2265–2274. [Google Scholar] [CrossRef]
- Leung, V.; Ajdic, D.; Koyanagi, S.; Lévesque, C.M. The formation of Streptococcus mutans persisters induced by the quorum-sensing peptide pheromone is affected by the LexA regulator. J. Bacteriol. 2015, 197, 1083–1094. [Google Scholar] [CrossRef]
- Dufour, D.; Zhao, H.; Gong, S.-G.; Lévesque, C.M. A DNA-damage inducible gene promotes the formation of antibiotic persisters in response to the quorum sensing signaling peptide in Streptococcus mutans. Genes 2022, 13, 1434. [Google Scholar] [CrossRef]
- Lévesque, C.M.; Mair, R.W.; Perry, J.A.; Lau, P.C.; Li, Y.H.; Cvitkovitch, D.G. Systemic inactivation and phenotypic characterization of two-component systems in expression of Streptococcus mutans virulence properties. Lett. Appl. Microbiol. 2007, 45, 398–404. [Google Scholar] [CrossRef]
- Biswas, I.; Jha, J.K.; Fromm, N. Shuttle expression plasmids for genetic studies in Streptococcus mutans. Microbiology 2008, 154, 2275–2282. [Google Scholar] [CrossRef]
- Dufour, D.; Villemin, C.; Perry, J.A.; Lévesque, C.M. Escape from the competence state in Streptococcus mutans is governed by the bacterial population density. Mol. Oral Microbiol. 2016, 31, 501–514. [Google Scholar] [CrossRef] [PubMed]
- Kaldalu, N.; Hauryliuk, V.; Tenson, T. Persisters-as elusive as ever. Appl. Microbiol. Biotechnol. 2016, 100, 6545–6553. [Google Scholar] [CrossRef] [PubMed]
- Spoering, A.L.; Lewis, K. Biofilms and planktonic cells of Pseudomonas aeruginosa have similar resistance to killing by antimicrobials. J. Bacteriol. 2001, 183, 6746–6751. [Google Scholar] [CrossRef] [PubMed]
- Eng, R.H.; Padberg, F.T.; Smith, S.M.; Tan, E.N.; Cherubin, C.E. Bactericidal effects of antibiotics on slowly growing and nongrowing bacteria. Antimicrob. Agents Chemother. 1991, 35, 1824–1828. [Google Scholar] [CrossRef]
- Ajdić, D.; McShan, W.M.; McLaughlin, R.E.; Savić, G.; Chang, J.; Carson, M.B.; Primeaux, C.; Tian, R.; Kenton, S.; Jia, H.; et al. Genome sequence of Streptococcus mutans UA159, a cariogenic dental pathogen. Proc. Natl. Acad. Sci. USA 2002, 99, 14434–14439. [Google Scholar] [CrossRef]
- Syed, M.A.; Koyanagi, S.; Sharma, E.; Jobin, M.-C.; Yakunin, A.F.; Lévesque, C.M. The chromosomal mazEF locus of Streptococcus mutans encodes a functional type II toxin-antitoxin addiction system. J. Bacteriol. 2011, 193, 1122–1130. [Google Scholar] [CrossRef]
- Zhou, Y.; Liao, H.; Pei, L.; Pu, Y. Combatting persister cells: The daunting task in post-antibiotics era. Cell Insight 2023, 2, 100104. [Google Scholar] [CrossRef]
- Vallenet, D.; Engelen, S.; Mornico, D.; Cruveiller, S.; Fleury, L.; Lajus, A.; Rouy, Z.; Roche, D.; Salvignol, G.; Scarpelli, C.; et al. MicroScope: A platform for microbial genome annotation and comparative genomics. Database 2009, 2009, bap021. [Google Scholar] [CrossRef]
- Jurėnas, D.; Fraikin, N.; Goormaghtigh, F.; Van Melderen, L. Biology and evolution of bacterial toxin-antitoxin systems. Nat. Rev. Microbiol. 2022, 20, 335–350. [Google Scholar] [CrossRef]
- Dirix, G.; Monsieurs, P.; Dombrecht, B.; Daniels, R.; Marchal, K.; Vanderleyden, J.; Michiels, J. Peptide signal molecules and bacteriocins in Gram-negative bacteria: A genome-wide in silico screening for peptides containing a double-glycine leader sequence and their cognate transporters. Peptides 2004, 25, 1425–1440. [Google Scholar] [CrossRef] [PubMed]
- Lemos, J.A.; Palmer, S.R.; Zeng, L.; Wen, Z.T.; Kajfasz, J.K.; Freires, I.A.; Abranches, J.; Brady, L.J. The Biology of Streptococcus mutans. Microbiol. Spectr. 2019, 7, 10–1128. [Google Scholar] [CrossRef] [PubMed]
