Genome-Wide Association Analysis across Endophenotypes in Alzheimer’s Disease: Main Effects and Disease Stage-Specific Interactions
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Genotyping and Imputation
2.3. Selected Phenotypes
2.4. Genetic Association Analysis
CN MRI Field Strength))
2.5. SNP × DX Interaction Analysis
2.6. Functional Analysis
3. Results
3.1. Genome-Wide Association Results
3.2. Known AD-Associated Genetic Regions
3.3. SNPxDiagnosis Results
3.4. Functional Results
4. Discussion
| Gene Region | Cross-Sectional | SNP × DX | Previous Link to AD | Potential Biological Relevance in AD |
|---|---|---|---|---|
| APOE | ● | Major AD Risk locus; Multiple GWAS | Lipid Transporter involved in CNS Maintenance | |
| XKR3 | ● | - | Apoptotic signaling | |
| SRSF10 | ● | - | Alternative splicing; enhanced lipogenesis | |
| RCAN3 | ● | Differentially expressed in AD [26] | Immune, T Cell development | |
| HLA-B | ● | Multiple GWAS [33,34] | Immune | |
| KIAA1671 | ● | AD Multi-Omic Weighted GWAS [53] | Immune | |
| HLA-DQB1 | ● | Multiple GWAS [20,30,31,32] | Immune | |
| HLA-DQA1 | ● | Multiple GWAS [20,30,31,32] | Immune | |
| ZNF826P | ● | - | Unknown | |
| BACH2 | ● | Upregulated in β-amyloid-treated SH-SY5Y neuroblastoma cells [48] | Apoptosis; Nuclear import of actin; Transcriptional regulator of NF-κB | |
| EP300 | ● | Associated with altered bile acids in AD; shown to be involved in AD regulatory network [49] | Mediates cAMP gene regulation | |
| PACRG-AS1 | ● | - | Lewy bodies; Heat shock, apoptosis |
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Adam, N.; Gerard, S. Genomic variants, genes, and pathways of Alzheimer’s disease: An overview. Am. J. Med. Genet. Part B 2017, 174, 5–26. [Google Scholar] [CrossRef]
- Karch, C.M.; Goate, A.M. Alzheimer’s disease risk genes and mechanisms of disease pathogenesis. Biol. Psychiatry 2015, 77, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Jack, C.R., Jr.; Bennett, D.A.; Blennow, K.; Carrillo, M.C.; Dunn, B.; Haeberlein, S.B.; Holtzman, D.M.; Jagust, W.; Jessen, F.; Karlawish, J.; et al. NIA-AA Research Framework: Toward a biological definition of Alzheimer’s disease. Alzheimers Dement. 2018, 14, 535–562. [Google Scholar] [CrossRef] [PubMed]
- Almasy, L.; Blangero, J. Endophenotypes as quantitative risk factors for psychiatric disease: Rationale and study design. Am. J. Med. Genet. 2001, 105, 42–44. [Google Scholar] [CrossRef] [PubMed]
