Identification of the Porcine Vascular Endothelial Cell-Specific Promoter ESAM1.0 Using Transcriptome Analysis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cells and Transfection
2.2. RNA Sequencing
2.3. DEG Analysis
2.4. Real-Time Quantitative PCR
2.5. Promoter Isolation and Vector Construction
2.6. Luciferase Assay
2.7. Statistical Analysis
3. Results
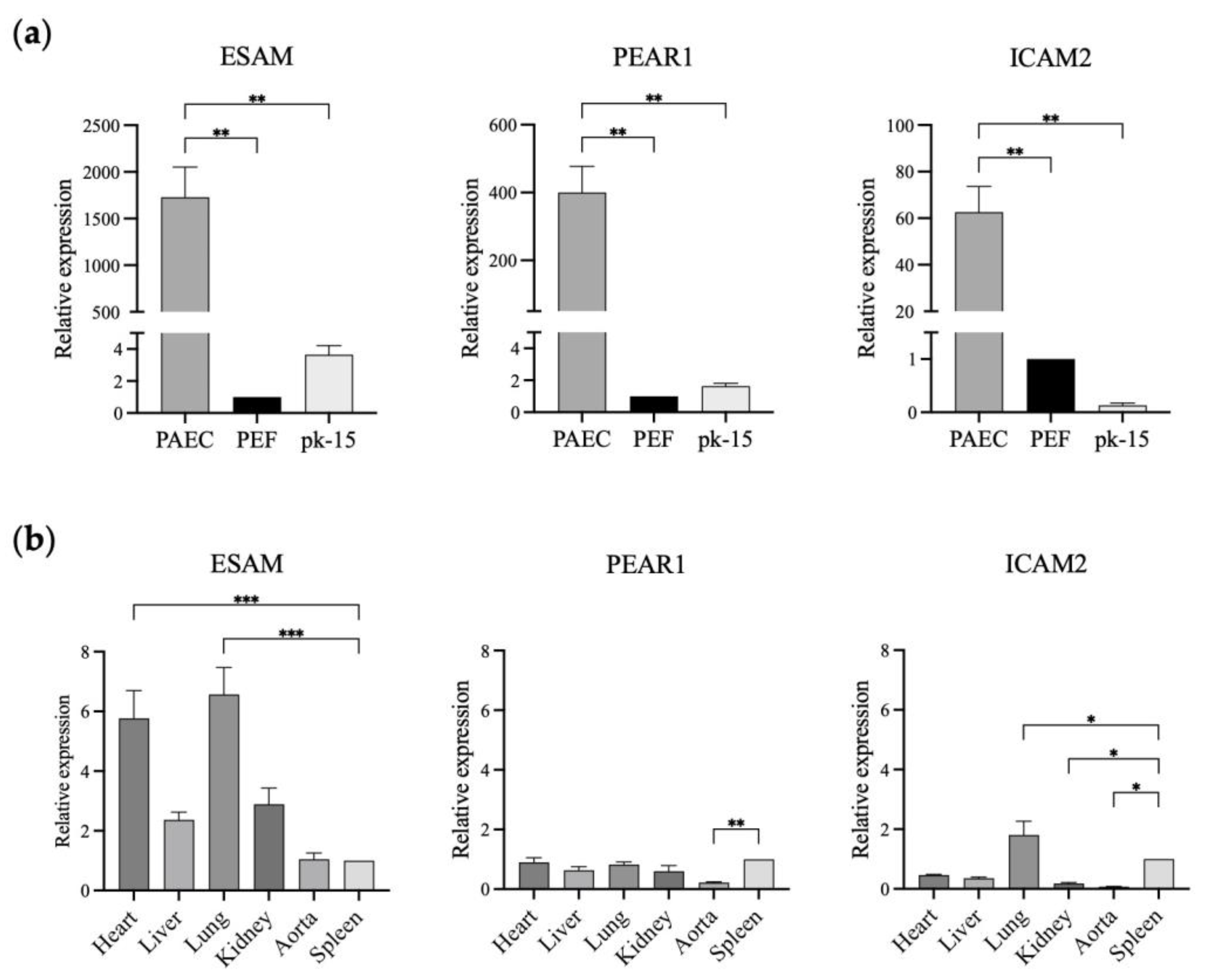
3.1. Identification of Genes with Porcine Endothelial Cell-Specific Expression
3.2. Potential of ESAM as an Endothelial Cell-Specific Promoter
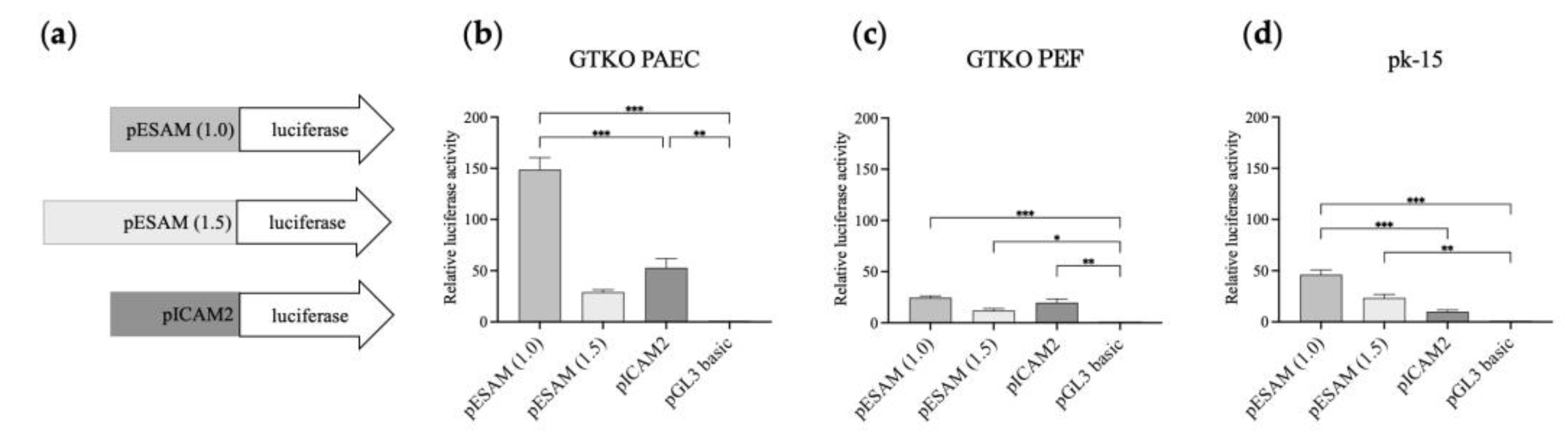
3.3. Isolation of the Putative Promoter Region of Porcine ESAM
3.4. Distinct Activity of the pESAM1.0 Promoter Leads to Specific Expression in Porcine Vascular Endothelial Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tritschler, H.; Fischer, K.; Seissler, J.; Fiedler, J.; Halbgebauer, R.; Huber-Lang, M.; Schnieke, A.; Brenner, R.E. New Insights into Xenotransplantation for Cartilage Repair: Porcine Multi-Genetically Modified Chondrocytes as a Promising Cell Source. Cells 2021, 10, 2152. [Google Scholar] [CrossRef] [PubMed]
- Lu, T.-Y.; Xu, X.-L.; Du, X.-G.; Wei, J.-H.; Yu, J.-N.; Deng, S.-L.; Qin, C. Advances in Innate Immunity to Overcome Immune Rejection during Xenotransplantation. Cells 2022, 11, 3865. [Google Scholar] [CrossRef]
- Aschheim, K.; DeFrancesco, L. Xenotransplantation: How close are we? Nat. Biotechnol. 2023, 41, 452–460. [Google Scholar] [CrossRef]
- Hryhorowicz, M.; Lipinski, D.; Hryhorowicz, S.; Nowak-Terpilowska, A.; Ryczek, N.; Zeyland, J. Application of Genetically Engineered Pigs in Biomedical Research. Genes 2020, 11, 670. [Google Scholar] [CrossRef]
- Good, A.H.; Cooper, D.K.; Malcolm, A.J.; Ippolito, R.M.; Koren, E.; Neethling, F.A.; Ye, Y.; Zuhdi, N.; Lamontagne, L.R. Identification of carbohydrate structures that bind human antiporcine antibodies: Implications for discordant xenografting in humans. Transplant. Proc. 1992, 24, 559–562. [Google Scholar] [PubMed]
