Effects of Mithramycin on BCL11A Gene Expression and on the Interaction of the BCL11A Transcriptional Complex to γ-Globin Gene Promoter Sequences
Abstract
:1. Introduction
2. Materials and Methods
2.1. Human K562 Cell Lines and Culture Conditions
2.2. Recruitment of β-Thalassemia Patients and In Vitro Culture of Erythroid Precursor Cells (ErPCs)
2.3. Preparation of Protein Extracts
2.4. Western Blotting
2.5. Preparation of Nuclear Extracts and Electrophoretic Mobility Shift Assays (EMSA)
2.6. Reverse Transcription and Quantitative Real-Time PCR (RT-qPCR)
2.7. Statistical Analysis
3. Results
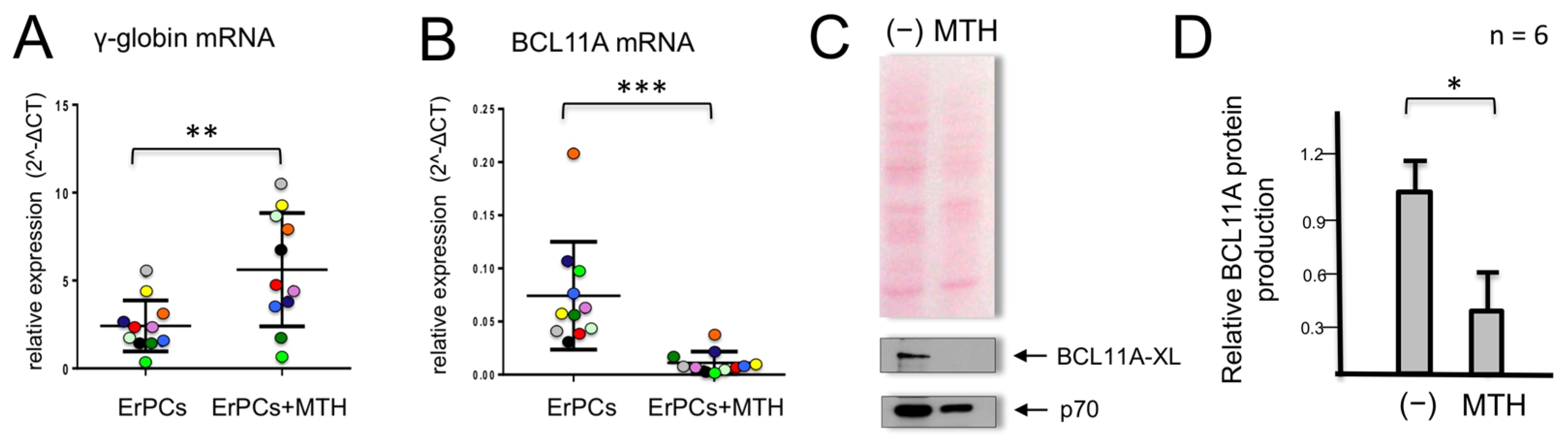
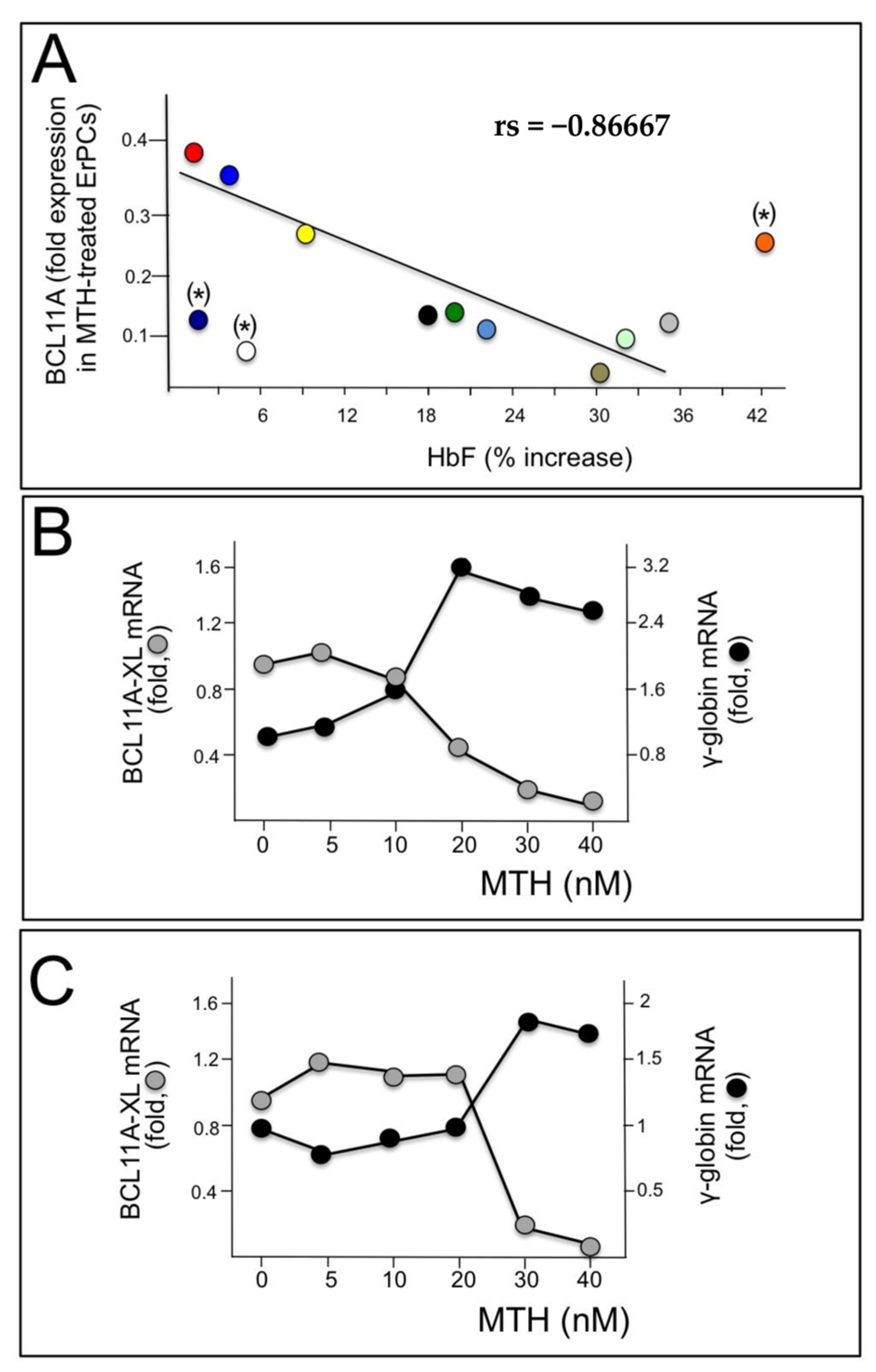
3.1. Mithramycin-Mediated Induction of γ-Globin Genes in Erythroid Precursor Cells (ErPC) Is Associated with a Dramatic Decrease in BCL11A mRNA and Protein Content
3.2. Analysis of the BCL11A Gene Promoter: Presence of Sp1 Transcription Factors Binding Sites Possibly Recognized by MTH
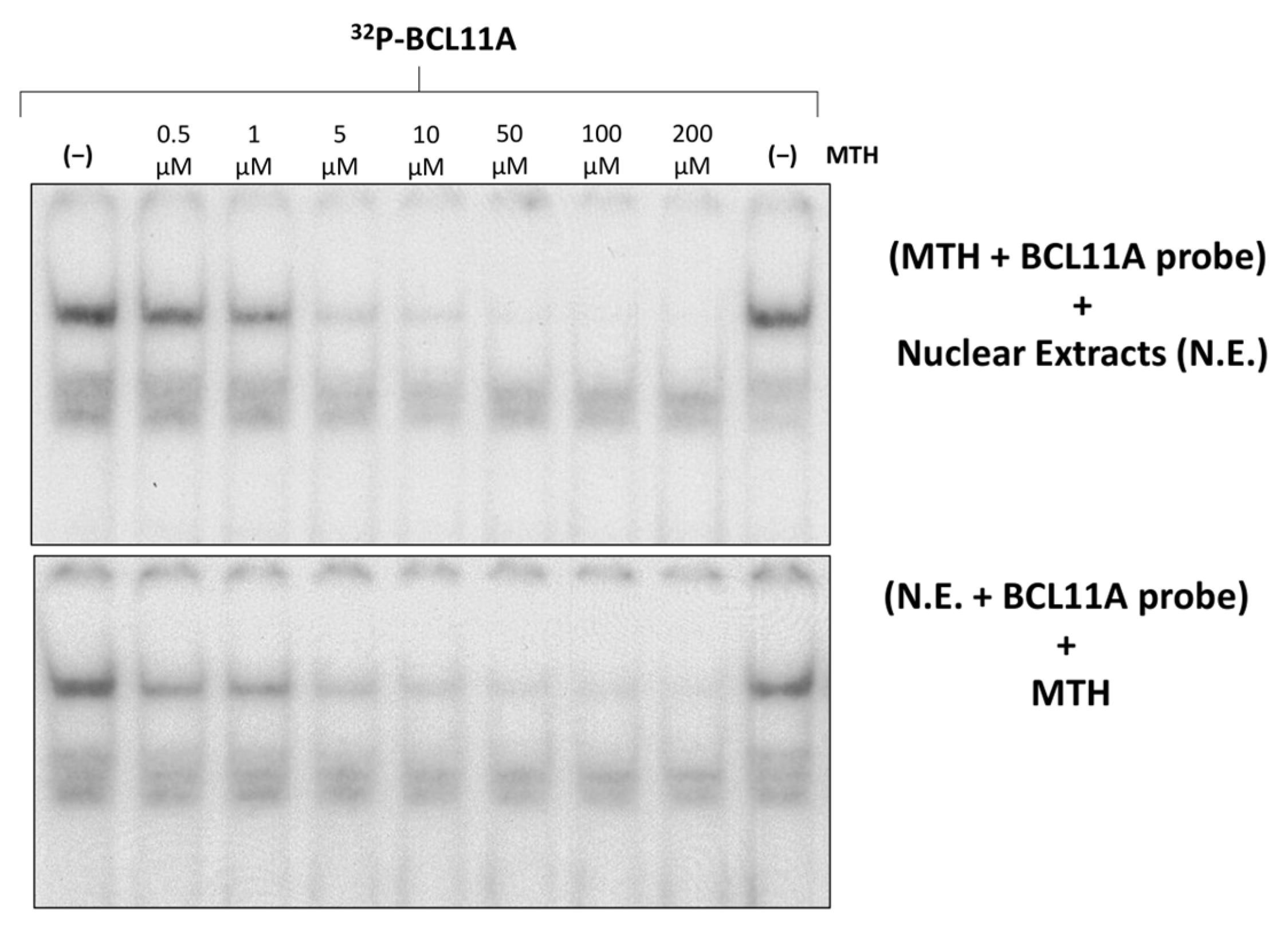
3.3. A Region of the Human γ-Globin Gene Promoter Recognized by Mithramycin Corresponds to a BCL11A Binding Site: A Second Level of MTH Action?
