Characteristics and Functions of MYB (v-Myb avivan myoblastsis virus oncogene homolog)-Related Genes in Arabidopsis thaliana
Abstract
:1. Introduction
2. Survey Methodology
3. Identification and Characterization of the MYB Genes
4. Structures and Main Functions of the MYB Gene Family
5. Functions of MYB-Related Genes
6. Light Response and Circadian Rhythm Regulation
7. The Development of Trichomes and Root Hairs
8. Telomere Metabolism
9. Plant Hormone Response
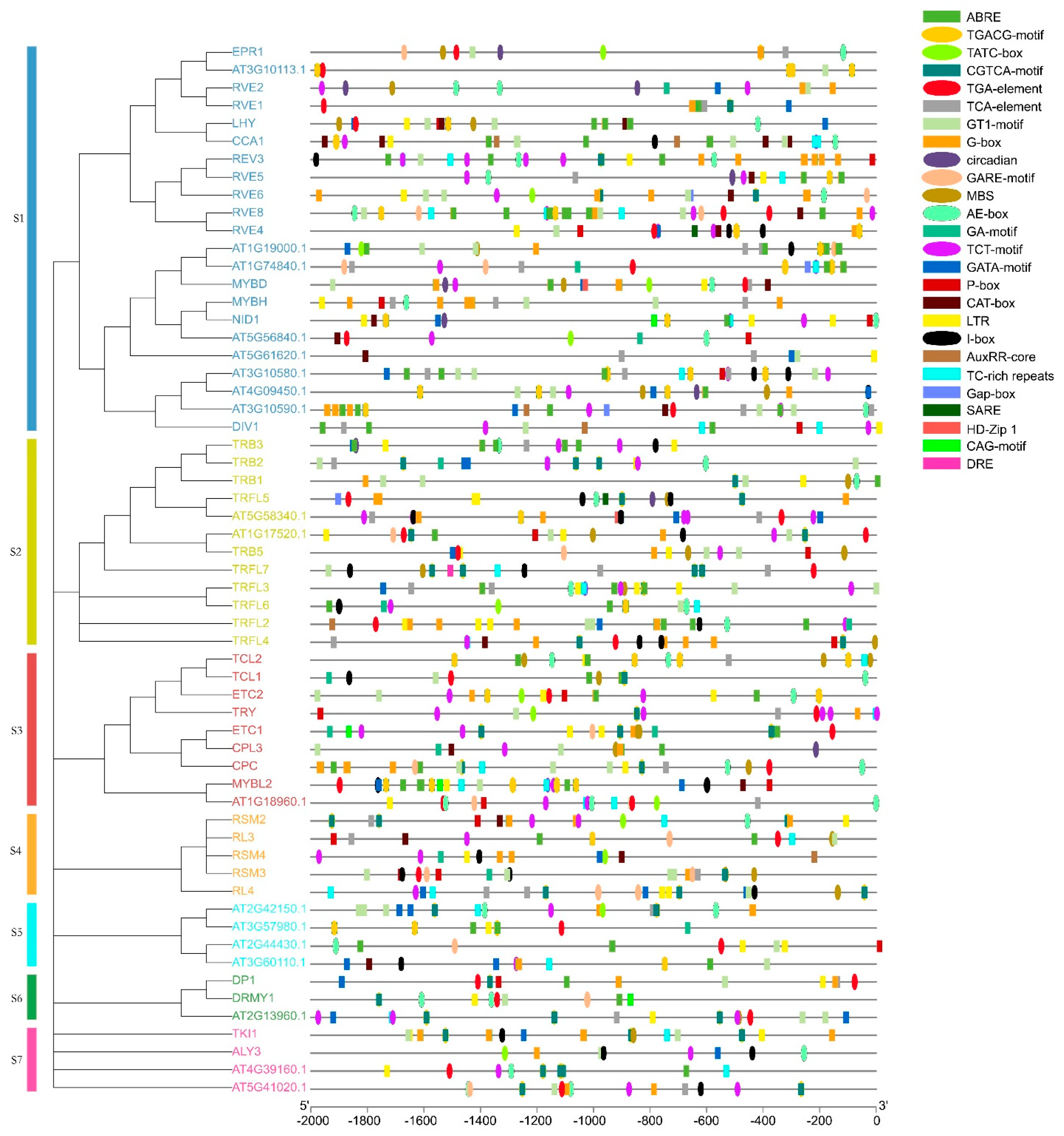
10. Promoter Analysis of AtMYB-Related Genes
11. Discussion
12. Conclusions and Outlook
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feller, A.; Machemer, K.; Braun, E.L.; Grotewold, E. Evolutionary and comparative analysis of MYB and bHLH plant transcription factors. Plant J. 2011, 66, 94–116. [Google Scholar] [CrossRef] [PubMed]
- Stracke, R.; Werber, M.; Weisshaar, B. The R2R3-MYB gene family in Arabidopsis thaliana. Curr. Opin. Plant Biol. 2001, 4, 447–456. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Li, K.; Li, Y.; Zhao, X.; Wang, L. MYB Transcription Factors as Regulators of Secondary Metabolism in Plants. Biology 2020, 9, 61. [Google Scholar] [CrossRef] [PubMed]
- Lal, M.; Bhardwaj, E.; Chahar, N.; Yadav, S.; Das, S. Comprehensive analysis of 1R- and 2R-MYBs reveals novel genic and protein features, complex organisation, selective expansion and insights into evolutionary tendencies. Funct. Integr. Genom. 2022, 22, 371–405. [Google Scholar] [CrossRef]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Kuraku, S.; Zmasek, C.M.; Nishimura, O.; Katoh, K. aLeaves facilitates on-demand exploration of metazoan gene family trees on MAFFT sequence alignment server with enhanced interactivity. Nucleic Acids Res. 2013, 41, W22–W28. [Google Scholar] [CrossRef]
- Katoh, K.; Rozewicki, J.; Yamada, K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2019, 20, 1160–1166. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Kumar, S. MEGA11: Molecular Evolutionary Genetics Analysis Version 11. Mol. Biol. Evol. 2021, 38, 3022–3027. [Google Scholar] [CrossRef]
- Yang, X.; Guo, T.; Li, J.; Chen, Z.; Guo, B.; An, X. Genome-wide analysis of the MYB-related transcription factor family and associated responses to abiotic stressors in Populus. Int. J. Biol. Macromol. 2021, 191, 359–376. [Google Scholar] [CrossRef]
- Bailey, T.L.; Boden, M.; Buske, F.A.; Frith, M.; Grant, C.E.; Clementi, L.; Ren, J.; Li, W.W.; Noble, W.S. MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res. 2009, 37, W202–W208. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef] [PubMed]
- Graf, T. Myb: A transcriptional activator linking proliferation and differentiation in hematopoietic cells. Curr. Opin. Genet. Dev. 1992, 2, 249–255. [Google Scholar] [CrossRef] [PubMed]