- Cooper, D.K.; Good, A.H.; Koren, E.; Oriol, R.; Malcolm, A.J.; Ippolito, R.M.; Neethling, F.A.; Ye, Y.; Romano, E.; Zuhdi, N. Identification of α-galactosyl and other carbohydrate epitopes that are bound by human anti-pig antibodies: Relevance to discordant xenografting in man. Transpl. Immunol. 1993, 1, 198–205. [Google Scholar] [CrossRef] [PubMed]
- Lai, L.; Kolber-Simonds, D.; Park, K.W.; Cheong, H.T.; Greenstein, J.L.; Im, G.S.; Samuel, M.; Bonk, A.; Rieke, A.; Day, B.N.; et al. Production of α-1,3-galactosyltransferase knockout pigs by nuclear transfer cloning. Science 2002, 295, 1089–1092. [Google Scholar] [CrossRef]
- Dai, Y.; Vaught, T.D.; Boone, J.; Chen, S.-H.; Phelps, C.J.; Ball, S.; Monahan, J.A.; Jobst, P.M.; McCreath, K.J.; Lamborn, A.E.; et al. Targeted disruption of the α1,3-galactosyltransferase gene in cloned pigs. Nat. Biotechnol. 2002, 20, 251–255. [Google Scholar] [CrossRef]
- Ahn, K.S.; Kim, Y.J.; Kim, M.; Lee, B.H.; Heo, S.Y.; Kang, M.-J.; Kang, Y.-K.; Lee, J.W.; Lee, K.-K.; Kim, J.-H.; et al. Resurrection of an alpha-1,3-galactosyltransferase gene-targeted miniature pig by recloning using postmortem ear skin fibroblasts. Theriogenology 2011, 75, 933–939. [Google Scholar] [CrossRef]
- Ryczek, N.; Hryhorowicz, M.; Zeyland, J.; Lipinski, D.; Slomski, R. CRISPR/Cas Technology in Pig-to-Human Xenotransplantation Research. Int. J. Mol. Sci. 2021, 22, 3196. [Google Scholar] [CrossRef]
- You, W.; Li, M.; Qi, Y.; Wang, Y.; Chen, Y.; Liu, Y.; Li, L.; Ouyang, H.; Pang, D. CRISPR/Cas9-Mediated Specific Integration of Fat-1 and IGF-1 at the pRosa26 Locus. Genes 2021, 12, 1027. [Google Scholar] [CrossRef]
- Yue, Y.; Xu, W.; Kan, Y.; Zhao, H.Y.; Zhou, Y.; Song, X.; Wu, J.; Xiong, J.; Goswami, D.; Yang, M.; et al. Extensive germline genome engineering in pigs. Nat. Biomed. Eng. 2021, 5, 134–143. [Google Scholar] [CrossRef]
- Aneesh Kumar, A.; Ajith Kumar, G.S.; Satheesh, G.; Surendran, A.; Chandran, M.; Kartha, C.C.; Jaleel, A. Proteomics Analysis Reveals Diverse Molecular Characteristics between Endocardial and Aortic-Valvular Endothelium. Genes 2021, 12, 1005. [Google Scholar] [CrossRef]