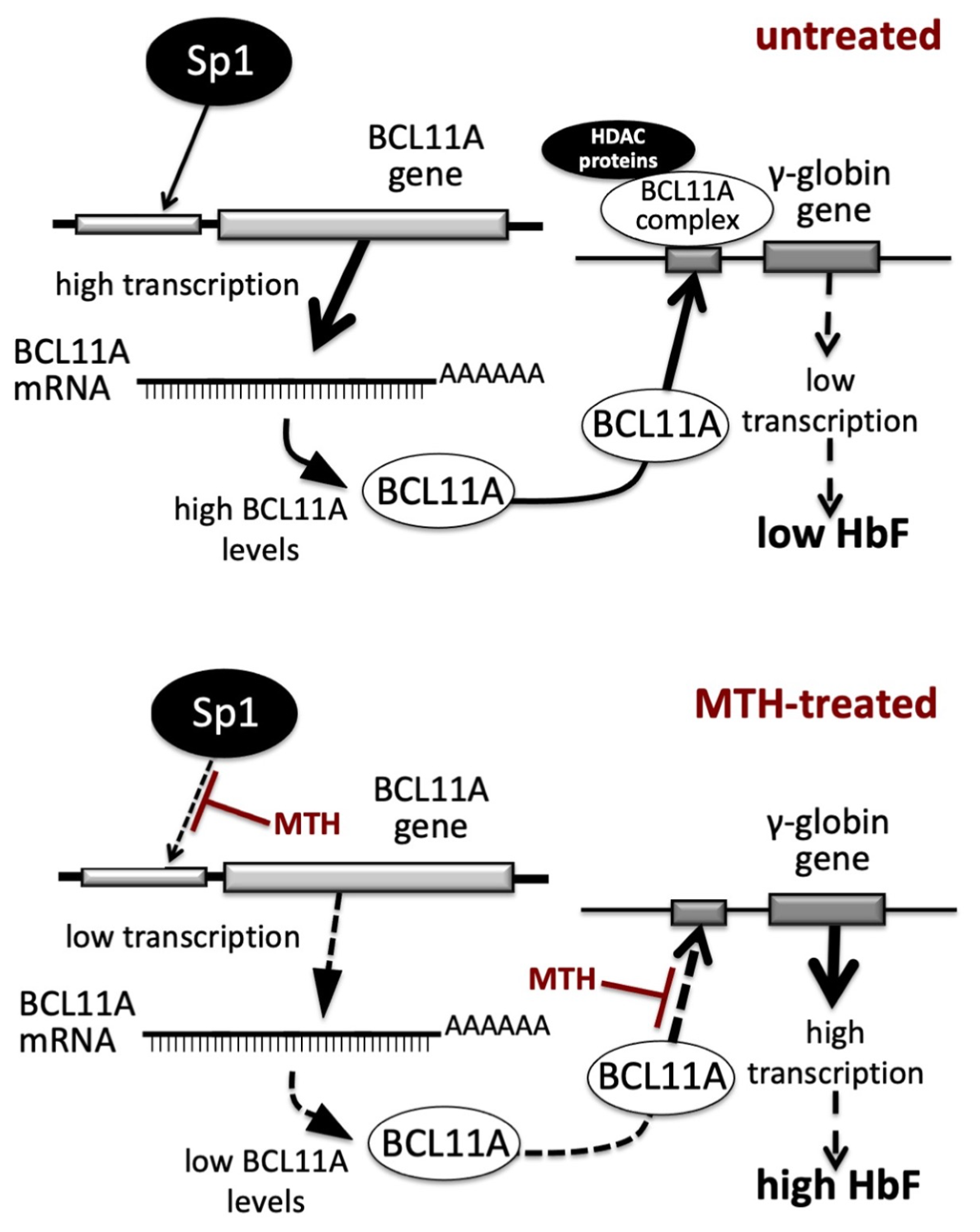
4. Discussion
5. Conclusions and Future Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Weatherall, D.J. Phenotype-genotype relationships in monogenic disease: Lessons from the thalassaemias. Nat. Rev. Genet. 2001, 2, 245–255. [Google Scholar] [CrossRef]
- Origa, R. β-Thalassemia. Genet. Med. 2017, 19, 609–619. [Google Scholar] [CrossRef]
- Lu, H.Y.; Orkin, S.H.; Sankaran, V.G. Fetal Hemoglobin Regulation in β-Thalassemia. Hematol. Oncol. Clin. N. Am. 2023, 37, 301–312. [Google Scholar] [CrossRef]
- Bou-Fakhredin, R.; De Franceschi, L.; Motta, I.; Cappellini, M.D.; Taher, A.T. Pharmacological Induction of Fetal Hemoglobin in β-Thalassemia and Sickle Cell Disease: An Updated Perspective. Pharmaceuticals 2022, 15, 753. [Google Scholar] [CrossRef]
- Sripichai, O.; Fucharoen, S. Fetal hemoglobin regulation in β-thalassemia: Heterogeneity, modifiers and therapeutic approaches. Expert. Rev. Hematol. 2016, 9, 1129–1137. [Google Scholar] [CrossRef]
- Gambari, R.; Fibach, E. Medicinal chemistry of fetal hemoglobin inducers for treatment of β-thalassemia. Curr. Med. Chem. 2007, 14, 199–212. [Google Scholar] [CrossRef] [PubMed]
- Prosdocimi, M.; Zuccato, C.; Cosenza, L.C.; Borgatti, M.; Lampronti, I.; Finotti, A.; Gambari, R. A Rational Approach to Drug Repositioning in β-thalassemia: Induction of Fetal Hemoglobin by Established Drugs. Wellcome Open Res. 2022, 7, 150. [Google Scholar] [CrossRef]
- Pavan, A.R.; Lopes, J.R.; Dos Santos, J.L. The state of the art of fetal hemoglobin-inducing agents. Expert. Opin. Drug Discov. 2022, 17, 1279–1293. [Google Scholar] [CrossRef] [PubMed]
- Feng, R.; Mayuranathan, T.; Huang, P.; Doerfler, P.A.; Li, Y.; Yao, Y.; Zhang, J.; Palmer, L.E.; Mayberry, K.; Christakopoulos, G.E.; et al. Activation of γ-globin expression by hypoxia-inducible factor 1α. Nature 2022, 610, 783–790. [Google Scholar] [CrossRef] [PubMed]
- Yu, L.; Myers, G.; Schneider, E.; Wang, Y.; Mathews, R.; Lim, K.C.; Siemieniak, D.; Tang, V.; Ginsburg, D.; Balbin-Cuesta, G.; et al. Identification of novel γ-globin inducers among all potential erythroid druggable targets. Blood Adv. 2022, 6, 3280–3285. [Google Scholar] [CrossRef]
- Finotti, A.; Gambari, R. Combined approaches for increasing fetal hemoglobin (HbF) and de novo production of adult hemoglobin (HbA) in erythroid cells from β-thalassemia patients: Treatment with HbF inducers and CRISPR-Cas9 based genome editing. Front. Genome Ed. 2023, 5, 1204536. [Google Scholar] [CrossRef]
- Sankaran, V.G.; Xu, J.; Orkin, S.H. Transcriptional silencing of fetal hemoglobin by BCL11A. Ann. N. Y. Acad. Sci. 2010, 1202, 64–68. [Google Scholar] [CrossRef]
- Li, H.; Lin, R.; Li, H.; Ou, R.; Wang, K.; Lin, J.; Li, C. MicroRNA-92a-3p-mediated inhibition of BCL11A upregulates γ-globin expression and inhibits oxidative stress and apoptosis in erythroid precursor cells. Hematology 2022, 27, 1152–1162. [Google Scholar] [CrossRef]
- Gasparello, J.; Fabbri, E.; Bianchi, N.; Breveglieri, G.; Zuccato, C.; Borgatti, M.; Gambari, R.; Finotti, A. BCL11A mRNA Targeting by miR-210: A Possible Network Regulating γ-Globin Gene Expression. Int. J. Mol. Sci. 2017, 18, 2530. [Google Scholar] [CrossRef]
- Mahmoud Ahmed, N.H.; Lai, M.I. The Novel Role of the B-Cell Lymphoma/Leukemia 11A (BCL11A) Gene in β-Thalassaemia Treatment. Cardiovasc. Hematol. Disord. Drug Targets 2023, 22, 226–236. [Google Scholar]
- Zhou, D.; Liu, K.; Sun, C.W.; Pawlik, K.M.; Townes, T.M. KLF1 regulates BCL11A expression and γ- to β-globin gene switching. Nat. Genet. 2010, 42, 742–744. [Google Scholar] [CrossRef]
- Khamphikham, P.; Sripichai, O.; Munkongdee, T.; Fucharoen, S.; Tongsima, S.; Smith, D.R. Genetic variation of Krüppel-like factor 1 (KLF1) and fetal hemoglobin (HbF) levels in β0-thalassemia/HbE disease. Int. J. Hematol. 2018, 107, 297–310. [Google Scholar] [CrossRef] [PubMed]
- Caria, C.A.; Faà, V.; Ristaldi, M.S. Krüppel-Like Factor 1: A Pivotal Gene Regulator in Erythropoiesis. Cells 2022, 11, 3069. [Google Scholar] [CrossRef] [PubMed]