- Saykin, A.J.; Shen, L.; Yao, X.; Kim, S.; Nho, K.; Risacher, S.L.; Ramanan, V.K.; Foroud, T.M.; Faber, K.M.; Sarwar, N.; et al. Genetic Studies of Quantitative MCI and AD Phenotypes in ADNI: Progress, Opportunities, and Plans. Alzheimers Dement. 2015, 11, 792–814. [Google Scholar] [CrossRef] [PubMed]
- Farrer, L.A.; Cupples, L.A.; Duijn, C.M.V.; Connor-Lacke, L.; Kiely, D.K.; Growdon, J.H. Rate of Progression of Alzheimer’s Disease Is Associated With Genetic Risk. Arch. Neurol. 2019, 52, 918–923. [Google Scholar] [CrossRef]
- Thalhauser, C.J.; Komarova, N.L. Alzheimer’s disease: Rapid and slow progression. J. R. Soc. Interface 2012, 9, 119–126. [Google Scholar] [CrossRef] [PubMed]
- Saykin, A.J.; Shen, L.; Foroud, T.M.; Potkin, S.G.; Swaminathan, S.; Kim, S.; Risacher, S.L.; Nho, K.; Huentelman, M.J.; Craig, D.W.; et al. Alzheimer’s Disease Neuroimaging Initiative biomarkers as quantitative phenotypes: Genetics core aims, progress, and plans. Alzheimers Dement. 2010, 6, 265–273. [Google Scholar] [CrossRef]
- Weiner, M.W.; Aisen, P.S.; Jack, C.R., Jr.; Jagust, W.J.; Trojanowski, J.Q.; Shaw, L.; Saykin, A.J.; Morris, J.C.; Cairns, N.; Beckett, L.A.; et al. The Alzheimer’s disease neuroimaging initiative: Progress report and future plans. Alzheimers Dement. 2010, 6, 202–211.e207. [Google Scholar] [CrossRef]
- Park, Y.H.; Hodges, A.; Risacher, S.L.; Lin, K.; Jang, J.-W.; Ahn, S.; Kim, S.; Lovestone, S.; Simmons, A.; Weiner, M.W.; et al. Dysregulated Fc γ receptor-mediated phagocytosis pathway in Alzheimer’s disease: Network-based gene expression analysis. Neurobiol. Aging 2020, 88, 24–32. [Google Scholar] [CrossRef]
- Hellwege, J.N.; Keaton, J.M.; Giri, A.; Gao, X.; Velez Edwards, D.R.; Edwards, T.L. Population Stratification in Genetic Association Studies. Curr. Protoc. Hum. Genet. 2017, 95, 1–22. [Google Scholar] [CrossRef] [PubMed]
- Freedman, M.L.; Reich, D.; Penney, K.L.; McDonald, G.J.; Mignault, A.A.; Patterson, N.; Gabriel, S.B.; Topol, E.J.; Smoller, J.W.; Pato, C.N.; et al. Assessing the impact of population stratification on genetic association studies. Nat. Genet. 2004, 36, 388–393. [Google Scholar] [CrossRef] [PubMed]
- Risacher, S.L.; Saykin, A.J. Neuroimaging and Other Biomarkers for Alzheimer’s Disease: The Changing Landscape of Early Detection. Annu. Rev. Clin. Psychol. 2013, 9, 621–648. [Google Scholar] [CrossRef]