- Klempnauer, K.-H.; Gonda, T.J.; Michael Bishop, J. Nucleotide sequence of the retroviral leukemia gene v-myb and its cellular progenitor c-myb: The architecture of a transduced oncogene. Cell 1982, 31, 453–463. [Google Scholar] [CrossRef] [PubMed]
- Paz-Ares, J.; Ghosal, D.; Wienand, U.; Peterson, P.A.; Saedler, H. The regulatory c1 locus of Zea mays encodes a protein with homology to myb proto-oncogene products and with structural similarities to transcriptional activators. EMBO J. 1987, 6, 3553–3558. [Google Scholar] [CrossRef]
- Tice-Baldwin, K.; Fink, G.R.; Arndt, K.T. BAS1 has a Myb motif and activates HIS4 transcription only in combination with BAS2. Science 1989, 246, 931–935. [Google Scholar] [CrossRef]
- Chen, Y.; Yang, X.; He, K.; Liu, M.; Li, J.; Gao, Z.; Lin, Z.; Zhang, Y.; Wang, X.; Qiu, X.; et al. The MYB transcription factor superfamily of Arabidopsis: Expression analysis and phylogenetic comparison with the rice MYB family. Plant Mol. Biol. 2006, 60, 107–124. [Google Scholar] [CrossRef]
- Katiyar, A.; Smita, S.; Lenka, S.K.; Rajwanshi, R.; Chinnusamy, V.; Bansal, K.C. Genome-wide classification and expression analysis of MYB transcription factor families in rice and Arabidopsis. BMC Genom. 2012, 13, 544. [Google Scholar] [CrossRef]
- Chen, S.; Niu, X.; Guan, Y.; Li, H. Genome-Wide Analysis and Expression Profiles of the MYB Genes in Brachypodium distachyon. Plant Cell Physiol. 2017, 58, 1777–1788. [Google Scholar] [CrossRef]
- Qing, J.; Dawei, W.; Jun, Z.; Yulan, X.; Bingqi, S.; Fan, Z. Genome-wide characterization and expression analyses of the MYB superfamily genes during developmental stages in Chinese jujube. PeerJ 2019, 7, e6353. [Google Scholar] [CrossRef]
- Arce-Rodriguez, M.L.; Martinez, O.; Ochoa-Alejo, N. Genome-Wide Identification and Analysis of the MYB Transcription Factor Gene Family in Chili Pepper (Capsicum spp.). Int. J. Mol. Sci. 2021, 22, 222–229. [Google Scholar] [CrossRef]
- Liu, L.; Chao, N.; Yidilisi, K.; Kang, X.; Cao, X. Comprehensive analysis of the MYB transcription factor gene family in Morus alba. BMC Plant Biol. 2022, 22, 281. [Google Scholar] [CrossRef]
- Du, Y.T.; Zhao, M.J.; Wang, C.T.; Gao, Y.; Wang, Y.X.; Liu, Y.W.; Chen, M.; Chen, J.; Zhou, Y.B.; Xu, Z.S.; et al. Identification and characterization of GmMYB118 responses to drought and salt stress. BMC Plant Biol. 2018, 18, 320. [Google Scholar] [CrossRef]
- Liu, Y.; Zeng, Y.; Li, Y.; Liu, Z.; Lin-Wang, K.; Espley, R.V.; Allan, A.C.; Zhang, J. Genomic survey and gene expression analysis of the MYB-related transcription factor superfamily in potato (Solanum tuberosum L.). Int. J. Biol. Macromol. 2020, 164, 2450–2464. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Lin, K.; Zhang, S.; Wu, J.; Fang, Y.; Wang, Y. Genome-Wide Analysis of Myeloblastosis-Related Genes in Brassica napus L. and Positive Modulation of Osmotic Tolerance by BnMRD107. Front. Plant Sci. 2021, 12, 678202. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Zhang, Z.; Fang, K.; Shan, Q.; He, L.; Dai, X.; Zou, X.; Liu, F. Genome-Wide Analysis of the MYB-Related Transcription Factor Family in Pepper and Functional Studies of CaMYB37 Involvement in Capsaicin Biosynthesis. Int. J. Mol. Sci. 2022, 23, 11667. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Ding, W.; Wu, X.; Wang, L.; Yang, X.; Yue, Y. Insights Into the MYB-Related Transcription Factors Involved in Regulating Floral Aroma Synthesis in Sweet Osmanthus. Front. Plant Sci. 2022, 13, 765213. [Google Scholar] [CrossRef] [PubMed]
- Dubos, C.; Stracke, R.; Grotewold, E.; Weisshaar, B.; Martin, C.; Lepiniec, L. MYB transcription factors in Arabidopsis. Trends Plant Sci. 2010, 15, 573–581. [Google Scholar] [CrossRef]
- Du, H.; Wang, Y.B.; Xie, Y.; Liang, Z.; Jiang, S.J.; Zhang, S.S.; Huang, Y.B.; Tang, Y.X. Genome-wide identification and evolutionary and expression analyses of MYB-related genes in land plants. DNA Res. 2013, 20, 437–448. [Google Scholar] [CrossRef]
- Rosinski, J.A.; Atchley, W.R. Molecular evolution of the Myb family of transcription factors: Evidence for polyphyletic origin. J. Mol. Evol. 1998, 46, 74–83. [Google Scholar] [CrossRef]
- Jiang, C.; Gu, J.; Chopra, S.; Gu, X.; Peterson, T. Ordered origin of the typical two- and three-repeat Myb genes. Gene 2004, 326, 13–22. [Google Scholar] [CrossRef]
- Feng, G.; Burleigh, J.G.; Braun, E.L.; Mei, W.; Barbazuk, W.B. Evolution of the 3R-MYB Gene Family in Plants. Genome Biol. Evol. 2017, 9, 1013–1029. [Google Scholar] [CrossRef] [PubMed]
- Yan, H.; Pei, X.; Zhang, H.; Li, X.; Zhang, X.; Zhao, M.; Chiang, V.L.; Sederoff, R.R.; Zhao, X. MYB-Mediated Regulation of Anthocyanin Biosynthesis. Int. J. Mol. Sci. 2021, 22, 3103. [Google Scholar] [CrossRef] [PubMed]
- Du, H.; Zhang, L.; Liu, L.; Tang, X.F.; Yang, W.J.; Wu, Y.M.; Huang, Y.B.; Tang, Y.X. Biochemical and molecular characterization of plant MYB transcription factor family. Biochemistry 2009, 74, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Roy, S. Function of MYB domain transcription factors in abiotic stress and epigenetic control of stress response in plant genome. Plant Signal. Behav. 2016, 11, e1117723. [Google Scholar] [CrossRef]
- Baldoni, E.; Genga, A.; Cominelli, E. Plant MYB Transcription Factors: Their Role in Drought Response Mechanisms. Int. J. Mol. Sci. 2015, 16, 15811–15851. [Google Scholar] [CrossRef]
- Ma, D.; Constabel, C.P. MYB Repressors as Regulators of Phenylpropanoid Metabolism in Plants. Trends Plant Sci. 2019, 24, 275–289. [Google Scholar] [CrossRef]
- Li, J.; Han, G.; Sun, C.; Sui, N. Research advances of MYB transcription factors in plant stress resistance and breeding. Plant Signal. Behav. 2019, 14, 1613131. [Google Scholar] [CrossRef]