- Gu, Q.; Palani, C.D.; Smith, A.; Li, B.; Amos-Abanyie, E.K.; Ogu, U.; Lu, L.; Pace, B.S.; Starlard-Davenport, A. MicroRNA29B induces fetal hemoglobin via inhibition of the HBG repressor protein MYB in vitro and in humanized sickle cell mice. Front. Med. 2022, 9, 1043686. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.S.; Hong, X.; Wang, G. Induction of endogenous γ-globin gene expression with decoy oligonucleotide targeting Oct-1 transcription factor consensus sequence. J. Hematol. Oncol. 2009, 2, 15. [Google Scholar] [CrossRef] [PubMed]
- Ju, J.; Wang, Y.; Liu, R.; Zhang, Y.; Xu, Z.; Wang, Y.; Wu, Y.; Liu, M.; Cerruti, L.; Zou, F.; et al. Human fetal globin gene expression is regulated by LYAR. Nucleic Acids Res. 2014, 42, 9740–9752. [Google Scholar] [CrossRef] [PubMed]
- Bianchi, N.; Cosenza, L.C.; Lampronti, I.; Finotti, A.; Breveglieri, G.; Zuccato, C.; Fabbri, E.; Marzaro, G.; Chilin, A.; De Angelis, G.; et al. Structural and Functional Insights on an Uncharacterized Aγ-Globin-Gene Polymorphism Present in Four β0-Thalassemia Families with High Fetal Hemoglobin Levels. Mol. Diagn. Ther. 2016, 20, 161–173. [Google Scholar] [CrossRef] [PubMed]
- Breveglieri, G.; Bianchi, N.; Cosenza, L.C.; Gamberini, M.R.; Chiavilli, F.; Zuccato, C.; Montagner, G.; Borgatti, M.; Lampronti, I.; Finotti, A.; et al. An Aγ-globin G->A gene polymorphism associated with β039 thalassemia globin gene and high fetal hemoglobin production. BMC Med. Genet. 2017, 18, 93. [Google Scholar] [CrossRef] [PubMed]
- Chen, D.; Zuo, Y.; Zhang, X.; Ye, Y.; Bao, X.; Huang, H.; Tepakhan, W.; Wang, L.; Ju, J.; Chen, G.; et al. A Genetic Variant Ameliorates β-Thalassemia Severity by Epigenetic-Mediated Elevation of Human Fetal Hemoglobin Expression. Am. J. Hum. Genet. 2017, 101, 130–138. [Google Scholar] [CrossRef]
- Wongborisuth, C.; Chumchuen, S.; Sripichai, O.; Anurathaphan, U.; Sathirapongsasuti, N.; Songdej, D.; Tangprasittipap, A.; Hongeng, S. Down-regulation of the transcriptional repressor ZNF802 (JAZF1) reactivates fetal hemoglobin in β0-thalassemia/HbE. Sci. Rep. 2022, 12, 4952. [Google Scholar] [CrossRef]
- Pule, G.D.; Mowla, S.; Novitzky, N.; Wonkam, A. Hydroxyurea down-regulates BCL11A, KLF-1 and MYB through miRNA-mediated actions to induce γ-globin expression: Implications for new therapeutic approaches of sickle cell disease. Clin. Transl. Med. 2016, 5, 15. [Google Scholar] [CrossRef]
- Mehta, S.; Buyanbat, A.; Kai, Y.; Karayel, O.; Goldman, S.R.; Seruggia, D.; Zhang, K.; Fujiwara, Y.; Donovan, K.A.; Zhu, Q.; et al. Temporal resolution of gene derepression and proteome changes upon PROTAC-mediated degradation of BCL11A protein in erythroid cells. Cell Chem. Biol. 2022, 29, 1273.e8–1287.e8. [Google Scholar] [CrossRef]
- Iftikhar, F.; Khan, M.B.N.; Tehreem, S.; Kanwal, N.; Musharraf, S.G. BCL11A-targeted γ-globin gene induction by triterpenoid glycosides of Fagonia indica: A preclinical scientific validation of indigenous herb for the treatment of β-hemoglobinopathies. Bioorg Chem. 2023, 140, 106768. [Google Scholar] [CrossRef]
- Khan, F.; Ali, H.; Musharraf, S.G. Tenofovir disoproxil fumarate-mediated γ-globin induction is correlated with the suppression of trans-acting factors in CD34+ progenitor cells: A role in the reactivation of fetal hemoglobin. Eur. J. Pharmacol. 2022, 927, 175036. [Google Scholar] [CrossRef]
- Liu, B.; Brendel, C.; Vinjamur, D.S.; Zhou, Y.; Harris, C.; McGuinness, M.; Manis, J.P.; Bauer, D.E.; Xu, H.; Williams, D.A. Development of a double shmiR lentivirus effectively targeting both BCL11A and ZNF410 for enhanced induction of fetal hemoglobin to treat β-hemoglobinopathies. Mol. Ther. 2022, 30, 2693–2708. [Google Scholar] [CrossRef]
- Uda, M.; Galanello, R.; Sanna, S.; Lettre, G.; Sankaran, V.G.; Chen, W.; Usala, G.; Busonero, F.; Maschio, A.; Albai, G.; et al. Genome wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of β-thalassemia. Proc. Natl. Acad. Sci. USA 2008, 105, 1620–1625. [Google Scholar] [CrossRef] [PubMed]