- Crane, P.K.; Carle, A.; Gibbons, L.E.; Insel, P.; Mackin, R.S.; Gross, A.; Jones, R.N.; Mukherjee, S.; Curtis, S.M.; Harvey, D.; et al. Development and assessment of a composite score for memory in the Alzheimer’s Disease Neuroimaging Initiative (ADNI). Brain Imaging Behav. 2012, 6, 502–516. [Google Scholar] [CrossRef] [PubMed]
- Risacher, S.L.; Shen, L.; West, J.D.; Kim, S.; McDonald, B.C.; Beckett, L.A.; Harvey, D.J.; Jack, C.R., Jr.; Weiner, M.W.; Saykin, A.J. Longitudinal MRI atrophy biomarkers: Relationship to conversion in the ADNI cohort. Neurobiol. Aging 2010, 31, 1401–1418. [Google Scholar] [CrossRef] [PubMed]
- Risacher, S.L.; Kim, S.; Nho, K.; Foroud, T.; Shen, L.; Petersen, R.C.; Jack, C.R.; Beckett, L.A.; Aisen, P.S.; Koeppe, R.A.; et al. APOE effect on Alzheimer’s biomarkers in older adults with significant memory concern. Alzheimers Dement. 2015, 11, 1417–1429. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.; Daly, M.J.; et al. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef] [PubMed]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
- Shabalin, A.A. Matrix eQTL: Ultra fast eQTL analysis via large matrix operations. Bioinformatics 2012, 28, 1353–1358. [Google Scholar] [CrossRef]
- Lambert, J.C.; Ibrahim-Verbaas, C.A.; Harold, D.; Naj, A.C.; Sims, R.; Bellenguez, C.; DeStafano, A.L.; Bis, J.C.; Beecham, G.W.; Grenier-Boley, B.; et al. Meta-analysis of 74,046 individuals identifies 11 new susceptibility loci for Alzheimer’s disease. Nat. Genet. 2013, 45, 1452–1458. [Google Scholar] [CrossRef]
- Raj, T.; Li, Y.I.; Wong, G.; Humphrey, J.; Wang, M.; Ramdhani, S.; Wang, Y.C.; Ng, B.; Gupta, I.; Haroutunian, V.; et al. Integrative transcriptome analyses of the aging brain implicate altered splicing in Alzheimer’s disease susceptibility. Nat. Genet. 2018, 50, 1584–1592. [Google Scholar] [CrossRef] [PubMed]
- Shkreta, L.; Delannoy, A.; Salvetti, A.; Chabot, B. SRSF10: An atypical splicing regulator with critical roles in stress response, organ development, and viral replication. RNA 2021, 27, 1302–1317. [Google Scholar] [CrossRef] [PubMed]
- Pihlajamaki, J.; Lerin, C.; Itkonen, P.; Boes, T.; Floss, T.; Schroeder, J.; Dearie, F.; Crunkhorn, S.; Burak, F.; Jimenez-Chillaron, J.C.; et al. Expression of the splicing factor gene SFRS10 is reduced in human obesity and contributes to enhanced lipogenesis. Cell Metab. 2011, 14, 208–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.; Dai, M.; Xu, Q.; Zhu, X.; Zhou, Y.; Jiang, S.; Wang, Y.; Ai, Z.; Ma, L.; Zhang, Y.; et al. SRSF10-mediated IL1RAP alternative splicing regulates cervical cancer oncogenesis via mIL1RAP-NF-κB-CD47 axis. Oncogene 2018, 37, 2394–2409. [Google Scholar] [CrossRef]