- Banz, Y.; Rieben, R. Endothelial cell protection in xenotransplantation: Looking after a key player in rejection. Xenotransplantation 2006, 13, 19–30. [Google Scholar] [CrossRef] [PubMed]
- Iwase, H.; Liu, H.; Wijkstrom, M.; Zhou, H.; Singh, J.; Hara, H.; Ezzelarab, M.; Long, C.; Klein, E.; Wagner, R.; et al. Pig kidney graft survival in a baboon for 136 days: Longest life-supporting organ graft survival to date. Xenotransplantation 2015, 22, 302–309. [Google Scholar] [CrossRef]
- Zou, L.; Zhang, Y.; He, Y.; Yu, H.; Chen, J.; Liu, D.; Lin, S.; Gao, M.; Zhong, G.; Lei, W.; et al. Selective Germline Genome Edited Pigs and Their Long Immune Tolerance in Non Human Primates. bioRxiv 2020. [Google Scholar] [CrossRef]
- Chen, T.; Wang, J.; Xue, B.; Kong, Q.; Liu, Z.; Yu, B. Identification and characterization of a novel porcine endothelial cell-specific Tie1 promoter. Xenotransplantation 2013, 20, 438–448. [Google Scholar] [CrossRef]
- Lee, S.C.; Lee, H.; Oh, K.B.; Hwang, I.S.; Yang, H.; Park, M.R.; Ock, S.A.; Woo, J.S.; Im, G.S.; Hwang, S. Production and Breeding of Transgenic Cloned Pigs Expressing Human CD73. Dev. Reprod. 2017, 21, 157–165. [Google Scholar] [CrossRef]
- Ock, S.A.; Lim, M.; Kim, Y.; Ullah, I.; Shin, Y.; Kim, Y.; Oh, K.B.; Hwang, S.; Hur, T.-Y.; Lee, S.; et al. Isolation and Culture of Purified Aortic Endothelial Cells Derived from Alpha 1, 3-Galactosyltransferase-Deficient Pigs. J. Embryo Transf. 2017, 32, 87–94. [Google Scholar] [CrossRef]
- Bella, K.; Ko, N.; Hwang, S.-s.; Im, G.-S.; Kim, D.-H.; Park, J.-K.; Ryoo, Z.Y.; Oh, K.B. Growth Factors Supplementation in Culture Medium Leads to Active Proliferation of Porcine Fibroblasts. J. Reprod. Dev. Biol. 2011, 35, 301–306. [Google Scholar]
- Sun, W.S.; Yang, H.; No, J.G.; Lee, H.; Lee, N.; Lee, M.; Kang, M.J.; Oh, K.B. Select Porcine Elongation Factor 1alpha Sequences Mediate Stable High-Level and Upregulated Expression of Heterologous Genes in Porcine Cells in Response to Primate Serum. Genes 2021, 12, 1046. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2009, 26, 139–140. [Google Scholar] [CrossRef] [PubMed]