- Shariati, L.; Khanahmad, H.; Salehi, M.; Hejazi, Z.; Rahimmanesh, I.; Tabatabaiefar, M.A.; Modarressi, M.H. Genetic disruption of the KLF1 gene to overexpress the γ-globin gene using the CRISPR/Cas9 system. J. Gene Med. 2016, 18, 294–301. [Google Scholar] [CrossRef] [PubMed]
- Khosravi, M.A.; Abbasalipour, M.; Concordet, J.P.; Berg, J.V.; Zeinali, S.; Arashkia, A.; Azadmanesh, K.; Buch, T.; Karimipoor, M. Targeted deletion of BCL11A gene by CRISPR-Cas9 system for fetal hemoglobin reactivation: A promising approach for gene therapy of β thalassemia disease. Eur. J. Pharmacol. 2019, 854, 398–405. [Google Scholar] [CrossRef]
- Shariati, L.; Rohani, F.; Heidari-Hafshejani, N.; Kouhpayeh, S.; Boshtam, M.; Mirian, M.; Rahimmanesh, I.; Hejazi, Z.; Modarres, M.; Pieper, I.L.; et al. Disruption of SOX6 gene using CRISPR/Cas9 technology for γ-globin reactivation: An approach towards gene therapy of β-thalassemia. J. Cell Biochem. 2018, 119, 9357–9363. [Google Scholar] [CrossRef] [PubMed]
- Fu, B.; Liao, J.; Chen, S.; Li, W.; Wang, Q.; Hu, J.; Yang, F.; Hsiao, S.; Jiang, Y.; Wang, L.; et al. CRISPR-Cas9-mediated gene editing of the BCL11A enhancer for pediatric β0/β0 transfusion-dependent β-thalassemia. Nat. Med. 2022, 28, 1573–1580. [Google Scholar] [CrossRef]
- Han, Y.; Tan, X.; Jin, T.; Zhao, S.; Hu, L.; Zhang, W.; Kurita, R.; Nakamura, Y.; Liu, J.; Li, D.; et al. CRISPR/Cas9-based multiplex genome editing of BCL11A and HBG efficiently induces fetal hemoglobin expression. Eur. J. Pharmacol. 2022, 918, 174788. [Google Scholar] [CrossRef]
- Weber, L.; Frati, G.; Felix, T.; Hardouin, G.; Casini, A.; Wollenschlaeger, C.; Meneghini, V.; Masson, C.; De Cian, A.; Chalumeau, A.; et al. Editing a γ-globin repressor binding site restores fetal hemoglobin synthesis and corrects the sickle cell disease phenotype. Sci. Adv. 2020, 6, eaay9392. [Google Scholar] [CrossRef]
- Frangoul, H.; Altshuler, D.; Cappellini, M.D.; Chen, Y.S.; Domm, J.; Eustace, B.K.; Foell, J.; de la Fuente, J.; Grupp, S.; Handgretinger, R.; et al. CRISPR-Cas9 Gene Editing for Sickle Cell Disease and β-Thalassemia. N. Engl. J. Med. 2021, 384, 252–260. [Google Scholar] [CrossRef]
- Méndez, C.; González-Sabín, J.; Morís, F.; Salas, J.A. Expanding the Chemical Diversity of the Antitumoral Compound Mithramycin by Combinatorial Biosynthesis and Biocatalysis: The Quest for Mithralogs with Improved Therapeutic Window. Planta Med. 2015, 81, 1326–1338. [Google Scholar] [CrossRef]
- Kormanec, J.; Novakova, R.; Csolleiova, D.; Feckova, L.; Rezuchova, B.; Sevcikova, B.; Homerova, D. The antitumor antibiotic mithramycin: New advanced approaches in modification and production. Appl. Microbiol. Biotechnol. 2020, 104, 7701–7721. [Google Scholar] [CrossRef]
- Sastry, M.; Fiala, R.; Patel, D.J. Solution structure of mithramycin dimers bound to partially overlapping sites on DNA. J. Mol. Biol. 1995, 251, 674–689. [Google Scholar] [CrossRef]
- Daniels, E.; Sakakeeny, C. Hypercalcemia: Pathophysiology, Clinical Signs, and Emergent Treatment. J. Am. Anim. Hosp. Assoc. 2015, 51, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Kennedy, B.J.; Torkelson, J.L. Long-term follow-up of stage III testicular carcinoma treated with mithramycin (plicamycin). Med. Pediatr. Oncol. 1995, 24, 327–328. [Google Scholar] [CrossRef] [PubMed]
- Seznec, J.; Silkenstedt, B.; Naumann, U. Therapeutic effects of the Sp1 inhibitor mithramycin A in glioblastoma. J. Neurooncol 2011, 101, 365–377. [Google Scholar] [CrossRef] [PubMed]
- Grohar, P.J.; Glod, J.; Peer, C.J.; Sissung, T.M.; Arnaldez, F.I.; Long, L.; Figg, W.D.; Whitcomb, P.; Helman, L.J.; Widemann, B.C. A phase I/II trial and pharmacokinetic study of mithramycin in children and adults with refractory Ewing sarcoma and EWS-FLI1 fusion transcript. Cancer Chemother. Pharmacol. 2017, 80, 645–652. [Google Scholar] [CrossRef]