- Thakur, S.; Vasudev, P.G. MYB transcription factors and their role in Medicinal plants. Mol. Biol. Rep. 2022, 49, 10995–11008. [Google Scholar] [CrossRef]
- Erpen, L.; Devi, H.S.; Grosser, J.W.; Dutt, M. Potential use of the DREB/ERF, MYB, NAC and WRKY transcription factors to improve abiotic and biotic stress in transgenic plants. Plant Cell Tissue Organ Cult. (PCTOC) 2017, 132, 1–25. [Google Scholar] [CrossRef]
- Zhang, Z.; Chen, J.; Su, Y.; Liu, H.; Chen, Y.; Luo, P.; Du, X.; Wang, D.; Zhang, H. TaLHY, a 1R-MYB Transcription Factor, Plays an Important Role in Disease Resistance against Stripe Rust Fungus and Ear Heading in Wheat. PLoS ONE 2015, 10, e0127723. [Google Scholar] [CrossRef]
- Shin, D.; Moon, S.J.; Han, S.; Kim, B.G.; Park, S.R.; Lee, S.K.; Yoon, H.J.; Lee, H.E.; Kwon, H.B.; Baek, D.; et al. Expression of StMYB1R-1, a novel potato single MYB-like domain transcription factor, increases drought tolerance. Plant Physiol. 2011, 155, 421–432. [Google Scholar] [CrossRef]
- Xiong, H.; Li, J.; Liu, P.; Duan, J.; Zhao, Y.; Guo, X.; Li, Y.; Zhang, H.; Ali, J.; Li, Z. Overexpression of OsMYB48-1, a novel MYB-related transcription factor, enhances drought and salinity tolerance in rice. PLoS ONE 2014, 9, e92913. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Q.; Liu, M.; Bo, C.; Wang, X.; Ma, Q.; Cheng, B.; Cai, R. Overexpression of a maize MYB48 gene confers drought tolerance in transgenic arabidopsis plants. J. Plant Biol. 2017, 60, 612–621. [Google Scholar] [CrossRef]
- Zhang, L.; Liu, G.; Jia, J.; Zhao, G.; Xia, C.; Zhang, L.; Li, F.; Zhang, Q.; Dong, C.; Gao, S.; et al. The wheat MYB-related transcription factor TaMYB72 promotes flowering in rice. J. Integr. Plant Biol. 2016, 58, 701–704. [Google Scholar] [CrossRef]
- Yong, Y.; Zhang, Y.; Lyu, Y. A MYB-Related Transcription Factor from Lilium lancifolium L. (LlMYB3) Is Involved in Anthocyanin Biosynthesis Pathway and Enhances Multiple Abiotic Stress Tolerance in Arabidopsis thaliana. Int. J. Mol. Sci. 2019, 20, 3195. [Google Scholar] [CrossRef]
- Tiwari, P.; Indoliya, Y.; Chauhan, A.S.; Singh, P.; Singh, P.K.; Singh, P.C.; Srivastava, S.; Pande, V.; Chakrabarty, D. Auxin-salicylic acid cross-talk ameliorates OsMYB-R1 mediated defense towards heavy metal, drought and fungal stress. J. Hazard. Mater. 2020, 399, 122811. [Google Scholar] [CrossRef] [PubMed]
- Harmer, S.L.; Hogenesch, J.B.; Straume, M.; Chang, H.S.; Han, B.; Zhu, T.; Wang, X.; Kreps, J.A.; Kay, S.A. Orchestrated transcription of key pathways in Arabidopsis by the circadian clock. Science 2000, 290, 2110–2113. [Google Scholar] [CrossRef] [PubMed]