- Raudvere, U.; Kolberg, L.; Kuzmin, I.; Arak, T.; Adler, P.; Peterson, H.; Vilo, J. g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 2019, 47, W191–W198. [Google Scholar] [CrossRef]
- Li, L.C.; Dahiya, R. MethPrimer: Designing primers for methylation PCRs. Bioinformatics 2002, 18, 1427–1431. [Google Scholar] [CrossRef]
- Sudo, T.; Yokota, T.; Oritani, K.; Satoh, Y.; Sugiyama, T.; Ishida, T.; Shibayama, H.; Ezoe, S.; Fujita, N.; Tanaka, H.; et al. The Endothelial Antigen ESAM Monitors Hematopoietic Stem Cell Status between Quiescence and Self-Renewal. J. Immunol. 2012, 189, 200–210. [Google Scholar] [CrossRef] [PubMed]
- Arabi, T.Z.; Sabbah, B.N.; Lerman, A.; Zhu, X.Y.; Lerman, L.O. Xenotransplantation: Current Challenges and Emerging Solutions. Cell Transplant. 2023, 32, 9636897221148771. [Google Scholar] [CrossRef]
- Griffith, B.P.; Goerlich, C.E.; Singh, A.K.; Rothblatt, M.; Lau, C.L.; Shah, A.; Lorber, M.; Grazioli, A.; Saharia, K.K.; Hong, S.N.; et al. Genetically Modified Porcine-to-Human Cardiac Xenotransplantation. N. Engl. J. Med. 2022, 387, 35–44. [Google Scholar] [CrossRef]
- Kummer, L.; Zaradzki, M.; Vijayan, V.; Arif, R.; Weigand, M.A.; Immenschuh, S.; Wagner, A.H.; Larmann, J. Vascular Signaling in Allogenic Solid Organ Transplantation—The Role of Endothelial Cells. Front. Physiol. 2020, 11, 443. [Google Scholar] [CrossRef]
- Chen, J.; Gao, H.; Chen, L.; Wang, X.; Song, Z.; Cooper, D.K.C.; Qu, Z.; Cai, Z.; Mou, L. A potential role of TLR2 in xenograft rejection of porcine iliac endothelial cells: An in vitro study. Xenotransplantation 2019, 26, e12526. [Google Scholar] [CrossRef] [PubMed]
- Ueda, T.; Yokota, T.; Okuzaki, D.; Uno, Y.; Mashimo, T.; Kubota, Y.; Sudo, T.; Ishibashi, T.; Shingai, Y.; Doi, Y.; et al. Endothelial Cell-Selective Adhesion Molecule Contributes to the Development of Definitive Hematopoiesis in the Fetal Liver. Stem Cell Rep. 2019, 13, 992–1005. [Google Scholar] [CrossRef] [PubMed]