- Bianchi, N.; Osti, F.; Rutigliano, C.; Corradini, F.G.; Borsetti, E.; Tomassetti, M.; Mischiati, C.; Feriotto, G.; Gambari, R. The DNA-binding drugs mithramycin and chromomycin are powerful inducers of erythroid differentiation of human K562 cells. Br. J. Haematol. 1999, 104, 258–265. [Google Scholar] [CrossRef]
- Carpenter, M.L.; Cassidy, S.A.; Fox, K.R. Interaction of mithramycin with isolated GC and CG sites. J. Mol. Recognit. 1994, 7, 189–197. [Google Scholar] [CrossRef]
- Fibach, E.; Bianchi, N.; Borgatti, M.; Prus, E.; Gambari, R. Mithramycin induces fetal hemoglobin production in normal and thalassemic human erythroid precursor cells. Blood 2003, 102, 1276–1281. [Google Scholar] [CrossRef]
- Andersson, L.C.; Jokinen, M.; Gahmberg, C.G. Induction of erythroid differentiation in the human leukaemia cell line K562. Nature 1979, 278, 364–365. [Google Scholar] [CrossRef]
- Rutherford, T.R.; Clegg, J.B.; Weatherall, D.J. K562 human leukaemic cells synthesise embryonic haemoglobin in response to haemin. Nature 1979, 280, 164–165. [Google Scholar] [CrossRef]
- Lampronti, I.; Bianchi, N.; Zuccato, C.; Dall’acqua, F.; Vedaldi, D.; Viola, G.; Potenza, R.; Chiavilli, F.; Breveglieri, G.; Borgatti, M.; et al. Increase in γ-globin mRNA content in human erythroid cells treated with angelicin analogs. Int. J. Hematol. 2009, 90, 318–327. [Google Scholar] [CrossRef] [PubMed]
- Borgatti, M.; Lampronti, I.; Romanelli, A.; Pedone, C.; Saviano, M.; Bianchi, N.; Mischiati, C.; Gambari, R. Transcription factor decoy molecules based on a peptide nucleic acid (PNA)-DNA chimera mimicking Sp1binding sites. J. Biol. Chem. 2002, 278, 7500–7509. [Google Scholar] [CrossRef] [PubMed]
- Finotti, A.; Treves, S.; Zorzato, F.; Gambari, R.; Feriotto, G. Upstream stimulatory factors are involved in the P1 promoter directed transcription of the A β H-J-J locus. BMC Mol. Biol. 2008, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Lozzio, B.B.; Lozzio, C.B. Properties of the K562 cell line derived from a patient with chronic myeloid leukemia. Int. J. Cancer 1977, 19, 136. [Google Scholar] [CrossRef]
- Finotti, A.; Gasparello, J.; Breveglieri, G.; Cosenza, L.C.; Montagner, G.; Bresciani, A.; Altamura, S.; Bianchi, N.; Martini, E.; Gallerani, E.; et al. Development and characterization of K562 cell clones expressing BCL11A-XL: Decreased hemoglobin production with fetal hemoglobin inducers and its rescue with mithramycin. Exp. Hematol. 2015, 43, 1062–1071. [Google Scholar] [CrossRef]
- Amoyal, I.; Goldfarb, A.; Fibach, E. Flow cytometric analysis of hydroxyurea effects on fetal hemoglobin production in cultures of β-thalassemia erythroid precursors. Hemoglobin 2003, 27, 77–87. [Google Scholar] [CrossRef]
- Fibach, E. Erythropoiesis In Vitro-A Research and Therapeutic Tool in Thalassemia. J. Clin. Med. 2019, 8, 2124. [Google Scholar] [CrossRef]
- Andrews, N.C.; Faller, D.V. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 1991, 19, 2499. [Google Scholar] [CrossRef]
- Choi, E.S.; Nam, J.S.; Jung, J.Y.; Cho, N.P.; Cho, S.D. Modulation of specificity protein 1 by mithramycin A as a novel therapeutic strategy for cervical cancer. Sci. Rep. 2014, 4, 7162. [Google Scholar] [CrossRef]
- Sleiman, S.F.; Langley, B.C.; Basso, M.; Berlin, J.; Xia, L.; Payappilly, J.B.; Kharel, M.K.; Guo, H.; Marsh, J.L.; Thompson, L.M.; et al. Mithramycin is a gene-selective Sp1 inhibitor that identifies a biological intersection between cancer and neurodegeneration. J. Neurosci. 2011, 31, 6858–6870. [Google Scholar] [CrossRef]
- Finotti, A.; Bianchi, N.; Fabbri, E.; Borgatti, M.; Breveglieri, G.; Gasparello, J.; Gambari, R. Erythroid induction of K562 cells treated with mithramycin is associated with inhibition of raptor gene transcription and mammalian target of rapamycin complex 1 (mTORC1) functions. Pharmacol. Res. 2015, 91, 57–68. [Google Scholar] [CrossRef] [PubMed]
- Miura, A.; Odahara, N.; Tominaga, A.; Inoue, K.; Kambe, Y.; Kurihara, T.; Miyata, A. Regulatory mechanism of PAC1 gene expression via Sp1 by nerve growth factor in PC12 cells. FEBS Lett. 2012, 586, 1731–1735. [Google Scholar] [CrossRef] [PubMed]