- Lu, S.X.; Knowles, S.M.; Andronis, C.; Ong, M.S.; Tobin, E.M. CIRCADIAN CLOCK ASSOCIATED1 and LATE ELONGATED HYPOCOTYL function synergistically in the circadian clock of Arabidopsis. Plant Physiol. 2009, 150, 834–843. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.Y.; Kenigsbuch, D.; Sun, L.; Harel, E.; Ong, M.S.; Tobin, E.M. A Myb-related transcription factor is involved in the phytochrome regulation of an Arabidopsis Lhcb gene. Plant Cell 1997, 9, 491–507. [Google Scholar] [CrossRef] [PubMed]
- Kyung, J.; Jeon, M.; Jeong, G.; Shin, Y.; Seo, E.; Yu, J.; Kim, H.; Park, C.M.; Hwang, D.; Lee, I. The two clock proteins CCA1 and LHY activate VIN3 transcription during vernalization through the vernalization-responsive cis-element. Plant Cell 2022, 34, 1020–1037. [Google Scholar] [CrossRef]
- Farre, E.M.; Harmer, S.L.; Harmon, F.G.; Yanovsky, M.J.; Kay, S.A. Overlapping and distinct roles of PRR7 and PRR9 in the Arabidopsis circadian clock. Curr. Biol. 2005, 15, 47–54. [Google Scholar] [CrossRef] [PubMed]
- Nakamichi, N.; Kiba, T.; Henriques, R.; Mizuno, T.; Chua, N.H.; Sakakibara, H. PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 2010, 22, 594–605. [Google Scholar] [CrossRef] [PubMed]
- Kuno, N.; Moller, S.G.; Shinomura, T.; Xu, X.; Chua, N.H.; Furuya, M. The Novel MYB Protein EARLY-PHYTOCHROME-RESPONSIVE1 Is a Component of a Slave Circadian Oscillator in Arabidopsis. Plant Cell 2003, 15, 2476–2488. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Chen, Y.; Wang, Z.Y.; Chen, Z.; Gu, H.; Qu, L.J. Constitutive expression of CIR1 (RVE2) affects several circadian-regulated processes and seed germination in Arabidopsis. Plant J. 2007, 51, 512–525. [Google Scholar] [CrossRef] [PubMed]
- Guan, Q.; Wu, J.; Zhang, Y.; Jiang, C.; Liu, R.; Chai, C.; Zhu, J. A DEAD box RNA helicase is critical for pre-mRNA splicing, cold-responsive gene regulation, and cold tolerance in Arabidopsis. Plant Cell 2013, 25, 342–356. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Xu, G.; Jing, Y.; Tang, W.; Lin, R. Phytochrome B and REVEILLE1/2-mediated signalling controls seed dormancy and germination in Arabidopsis. Nat. Commun. 2016, 7, 12377. [Google Scholar] [CrossRef]
- Rawat, R.; Schwartz, J.; Jones, M.A.; Sairanen, I.; Cheng, Y.; Andersson, C.R.; Zhao, Y.; Ljung, K.; Harmer, S.L. REVEILLE1, a Myb-like transcription factor, integrates the circadian clock and auxin pathways. Proc. Natl. Acad. Sci. USA 2009, 106, 16883–16888. [Google Scholar] [CrossRef]
- Meissner, M.; Orsini, E.; Ruschhaupt, M.; Melchinger, A.E.; Hincha, D.K.; Heyer, A.G. Mapping quantitative trait loci for freezing tolerance in a recombinant inbred line population of Arabidopsis thaliana accessions Tenela and C24 reveals REVEILLE1 as negative regulator of cold acclimation. Plant Cell Environ. 2013, 36, 1256–1267. [Google Scholar] [CrossRef]
- Xu, G.; Guo, H.; Zhang, D.; Chen, D.; Jiang, Z.; Lin, R. REVEILLE1 promotes NADPH: Protochlorophyllide oxidoreductase A expression and seedling greening in Arabidopsis. Photosynth. Res. 2015, 126, 331–340. [Google Scholar] [CrossRef]