- Katsumata, T.; Nguyen-Tra Le, M.; Kawada-Matsuo, M.; Taniguchi, Y.; Ouhara, K.; Oogai, Y.; Nakata, M.; Mizuno, N.; Nishitani, Y.; Komatsuzawa, H. Comprehensive characterization of sortase A-dependent surface proteins in Streptococcus mutans. Microbiol. Immunol. 2022, 66, 145–156. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, T.; Tsuchiya, T. Multidrug efflux transporters in the MATE family. Biochim. Biophys. Acta 2009, 1794, 763–768. [Google Scholar] [CrossRef] [PubMed]
- Matsui, R.; Cvitkovitch, D. Acid tolerance mechanisms utilized by Streptococcus mutans. Future Microbiol. 2010, 5, 403–417. [Google Scholar] [CrossRef]
- Kajfasz, J.K.; Ganguly, T.; Hardin, E.L.; Abranches, J.; Lemos, J.A. Transcriptome responses of Streptococcus mutans to peroxide stress: Identification of novel antioxidant pathways regulated by Spx. Sci. Rep. 2017, 7, 16018. [Google Scholar] [CrossRef]
- Abranches, J.; Lemos, J.A.; Burne, R.A. Osmotic stress responses of Streptococcus mutans UA159. FEMS Microbiol. Lett. 2006, 255, 240–246. [Google Scholar] [CrossRef]
- Griswold, A.R.; Jameson-Lee, M.; Burne, R.A. Regulation and physiologic significance of the agmatine deiminase system of Streptococcus mutans UA159. J. Bacteriol. 2006, 188, 834–841. [Google Scholar] [CrossRef]
- Stipp, R.N.; Boisvert, H.; Smith, D.J.; Höfling, J.F.; Duncan, M.J.; Mattos-Graner, R.O. CovR and VicRK regulate cell surface biogenesis genes required for biofilm formation in Streptococcus mutans. PLoS ONE 2013, 8, e58271. [Google Scholar] [CrossRef]
- Cohen, S.E.; Lewis, C.A.; Mooney, R.A.; Kohanski, M.A.; Collins, J.J.; Landick, R.; Walker, G.C. Roles for the transcription elongation factor NusA in both DNA repair and damage tolerance pathways in Escherichia coli. Proc. Natl. Acad. Sci. USA 2010, 107, 15517–15522. [Google Scholar] [CrossRef]
- Gonzalez, K.; Faustoferri, R.C.; Quivey, R.G., Jr. Role of DNA base excision repair in the mutability and virulence of Streptococcus mutans. Mol. Microbiol. 2012, 85, 361–377. [Google Scholar] [CrossRef] [PubMed]
- Senadheera, M.D.; Guggenheim, B.; Spatafora, G.A.; Huang, Y.C.; Choi, J.; Hung, D.C.; Treglown, J.S.; Goodman, S.D.; Ellen, R.P.; Cvitkovitch, D.G. A VicRK signal transduction system in Streptococcus mutans affects gtfBCD, gbpB, and ftf expression, biofilm formation, and genetic competence development. J. Bacteriol. 2005, 187, 4064–4076. [Google Scholar] [CrossRef] [PubMed]
- Wood, T.K.; Knabel, S.J.; Kwan, B.W. Bacterial persister cell formation and dormancy. Appl. Environ. Microbiol. 2013, 79, 7116–7121. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.L.; Bassler, B.L. Bacterial quorum-sensing network architectures. Annu. Rev. Genet. 2009, 43, 197–222. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zarkan, A. Bacterial survivors: Evaluating the mechanisms of antibiotic persistence. Microbiology 2022, 168, 001266. [Google Scholar] [CrossRef]
- Baty, J.J.; Stoner, S.N.; Scoffield, J.A. Oral commensal streptococci: Gatekeepers of the oral cavity. J. Bacteriol. 2022, 204, e0025722. [Google Scholar] [CrossRef]
- Lemos, J.A.; Quivey, R.G.; Koo, H.; Abranches, J. Streptococcus mutans: A new Gram-positive paradigm? Microbiology 2013, 159, 436–445. [Google Scholar] [CrossRef]
- Dhaked, H.P.S.; Biswas, I. Distribution of two-component signal transduction systems BlpRH and ComDE across streptococcal species. Front. Microbiol. 2022, 13, 960994. [Google Scholar] [CrossRef]
- Keren, I.; Minami, S.; Rubin, E.; Lewis, K. Characterization and transcriptome analysis of Mycobacterium tuberculosis persisters. mBio 2011, 2, e00100-11. [Google Scholar] [CrossRef]
- Pu, Y.; Zhao, Z.; Li, Y.; Zou, J.; Ma, Q.; Zhao, Y.; Ke, Y.; Zhu, Y.; Chen, H.; Baker, M.A.B.; et al. Enhanced efflux activity facilitates drug tolerance in dormant bacterial cells. Mol. Cell. 2016, 62, 284–294. [Google Scholar] [CrossRef]