- Reamon-Buettner, S.M.; Borlak, J. Epigenetic silencing of cell adhesion molecule 1 in different cancer progenitor cells of transgenic c-Myc and c-Raf mouse lung tumors. Cancer Res. 2008, 68, 7587–7596. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.A.; Chae, J.I.; Shim, J.H. Natural diterpenes from coffee, cafestol and kahweol induce apoptosis through regulation of specificity protein 1 expression in human malignant pleural mesothelioma. J. Biomed. Sci. 2012, 19, 60. [Google Scholar] [CrossRef]
- Lee, T.J.; Jung, E.M.; Lee, J.T.; Kim, S.; Park, J.W.; Choi, K.S.; Kwon, T.K. Mithramycin A sensitizes cancer cells to TRAIL-mediated apoptosis by down-regulation of XIAP gene promoter through Sp1 sites. Mol. Cancer Ther. 2006, 5, 2737–2746. [Google Scholar] [CrossRef]
- Eisermann, K.; Broderick, C.J.; Bazarov, A.; Moazam, M.M.; Fraizer, G.C. Androgen up-regulates vascular endothelial growth factor expression in prostate cancer cells via an Sp1 binding site. Mol. Cancer 2013, 12, 7. [Google Scholar] [CrossRef]
- Zuccato, C.; Bianchi, N.; Borgatti, M.; Lampronti, I.; Massei, F.; Favre, C.; Gambari, R. Everolimus is a potent inducer of erythroid differentiation and γ-globin gene expression in human erythroid cells. Acta Haematol. 2007, 117, 168–176. [Google Scholar] [CrossRef]
- Zuccato, C.; Cosenza, L.C.; Zurlo, M.; Lampronti, I.; Borgatti, M.; Scapoli, C.; Gambari, R.; Finotti, A. Treatment of Erythroid Precursor Cells from β-Thalassemia Patients with Cinchona Alkaloids: Induction of Fetal Hemoglobin Production. Int. J. Mol. Sci. 2021, 22, 13433. [Google Scholar] [CrossRef]
- Kurita, R.; Suda, N.; Sudo, K.; Miharada, K.; Hiroyama, T.; Miyoshi, H.; Tani, K.; Nakamura, Y. Establishment of immortalized human erythroid progenitor cell lines able to produce enucleated red blood cells. PLoS ONE 2013, 8, e59890. [Google Scholar] [CrossRef] [PubMed]
- Papasavva, P.L.; Papaioannou, N.Y.; Patsali, P.; Kurita, R.; Nakamura, Y.; Sitarou, M.; Christou, S.; Kleanthous, M.; Lederer, C.W. Distinct miRNA Signatures and Networks Discern Fetal from Adult Erythroid Differentiation and Primary from Immortalized Erythroid Cells. Int. J. Mol. Sci. 2021, 22, 3626. [Google Scholar] [CrossRef] [PubMed]
- Sissung, T.M.; Huang, P.A.; Hauke, R.J.; McCrea, E.M.; Peer, C.J.; Barbier, R.H.; Strope, J.D.; Ley, A.M.; Zhang, M.; Hong, J.A.; et al. Severe Hepatotoxicity of Mithramycin Therapy Caused by Altered Expression of Hepatocellular Bile Transporters. Mol. Pharmacol. 2019, 96, 158–167. [Google Scholar] [CrossRef]
- Albertini, V.; Jain, A.; Vignati, S.; Napoli, S.; Rinaldi, A.; Kwee, I.N.; Nur-e-Alam, M.; Bergant, J.; Bertoni, F.; Carbone, G.M.; et al. Novel GC-rich DNA-binding compound produced by a genetically engineered mutant of the mithramycin producer Streptomyces argillaceus exhibits improved transcriptional repressor activity: Implications for cancer therapy. Nucleic Acids Res. 2006, 34, 1721–1734. [Google Scholar] [CrossRef] [PubMed]
- Núñez, L.E.; Nybo, S.E.; González-Sabín, J.; Pérez, M.; Menéndez, N.; Braña, A.F.; Shaaban, K.A.; He, M.; Morís, F.; Salas, J.A.; et al. A novel mithramycin analogue with high antitumor activity and less toxicity generated by combinatorial biosynthesis. J. Med. Chem. 2012, 55, 5813–5825. [Google Scholar] [CrossRef] [PubMed]
- Fernández-Guizán, A.; Mansilla, S.; Barceló, F.; Vizcaíno, C.; Núñez, L.E.; Morís, F.; González, S.; Portugal, J. The activity of a novel mithramycin analog is related to its binding to DNA, cellular accumulation, and inhibition of Sp1-driven gene transcription. Chem. Biol. Interact. 2014, 219C, 123–132. [Google Scholar] [CrossRef] [PubMed]
- Estupiñán, Ó.; Niza, E.; Bravo, I.; Rey, V.; Tornín, J.; Gallego, B.; Clemente-Casares, P.; Moris, F.; Ocaña, A.; Blanco-Lorenzo, V.; et al. Mithramycin delivery systems to develop effective therapies in sarcomas. J. Nanobiotechnol. 2021, 19, 267. [Google Scholar] [CrossRef] [PubMed]