- Ramanan, V.K.; Risacher, S.L.; Nho, K.; Kim, S.; Shen, L.; McDonald, B.C.; Yoder, K.K.; Hutchins, G.D.; West, J.D.; Tallman, E.F.; et al. GWAS of longitudinal amyloid accumulation on 18F-florbetapir PET in Alzheimer’s disease implicates microglial activation gene IL1RAP. Brain 2015, 138, 3076–3088. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.S.; De Muynck, L. Differentially expressed genes in Alzheimer’s disease highlighting the roles of microglia genes including OLR1 and astrocyte gene CDK2AP1. Brain Behav. Immun. Health 2021, 13, 100227. [Google Scholar] [CrossRef] [PubMed]
- Fu, Q.; Wu, Y. RCAN1 in the inverse association between Alzheimer’s disease and cancer. Oncotarget 2018, 9, 54–66. [Google Scholar] [CrossRef]
- Wong, H.; Levenga, J.; Cain, P.; Rothermel, B.; Klann, E.; Hoeffer, C. RCAN1 overexpression promotes age-dependent mitochondrial dysregulation related to neurodegeneration in Alzheimer’s disease. Acta Neuropathol. 2015, 130, 829–843. [Google Scholar] [CrossRef]
- Kodigepalli, K.M.; Bowers, K.; Sharp, A.; Nanjundan, M. Roles and regulation of phospholipid scramblases. FEBS Lett. 2015, 589, 3–14. [Google Scholar] [CrossRef]
- Mansouri, L.; Messalmani, M.; Klai, S.; Bedoui, I.; Derbali, H.; Gritli, N.; Mrissa, R.; Fekih-Mrissa, N. Association of HLA-DR/DQ polymorphism with Alzheimer’s disease. Am. J. Med. Sci. 2015, 349, 334–337. [Google Scholar] [CrossRef]
- Mattiace, L.A.; Davies, P.; Dickson, D.W. Detection of HLA-DR on microglia in the human brain is a function of both clinical and technical factors. Am. J. Pathol. 1990, 136, 1101–1114. [Google Scholar] [PubMed]
- Zota, V.; Nemirovsky, A.; Baron, R.; Fisher, Y.; Selkoe, D.J.; Altmann, D.M.; Weiner, H.L.; Monsonego, A. HLA-DR alleles in amyloid β-peptide autoimmunity: A highly immunogenic role for the DRB1*1501 allele. J. Immunol. 2009, 183, 3522–3530. [Google Scholar] [CrossRef] [PubMed]
- Steele, N.Z.; Carr, J.S.; Bonham, L.W.; Geier, E.G.; Damotte, V.; Miller, Z.A.; Desikan, R.S.; Boehme, K.L.; Mukherjee, S.; Crane, P.K.; et al. Fine-mapping of the human leukocyte antigen locus as a risk factor for Alzheimer disease: A case-control study. PLoS Med. 2017, 14, e1002272. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, D.J.; Barnardo, M.C.; Fuggle, S.; Quiroga, I.; Sutherland, A.; Warden, D.R.; Barnetson, L.; Horton, R.; Beck, S.; Smith, A.D. Replication of the association of HLA-B7 with Alzheimer’s disease: A role for homozygosity? J. Neuroinflamm. 2006, 3, 33. [Google Scholar] [CrossRef] [PubMed]