- Barak, S.; Tobin, E.M.; Andronis, C.; Sugano, S.; Green, R.M. All in good time: The Arabidopsis circadian clock. Trends Plant Sci. 2000, 5, 517–522. [Google Scholar] [CrossRef]
- McClung, C.R. The Plant Circadian Oscillator. Biology 2019, 8, 14. [Google Scholar] [CrossRef] [PubMed]
- Xue, X.; Sun, K.; Zhu, Z. CIRCADIAN CLOCK ASSOCIATED 1 gates morning phased auxin response in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2020, 527, 935–940. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Grundy, J.; Veflingstad, S.R.; Dyer, N.P.; Hannah, M.A.; Ott, S.; Carre, I.A. Circadian control of abscisic acid biosynthesis and signalling pathways revealed by genome-wide analysis of LHY binding targets. New Phytol. 2018, 220, 893–907. [Google Scholar] [CrossRef]
- Bancos, S.; Szatmari, A.M.; Castle, J.; Kozma-Bognar, L.; Shibata, K.; Yokota, T.; Bishop, G.J.; Nagy, F.; Szekeres, M. Diurnal regulation of the brassinosteroid-biosynthetic CPD gene in Arabidopsis. Plant Physiol. 2006, 141, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.G.; Won, J.H.; Choi, Y.R.; Lee, K.; Seo, P.J. Brassinosteroids Regulate Circadian Oscillation via the BES1/TPL-CCA1/LHY Module in Arabidopsisthaliana. iScience 2020, 23, 101528. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Zhu-Salzman, K. LATE ELONGATED HYPOCOTYL potentiates resistance conferred by CIRCADIAN CLOCK ASSOCIATED1 to aphid by co-regulating the expression of indole glucosinolate biosynthetic genes. Plant Signal. Behav. 2021, 16, 1908708. [Google Scholar] [CrossRef]
- Kidokoro, S.; Hayashi, K.; Haraguchi, H.; Ishikawa, T.; Soma, F.; Konoura, I.; Toda, S.; Mizoi, J.; Suzuki, T.; Shinozaki, K.; et al. Posttranslational regulation of multiple clock-related transcription factors triggers cold-inducible gene expression in Arabidopsis. Proc. Natl. Acad. Sci. USA 2021, 118, e2021048118. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Tian, Y.Y.; Li, J.Y.; Yuan, L.; Zhang, L.L.; Wang, Z.Y.; Xu, X.; Davis, S.J.; Liu, J.X. A competition-attenuation mechanism modulates thermoresponsive growth at warm temperatures in plants. New Phytol. 2022, 237, 177–191. [Google Scholar] [CrossRef]
- Farinas, B.; Mas, P. Functional implication of the MYB transcription factor RVE8/LCL5 in the circadian control of histone acetylation. Plant J. 2011, 66, 318–329. [Google Scholar] [CrossRef]
- Hsu, P.Y.; Devisetty, U.K.; Harmer, S.L. Accurate timekeeping is controlled by a cycling activator in Arabidopsis. eLife 2013, 2, e00473. [Google Scholar] [CrossRef]