- Cain, J.A.; Montibus, B.; Oakey, R.J. Intragenic CpG Islands and Their Impact on Gene Regulation. Front. Cell Dev. Biol. 2022, 10, 832348. [Google Scholar] [CrossRef]
- Kaluscha, S.; Domcke, S.; Wirbelauer, C.; Stadler, M.B.; Durdu, S.; Burger, L.; Schübeler, D. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat. Genet. 2022, 54, 1895–1906. [Google Scholar] [CrossRef]
- Mohamed Sa’dom, S.A.F.; Raikundalia, S.; Shamsuddin, S.; See Too, W.C.; Few, L.L. DNA Methylation of Human Choline Kinase Alpha Promoter-Associated CpG Islands in MCF-7 Cells. Genes. 2021, 12, 853. [Google Scholar] [CrossRef] [PubMed]


| Primer | Sequence (5′→3′) | |
|---|---|---|
| ACTB | (F) | CATCACCATCGGCAACGAGC |
| (R) | TAGAGGTCCTTGCGGATGTC | |
| 18S rRNA | (F) | AAAGGAATTGACGGAAGGGC |
| (R) | CCCACGGAATCGAGAAAGAG | |
| ESAM | (F) | GCAAGGGGAGGTGTCTTCAA |
| (R) | TGTGGCTCCACCGATGTATG | |
| PEAR1 | (F) | ATCCCCGAGAATGGCAACTG |
| (R) | TTGCAGGGTACACAGCGTTT | |
| ICAM2 | (F) | ATCATCATCGCGGTCGTGTC |
| (R) | CATGTAGGAACCTGTCCGCC | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, S.E.; Sun, W.-S.; Oh, M.; Lee, S.; No, J.-G.; Lee, H.; Lee, P.; Oh, K.B. Identification of the Porcine Vascular Endothelial Cell-Specific Promoter ESAM1.0 Using Transcriptome Analysis. Genes 2023, 14, 1928. https://doi.org/10.3390/genes14101928
Kim SE, Sun W-S, Oh M, Lee S, No J-G, Lee H, Lee P, Oh KB. Identification of the Porcine Vascular Endothelial Cell-Specific Promoter ESAM1.0 Using Transcriptome Analysis. Genes. 2023; 14(10):1928. https://doi.org/10.3390/genes14101928
Chicago/Turabian StyleKim, Sang Eun, Wu-Sheng Sun, Miae Oh, Seunghoon Lee, Jin-Gu No, Haesun Lee, Poongyeon Lee, and Keon Bong Oh. 2023. "Identification of the Porcine Vascular Endothelial Cell-Specific Promoter ESAM1.0 Using Transcriptome Analysis" Genes 14, no. 10: 1928. https://doi.org/10.3390/genes14101928
APA StyleKim, S. E., Sun, W.-S., Oh, M., Lee, S., No, J.-G., Lee, H., Lee, P., & Oh, K. B. (2023). Identification of the Porcine Vascular Endothelial Cell-Specific Promoter ESAM1.0 Using Transcriptome Analysis. Genes, 14(10), 1928. https://doi.org/10.3390/genes14101928