- Martini, C.L.; Coronado, A.Z.; Melo, M.C.N.; Gobbi, C.N.; Lopez, Ú.S.; de Mattos, M.C.; Amorim, T.T.; Botelho, A.M.N.; Vasconcelos, A.T.R.; Almeida, L.G.P.; et al. Cellular growth arrest and efflux pumps are associated with antibiotic persisters in Streptococcus pyogenes induced in biofilm-like environments. Front. Microbiol. 2021, 12, 716628. [Google Scholar] [CrossRef] [PubMed]
- Shan, Y.; Lazinski, D.; Rowe, S.; Camilli, A.; Lewis, K. Genetic basis of persister tolerance to aminoglycosides in Escherichia coli. mBio 2015, 6, e00078-15. [Google Scholar] [CrossRef] [PubMed]
- Yadav, P.; Verma, S.; Bauer, R.; Kumari, M.; Dua, M.; Johri, A.K.; Yadav, V.; Spellerberg, B. Deciphering streptococcal biofilms. Microorganisms 2020, 8, 1835. [Google Scholar] [CrossRef] [PubMed]
- Ciofu, O.; Moser, C.; Jensen, P.Ø.; Høiby, N. Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 2022, 20, 621–635. [Google Scholar] [CrossRef] [PubMed]

| Biological Process | Gene | Functional Protein | Log2FC |
|---|---|---|---|
| Peptide toxin/antimicrobial | SMU.40 | RelE-like toxin | 6.10 |
| SMU.299 | Pep299 | 2.48 | |
| Biofilm | SMU.1396 | GbpC | 5.14 |
| SMU.610 | SpaP | 2.05 | |
| Efflux pump | SMU.1286 | Multidrug efflux pump | 3.09 |
| SMU.2109 | Multidrug efflux pump | 2.46 | |
| SMU.71 | Multidrug efflux pump | 2.42 | |
| Acid/starvation stress | SMU.262 | AguB; putrescine carbamoyl transferase | 4.21 |
| SMU.263 | AguD; agmatine/putrescine antiporter | 4.64 | |
| SMU.264 | AguA; agmatine deiminase | 4.43 | |
| SMU.265 | AguC; carbamate kinase | 4.81 | |
| Oxidative stress | SMU.1297 | 3′-phosphoadenosine phosphatase | 3.33 |
| SMU.924 | Tpx; thiol peroxidase | 2.55 | |
| SMU.629 | SodA; superoxide dismutase | 2.10 | |
| SMU.1117 | Nox; H2O-forming NADH oxidase | 2.00 | |
| Osmoprotectant | SMU.1062 | Proline/glycine betaine permease | 3.48 |
| SMU.1063 | ATP-binding protein | 4.15 | |
| SMU.2116 | ATP-binding protein | 3.31 | |
| SMU.2117 | Membrane permease | 2.91 | |
| SMU.2118 | Substrate-binding protein | 2.16 | |
| SMU.2119 | Membrane permease | 1.32 | |
| Global stress response | SMU.418 | NusA; transcription factor | 2.74 |
| SMU.1515 | VicX; hydrolase | 2.21 | |
| SMU.1516 | VicK; histidine kinase | 2.00 | |
| SMU.1517 | VicR; response regulator | 2.13 | |
| SMU.1649 | Smx; exonuclease | 2.70 | |
| SMU.1865 | MutY; DNA glycosylase/lyase | 2.00 | |
| SMU.562 | ClpE; Clp ATPase | 2.58 |
| Stress Condition | Fold-Change (Stress/No Stress) |
|---|---|
| Quinolone | 19.49 (±6.68) |
| Mitomycin C | 3.64 (±0.65) |
| Hydrogen peroxide | 2.07 (±0.31) |
| THYE broth at pH 5 | 3.54 (±0.52) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dufour, D.; Li, H.; Gong, S.-G.; Lévesque, C.M. Transcriptome Analysis of Streptococcus mutans Quorum Sensing-Mediated Persisters Reveals an Enrichment in Genes Related to Stress Defense Mechanisms. Genes 2023, 14, 1887. https://doi.org/10.3390/genes14101887
Dufour D, Li H, Gong S-G, Lévesque CM. Transcriptome Analysis of Streptococcus mutans Quorum Sensing-Mediated Persisters Reveals an Enrichment in Genes Related to Stress Defense Mechanisms. Genes. 2023; 14(10):1887. https://doi.org/10.3390/genes14101887
Chicago/Turabian StyleDufour, Delphine, Haowen Li, Siew-Ging Gong, and Céline M. Lévesque. 2023. "Transcriptome Analysis of Streptococcus mutans Quorum Sensing-Mediated Persisters Reveals an Enrichment in Genes Related to Stress Defense Mechanisms" Genes 14, no. 10: 1887. https://doi.org/10.3390/genes14101887
APA StyleDufour, D., Li, H., Gong, S.-G., & Lévesque, C. M. (2023). Transcriptome Analysis of Streptococcus mutans Quorum Sensing-Mediated Persisters Reveals an Enrichment in Genes Related to Stress Defense Mechanisms. Genes, 14(10), 1887. https://doi.org/10.3390/genes14101887