- Perez-Garcia, P.; Ma, Y.; Yanovsky, M.J.; Mas, P. Time-dependent sequestration of RVE8 by LNK proteins shapes the diurnal oscillation of anthocyanin biosynthesis. Proc. Natl. Acad. Sci. USA 2015, 112, 5249–5253. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, N.H.; Lee, H. MYB-related transcription factors function as regulators of the circadian clock and anthocyanin biosynthesis in Arabidopsis. Plant Signal. Behav. 2016, 11, e1139278. [Google Scholar] [CrossRef] [PubMed]
- Gray, J.A.; Shalit-Kaneh, A.; Chu, D.N.; Hsu, P.Y.; Harmer, S.L. The REVEILLE Clock Genes Inhibit Growth of Juvenile and Adult Plants by Control of Cell Size. Plant Physiol. 2017, 173, 2308–2322. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.; Kim, J.H.; Nguyen, H.N.; Jikumaru, Y.; Kamiya, Y.; Hong, S.W.; Lee, H. A novel Arabidopsis MYB-like transcription factor, MYBH, regulates hypocotyl elongation by enhancing auxin accumulation. J. Exp. Bot. 2013, 64, 3911–3922. [Google Scholar] [CrossRef] [PubMed]
- Huang, C.K.; Lo, P.C.; Huang, L.F.; Wu, S.J.; Yeh, C.H.; Lu, C.A. A single-repeat MYB transcription repressor, MYBH, participates in regulation of leaf senescence in Arabidopsis. Plant Mol. Biol. 2015, 88, 269–286. [Google Scholar] [CrossRef]
- Nguyen, N.H.; Jeong, C.Y.; Kang, G.H.; Yoo, S.D.; Hong, S.W.; Lee, H. MYBD employed by HY5 increases anthocyanin accumulation via repression of MYBL2 in Arabidopsis. Plant J 2015, 84, 1192–1205. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, Y.; Song, Z.; Zhang, H. Repression of MYBL2 by Both microRNA858a and HY5 Leads to the Activation of Anthocyanin Biosynthetic Pathway in Arabidopsis. Mol. Plant 2016, 9, 1395–1405. [Google Scholar] [CrossRef]
- Zhao, D.; Zheng, Y.; Yang, L.; Yao, Z.; Cheng, J.; Zhang, F.; Jiang, H.; Liu, D. The transcription factor AtGLK1 acts upstream of MYBL2 to genetically regulate sucrose-induced anthocyanin biosynthesis in Arabidopsis. BMC Plant Biol. 2021, 21, 242. [Google Scholar] [CrossRef]
- Wada, T.; Tachibana, T.; Shimura, Y.; Okada, K. Epidermal cell differentiation in Arabidopsis determined by a Myb homolog, CPC. Science 1997, 277, 1113–1116. [Google Scholar] [CrossRef]
- Zhao, M.; Morohashi, K.; Hatlestad, G.; Grotewold, E.; Lloyd, A. The TTG1-bHLH-MYB complex controls trichome cell fate and patterning through direct targeting of regulatory loci. Development 2008, 135, 1991–1999. [Google Scholar] [CrossRef]
- Serna, L. CAPRICE positively regulates stomatal formation in the Arabidopsis hypocotyl. Plant Signal. Behav. 2008, 3, 1077–1082. [Google Scholar] [CrossRef] [PubMed]
- Zhu, H.F.; Fitzsimmons, K.; Khandelwal, A.; Kranz, R.G. CPC, a single-repeat R3 MYB, is a negative regulator of anthocyanin biosynthesis in Arabidopsis. Mol. Plant 2009, 2, 790–802. [Google Scholar] [CrossRef] [PubMed]
- Kirik, V.; Simon, M.; Wester, K.; Schiefelbein, J.; Hulskamp, M. ENHANCER of TRY and CPC 2 (ETC2) reveals redundancy in the region-specific control of trichome development of Arabidopsis. Plant Mol. Biol. 2004, 55, 389–398. [Google Scholar] [CrossRef]