- Ferreira-Vieira, T.H.; Guimaraes, I.M.; Silva, F.R.; Ribeiro, F.M. Alzheimer’s Disease: Targeting the Cholinergic System. Curr. Neuropharmacol. 2016, 14, 101–115. [Google Scholar] [CrossRef] [PubMed]
- Yurov, Y.B.; Vorsanova, S.G.; Iourov, I.Y. The DNA Replication Stress Hypothesis of Alzheimer’s Disease. Sci. World J. 2011, 11, 2602–2612. [Google Scholar] [CrossRef]
- Hoe, H.S.; Cooper, M.J.; Burns, M.P.; Lewis, P.A.; van der Brug, M.; Chakraborty, G.; Cartagena, C.M.; Pak, D.T.S.; Cookson, M.R.; Rebeck, G.W. The Metalloprotease Inhibitor TIMP-3 Regulates Amyloid Precursor Protein and Apolipoprotein E Receptor Proteolysis. J. Neurosci. 2007, 27, 10895–10905. [Google Scholar] [CrossRef]
- Alldred, M.J.; Duff, K.E.; Ginsberg, S.D. Microarray analysis of CA1 pyramidal neurons in a mouse model of tauopathy reveals progressive synaptic dysfunction. Neurobiol. Dis. 2012, 45, 751–762. [Google Scholar] [CrossRef]
- Gao, Y.L.; Wang, N.; Sun, F.R.; Cao, X.P.; Zhang, W.; Yu, J.T. Tau in neurodegenerative disease. Ann. Transl. Med. 2018, 6, 175. [Google Scholar] [CrossRef]
- Pascale, E.; Di Battista, M.E.; Rubino, A.; Purcaro, C.; Valente, M.; Fattapposta, F.; Ferraguti, G.; Meco, G. Genetic Architecture of MAPT Gene Region in Parkinson Disease Subtypes. Front. Cell Neurosci. 2016, 10, 96. [Google Scholar] [CrossRef]
- Aiello Bowles, E.J.; Crane, P.K.; Walker, R.L.; Chubak, J.; LaCroix, A.Z.; Anderson, M.L.; Rosenberg, D.; Keene, C.D.; Larson, E.B. Cognitive Resilience to Alzheimer’s Disease Pathology in the Human Brain. J. Alzheimer’s Dis. JAD 2019, 68, 1071–1083. [Google Scholar] [CrossRef] [PubMed]
- Arenaza-Urquijo, E.M.; Vemuri, P. Resistance vs resilience to Alzheimer disease: Clarifying terminology for preclinical studies. Neurology 2018, 90, 695–703. [Google Scholar] [CrossRef] [PubMed]
- Lam, B.; Masellis, M.; Freedman, M.; Stuss, D.T.; Black, S.E. Clinical, imaging, and pathological heterogeneity of the Alzheimer’s disease syndrome. Alzheimers Res. Ther. 2013, 5, 1. [Google Scholar] [CrossRef] [PubMed]
- Komarova, N.L.; Thalhauser, C.J. High degree of heterogeneity in Alzheimer’s disease progression patterns. PLoS Comput. Biol. 2011, 7, e1002251. [Google Scholar] [CrossRef] [PubMed]