- Estupiñán, Ó.; Rendueles, C.; Suárez, P.; Rey, V.; Murillo, D.; Morís, F.; Gutiérrez, G.; Blanco-López, M.D.C.; Matos, M.; Rodríguez, R. Nano-Encapsulation of Mithramycin in Transfersomes and Polymeric Micelles for the Treatment of Sarcomas. J. Clin. Med. 2021, 10, 1358. [Google Scholar] [CrossRef]
- Capretto, L.; Mazzitelli, S.; Brognara, E.; Lampronti, I.; Carugo, D.; Hill, M.; Zhang, X.; Gambari, R.; Nastruzzi, C. Mithramycin encapsulated in polymeric micelles by microfluidic technology as novel therapeutic protocol for β-thalassemia. Int. J. Nanomed. 2012, 7, 307–324. [Google Scholar]
- Radmilovic, M.; Zukic, B.; Petrovic, M.S.; Bartsakoulia, M.; Stankovic, B.; Kotur, N.; Dokmanovic, L.; Georgitsi, M.; Patrinos, G.P.; Pavlovic, S. Functional analysis of a novel KLF1 gene promoter variation associated with hereditary persistence of fetal hemoglobin. Ann. Hematol. 2013, 92, 53–58. [Google Scholar] [CrossRef]
- Sala, A.; Saitta, B.; De Luca, P.; Cervellera, M.N.; Casella, I.; Lewis, R.E.; Watson, R.; Peschle, C. B-MYB transactivates its own promoter through SP1-binding sites. Oncogene 1999, 18, 1333–1339. [Google Scholar] [CrossRef]
- Sun, J.; Muto, A.; Hoshino, H.; Kobayashi, A.; Nishimura, S.; Yamamoto, M.; Hayashi, N.; Ito, E.; Igarashi, K. The promoter of mouse transcription repressor bach1 is regulated by Sp1 and trans-activated by Bach1. J. Biochem. 2001, 130, 385–392. [Google Scholar] [CrossRef]
- Zu, X.; Yu, L.; Sun, Q.; Liu, F.; Wang, J.; Xie, Z.; Wang, Y.; Xu, W.; Jiang, Y. SP1 enhances Zbtb7A gene expression via direct binding to GC box in HePG2 cells. BMC Res. Notes 2009, 2, 175. [Google Scholar] [CrossRef] [PubMed]
- Lu, K.; Wang, Q.; Hao, L.; Wei, G.; Wang, T.; Lu, W.W.; Xiao, G.; Tong, L.; Zhao, X.; Chen, D. MiR-204 ameliorates osteoarthritis pain by inhibiting SP1-LRP1 signaling and blocking neuro-cartilage interaction. Bioact. Mater. 2023, 26, 425–436. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Yang, Y.; Ji, Y.; Wang, G.; Xu, W.; Wang, B.; Guo, H.; Ren, J.; Yan, J.; Wang, N. MiR-135a-5p/SP1 Axis Regulates Spinal Astrocyte Proliferation and Migration. Neuroscience 2023, 515, 12–24. [Google Scholar] [CrossRef] [PubMed]
- Nalla, L.V.; Khairnar, A. Empagliflozin mediated miR-128-3p upregulation promotes differentiation of hypoxic cancer stem-like cells in breast cancer. Eur. J. Pharmacol. 2023, 943, 175565. [Google Scholar] [CrossRef]
- Ma, R.; Zhang, D.; Song, Y.; Kong, J.; Mu, C.; Shen, P.; Gui, W. miR-335-5p regulates the proliferation, migration and phenotypic switching of vascular smooth muscle cells in aortic dissection by directly regulating SP1. Acta Biochim. Biophys. Sin. 2022, 54, 961–973. [Google Scholar] [CrossRef] [PubMed]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Finotti, A.; Gasparello, J.; Zuccato, C.; Cosenza, L.C.; Fabbri, E.; Bianchi, N.; Gambari, R. Effects of Mithramycin on BCL11A Gene Expression and on the Interaction of the BCL11A Transcriptional Complex to γ-Globin Gene Promoter Sequences. Genes 2023, 14, 1927. https://doi.org/10.3390/genes14101927
Finotti A, Gasparello J, Zuccato C, Cosenza LC, Fabbri E, Bianchi N, Gambari R. Effects of Mithramycin on BCL11A Gene Expression and on the Interaction of the BCL11A Transcriptional Complex to γ-Globin Gene Promoter Sequences. Genes. 2023; 14(10):1927. https://doi.org/10.3390/genes14101927
Chicago/Turabian StyleFinotti, Alessia, Jessica Gasparello, Cristina Zuccato, Lucia Carmela Cosenza, Enrica Fabbri, Nicoletta Bianchi, and Roberto Gambari. 2023. "Effects of Mithramycin on BCL11A Gene Expression and on the Interaction of the BCL11A Transcriptional Complex to γ-Globin Gene Promoter Sequences" Genes 14, no. 10: 1927. https://doi.org/10.3390/genes14101927
APA StyleFinotti, A., Gasparello, J., Zuccato, C., Cosenza, L. C., Fabbri, E., Bianchi, N., & Gambari, R. (2023). Effects of Mithramycin on BCL11A Gene Expression and on the Interaction of the BCL11A Transcriptional Complex to γ-Globin Gene Promoter Sequences. Genes, 14(10), 1927. https://doi.org/10.3390/genes14101927