- Chételat, G.; La Joie, R.; Villain, N.; Perrotin, A.; de La Sayette, V.; Eustache, F.; Vandenberghe, R. Amyloid imaging in cognitively normal individuals, at-risk populations and preclinical Alzheimer’s disease. Neuroimage Clin. 2013, 2, 356–365. [Google Scholar] [CrossRef] [PubMed]
- Leonenko, G.; Shoai, M.; Bellou, E.; Sims, R.; Williams, J.; Hardy, J.; Escott-Price, V. Genetic risk for alzheimer disease is distinct from genetic risk for amyloid deposition. Ann. Neurol. 2019, 86, 427–435. [Google Scholar] [CrossRef] [PubMed]
- Wightman, D.P.; Jansen, I.E.; Savage, J.E.; Shadrin, A.A.; Bahrami, S.; Holland, D.; Rongve, A.; Børte, S.; Winsvold, B.S.; Drange, O.K.; et al. A genome-wide association study with 1,126,563 individuals identifies new risk loci for Alzheimer’s disease. Nat. Genet. 2021, 53, 1276–1282. [Google Scholar] [CrossRef] [PubMed]
- Martinez, T.; Pascual, A. Gene expression profile in β-amyloid-treated SH-SY5Y neuroblastoma cells. Brain Res. Bull. 2007, 72, 225–231. [Google Scholar] [CrossRef]
- Aubry, S.; Shin, W.; Crary, J.F.; Lefort, R.; Qureshi, Y.H.; Lefebvre, C.; Califano, A.; Shelanski, M.L. Assembly and Interrogation of Alzheimer’s Disease Genetic Networks Reveal Novel Regulators of Progression. PLoS ONE 2015, 10, e0120352. [Google Scholar] [CrossRef]
- Valor, L.M.; Viosca, J.; Lopez-Atalaya, J.P.; Barco, A. Lysine acetyltransferases CBP and p300 as therapeutic targets in cognitive and neurodegenerative disorders. Curr. Pharm. Des. 2013, 19, 5051–5064. [Google Scholar] [CrossRef]
- Chen, X.; Li, Y.; Wang, C.; Tang, Y.; Mok, S.-A.; Tsai, R.M.; Rojas, J.C.; Karydas, A.; Miller, B.L.; Boxer, A.L.; et al. Promoting tau secretion and propagation by hyperactive p300/CBP via autophagy-lysosomal pathway in tauopathy. Mol. Neurodegener. 2020, 15, 2. [Google Scholar] [CrossRef]
- Planche, V.; Manjon, J.V.; Mansencal, B.; Lanuza, E.; Tourdias, T.; Catheline, G.; Coupé, P. Structural progression of Alzheimer’s disease over decades: The MRI staging scheme. Brain Commun. 2022, 4, fcac109. [Google Scholar] [CrossRef]
- Ma, Y.; Vardarajan, B.N.; CHARGE ADSP FUS; Bennett, D.A.; Fornage, M.; Seshadri, S.; Destefano, A.L.; De Jager, P.L. Alzheimer’s disease GWAS weighted by multi-omics and endophenotypes identifies novel risk loci. Alzheimer’s Dement. 2020, 16, e043977. [Google Scholar] [CrossRef]
- Golanska, E.; Sieruta, M.; Gresner, S.M.; Hulas-Bigoszewska, K.; Corder, E.H.; Styczynska, M.; Peplonska, B.; Barcikowska, M.; Liberski, P.P. Analysis of APBB2 gene polymorphisms in sporadic Alzheimer’s disease. Neurosci. Lett. 2008, 447, 164–166. [Google Scholar] [CrossRef]
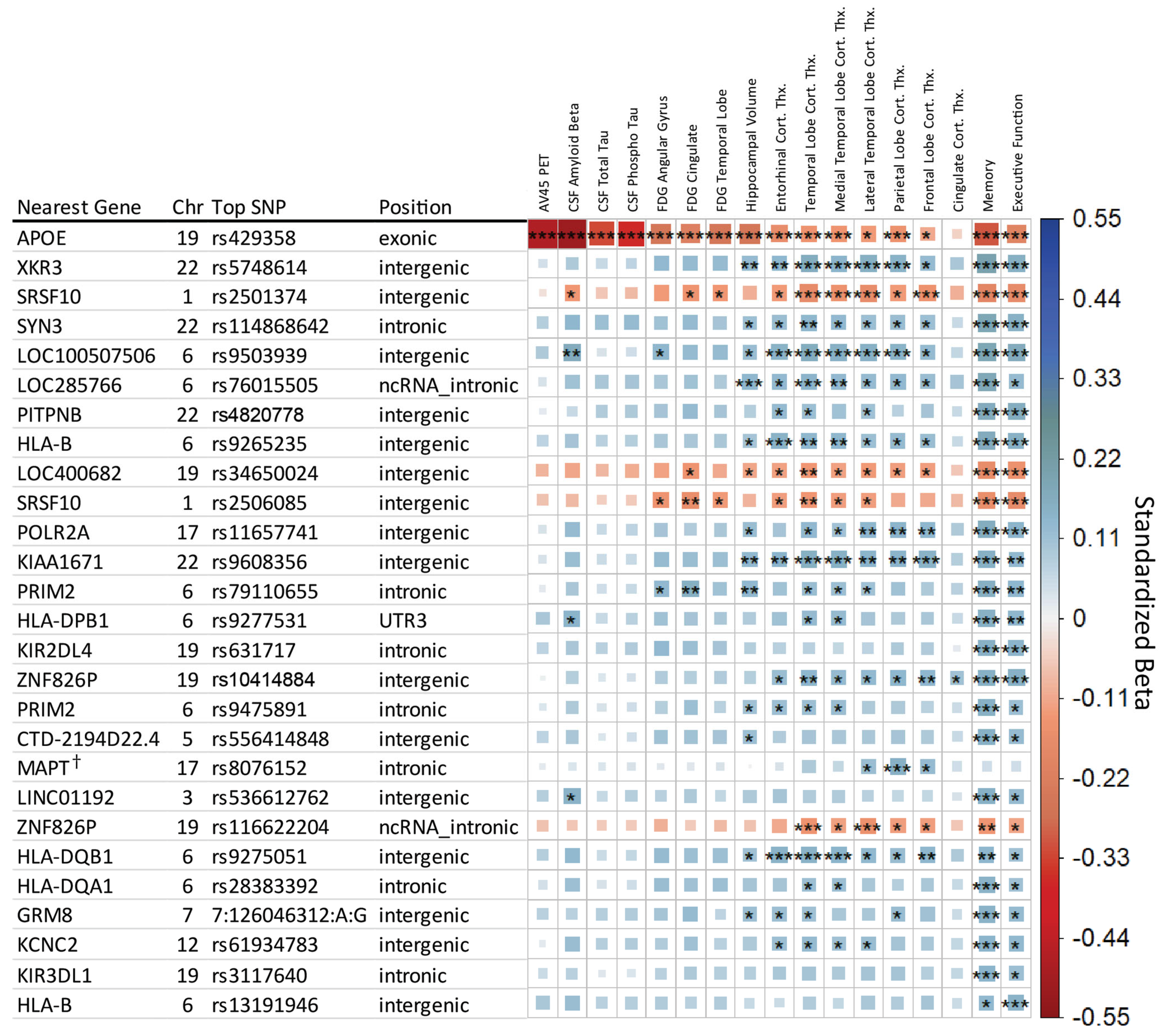
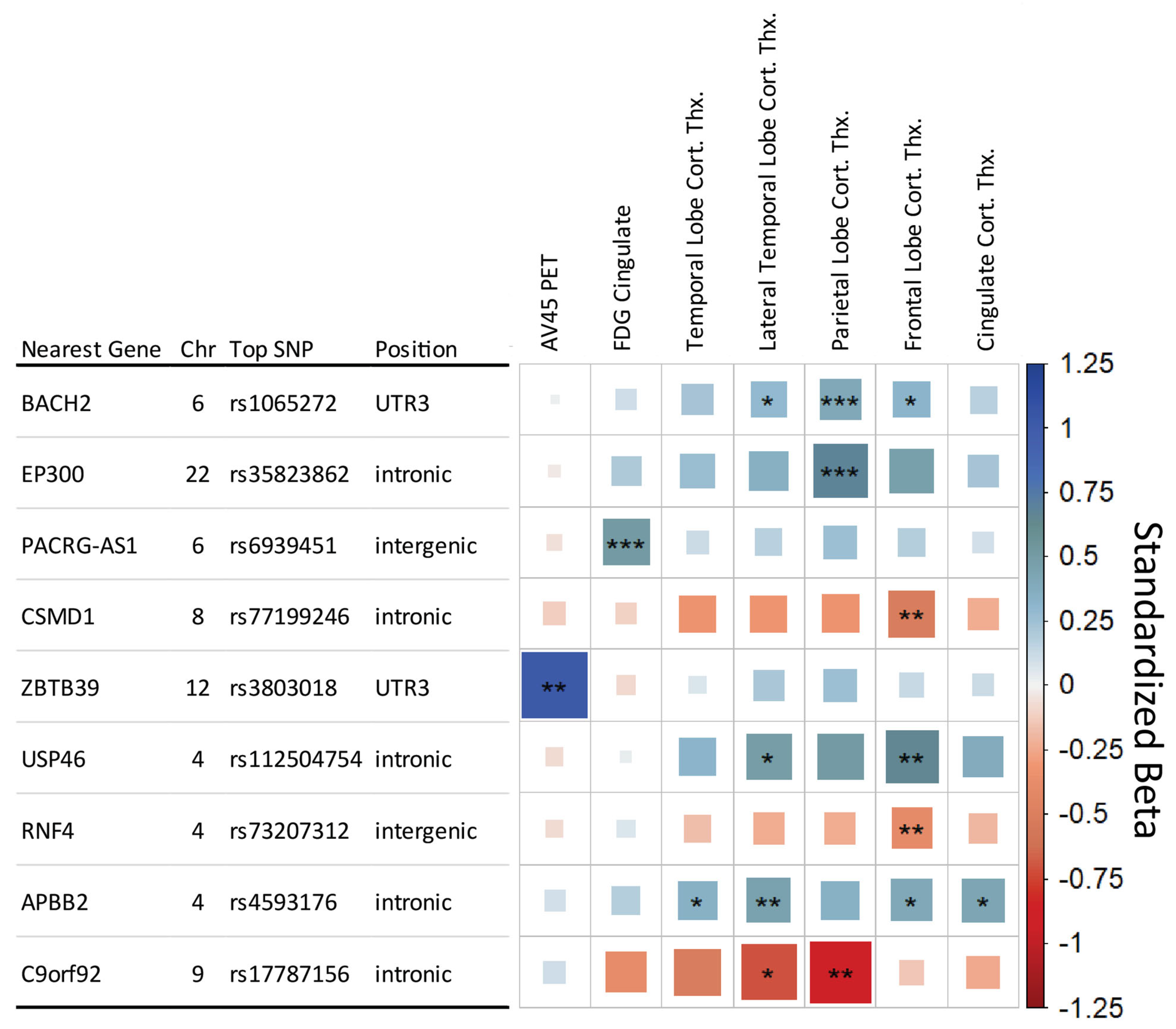

| Selected Endophenotype | N | Genetic PC1 | Genetic PC2 | APOE E2/E3/E4 Allele * | Age | Sex | Education | Intracranial Volume | MRI Field Strength † |
|---|---|---|---|---|---|---|---|---|---|
| Memory Composite Score | 1565 | • | • | • | • | • | • | ||
| Executive Function Composite Score | 1565 | • | • | • | • | • | • | ||
| Bilateral mean hippocampus volume | 1555 | • | • | • | • | • | • | • | • |
| Bilateral mean entorhinal cortex thickness | 1555 | • | • | • | • | • | • | • | • |
| Bilateral mean frontal lobe thickness | 1555 | • | • | • | • | • | • | • | • |
| Bilateral mean cingulate thickness | 1555 | • | • | • | • | • | • | • | • |
| Bilateral mean parietal lobe thickness | 1555 | • | • | • | • | • | • | • | • |
| Bilateral mean temp. lobe thickness | 1555 | • | • | • | • | • | • | • | • |
| Bilateral mean medial temp. lobe thickness | 1555 | • | • | • | • | • | • | • | • |
| Bilateral mean lateral temp. lobe thickness | 1555 | • | • | • | • | • | • | • | • |
| Mean FDG PET SUVR in Angular Gyrus | 1158 | • | • | • | • | • | |||
| Mean FDG PET SUVR in Cingulate | 1158 | • | • | • | • | • | |||
| Mean FDG PET SUVR in Bilateral Mean Temp. Lobe | 1158 | • | • | • | • | • | |||
| [18F]Florbetapir amyloid PET | 791 | • | • | • | • | • | |||
| CSF amyloid-β 1-42 peptide | 981 | • | • | • | • | • | |||
| CSF Total Tau | 1103 | • | • | • | • | • | |||
| CSF Phosphorylated Tau | 1103 | • | • | • | • | • |
| SNP | eQTL Gene | ADNI Blood eQTL FDR p-Value | GTEx Tissue | AMP-AD FDR Significant | |||||
|---|---|---|---|---|---|---|---|---|---|
| MSBB | Mayo | ||||||||
| BM-10 | BM-22 | BM-36 | BM-44 | Cerebellum | Temporal Cortex | ||||
| rs2506085 | RCAN3 | NA | NA | ● | |||||
| rs2506085 | FUCA1 | NA | Whole Blood | ||||||
| rs28383392 | HLA-DQA1 | 3.15 × 10−93 | - | ● | ● | ● | ● | ● | |
| rs28383392 | HLA-DQB1 | 9.15 × 10−92 | - | ● | ● | ● | ● | ● | ● |
| rs3117640 | FCAR | 1.38 × 10−3 | - | ||||||
| rs3117640 | KIR2DS4 | 7.93 × 10−19 | - | ||||||
| rs3117640 | KIR3DL1 | 6.53 × 10−4 | - | ||||||
| rs3803018 * | RDH16 | NA | Brain-cb | ||||||
| rs3803018 * | STAT6 | 3.92 × 10−4 | NA | ||||||
| rs5748614 | TPTEP1 | NA | Whole Blood | ||||||
| rs5748614 | XKR3 | 2.96 × 10−14 | Whole Blood | ||||||
| rs587750081 | ZNF826P | 1.19 × 10−18 | - | ||||||
| rs631717 | KIR2DS4 | 7.26 × 10−29 | Whole Blood | ||||||
| rs631717 | KIR3DL1 | 8.78 × 10−12 | Whole Blood | ||||||
| rs631717 | KIR3DL2 | 3.68 × 10−6 | Whole Blood | ||||||
| rs8076152 | KANSL1-AS1 | NA | Whole Blood/Brain-Hipp | ● | ● | ● | ● | ||
| rs8076152 | KANSL1-AS1 | NA | Brain-cb | ● | ● | ● | ● | ● | |
| rs8076152 | LRRC37A4P | 8.40 × 10−20 | Whole Blood/Brain-Hipp | ● | ● | ||||
| rs8076152 | ARL17B | NA | Brain-cb | ● | ● | ||||
| rs8076152 | MAPK8IP1 | 2.48 × 10−7 | Whole Blood/Brain-Hipp | ||||||
| rs9265235 | HLA-B | 9.87 × 10−6 | NA | ● | ● | ||||
| rs9265235 | C4A | NA | Brain-Hipp | ● | |||||
| rs9275051 | HLA-DQA1 | 1.23 × 10−50 | Whole Blood | ● | ● | ● | ● | ● | ● |
| rs9275051 | HLA-DQA2 | NA | Whole Blood/Brain-Hipp | ||||||
| rs9275051 | HLA-DQB2 | NA | Whole Blood/Brain-Hipp | ||||||
| rs9275051 | HLA-DQB1 | 7.66 × 10−45 | Whole Blood/Brain-Hipp | ● | ● | ● | ● | ● | |
| rs9275051 | HLA-DRB1 | NA | Brain-cb | ● | ● | ● | ● | ||
| rs35823862 * | ADSL | NA | NA | ● | |||||
| rs35823862 * | MCHR1 | NA | Brain-cb | ● | |||||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Rosewood, T.J.; Nho, K.; Risacher, S.L.; Gao, S.; Shen, L.; Foroud, T.; Saykin, A.J.; on behalf of the Alzheimer’s Disease Neuroimaging Initiative. Genome-Wide Association Analysis across Endophenotypes in Alzheimer’s Disease: Main Effects and Disease Stage-Specific Interactions. Genes 2023, 14, 2010. https://doi.org/10.3390/genes14112010
Rosewood TJ, Nho K, Risacher SL, Gao S, Shen L, Foroud T, Saykin AJ, on behalf of the Alzheimer’s Disease Neuroimaging Initiative. Genome-Wide Association Analysis across Endophenotypes in Alzheimer’s Disease: Main Effects and Disease Stage-Specific Interactions. Genes. 2023; 14(11):2010. https://doi.org/10.3390/genes14112010
Chicago/Turabian StyleRosewood, Thea J., Kwangsik Nho, Shannon L. Risacher, Sujuan Gao, Li Shen, Tatiana Foroud, Andrew J. Saykin, and on behalf of the Alzheimer’s Disease Neuroimaging Initiative. 2023. "Genome-Wide Association Analysis across Endophenotypes in Alzheimer’s Disease: Main Effects and Disease Stage-Specific Interactions" Genes 14, no. 11: 2010. https://doi.org/10.3390/genes14112010
APA StyleRosewood, T. J., Nho, K., Risacher, S. L., Gao, S., Shen, L., Foroud, T., Saykin, A. J., & on behalf of the Alzheimer’s Disease Neuroimaging Initiative. (2023). Genome-Wide Association Analysis across Endophenotypes in Alzheimer’s Disease: Main Effects and Disease Stage-Specific Interactions. Genes, 14(11), 2010. https://doi.org/10.3390/genes14112010