- Kirik, V.; Simon, M.; Huelskamp, M.; Schiefelbein, J. The ENHANCER OF TRY AND CPC1 gene acts redundantly with TRIPTYCHON and CAPRICE in trichome and root hair cell patterning in Arabidopsis. Dev. Biol. 2004, 268, 506–513. [Google Scholar] [CrossRef] [PubMed]
- Gan, L.; Xia, K.; Chen, J.G.; Wang, S. Functional characterization of TRICHOMELESS2, a new single-repeat R3 MYB transcription factor in the regulation of trichome patterning in Arabidopsis. BMC Plant Biol. 2011, 11, 176. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, R.; Iwata, M.; Sano, R.; Inoue, K.; Okada, K.; Wada, T. Arabidopsis CAPRICE-LIKE MYB 3 (CPL3) controls endoreduplication and flowering development in addition to trichome and root hair formation. Development 2008, 135, 1335–1345. [Google Scholar] [CrossRef]
- Wang, S.; Kwak, S.H.; Zeng, Q.; Ellis, B.E.; Chen, X.Y.; Schiefelbein, J.; Chen, J.G. TRICHOMELESS1 regulates trichome patterning by suppressing GLABRA1 in Arabidopsis. Development 2007, 134, 3873–3882. [Google Scholar] [CrossRef]
- Tominaga-Wada, R.; Wada, T. Extended C termini of CPC-LIKE MYB proteins confer functional diversity in Arabidopsis epidermal cell differentiation. Development 2017, 144, 2375–2380. [Google Scholar] [CrossRef]
- Schellmann, S.; Schnittger, A.; Kirik, V.; Wada, T.; Okada, K.; Beermann, A.; Thumfahrt, J.; Jurgens, G.; Hulskamp, M. TRIPTYCHON and CAPRICE mediate lateral inhibition during trichome and root hair patterning in Arabidopsis. EMBO J. 2002, 21, 5036–5046. [Google Scholar] [CrossRef]
- Wester, K.; Digiuni, S.; Geier, F.; Timmer, J.; Fleck, C.; Hulskamp, M. Functional diversity of R3 single-repeat genes in trichome development. Development 2009, 136, 1487–1496. [Google Scholar] [CrossRef]
- Wang, S.; Hubbard, L.; Chang, Y.; Guo, J.; Schiefelbein, J.; Chen, J.G. Comprehensive analysis of single-repeat R3 MYB proteins in epidermal cell patterning and their transcriptional regulation in Arabidopsis. BMC Plant Biol. 2008, 8, 81. [Google Scholar] [CrossRef] [PubMed]
- Tominaga-Wada, R.; Nukumizu, Y. Expression analysis of an R3-Type MYB transcription factor CPC-LIKE MYB4 (TRICHOMELESS2) and CPL4-Related transcripts in Arabidopsis. Int. J. Mol. Sci. 2012, 13, 3478–3491. [Google Scholar] [CrossRef] [PubMed]
- Sawa, S. Overexpression of the AtmybL2 gene represses trichome development in Arabidopsis. DNA Res. 2002, 9, 31–34. [Google Scholar] [CrossRef] [PubMed]
- Ohmagari, M.; Kono, Y.; Tominaga, R. Effect of phosphate starvation on CAPRICE homolog gene expression in the root of Arabidopsis. Plant Biotechnol. 2020, 37, 349–352. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.; Tian, H.; Hu, Q.; Guo, H.; Yang, L.; Cai, L.; Wang, X.; Liu, B.; Wang, S. Ectopic expression of R3 MYB transcription factor gene OsTCL1 in Arabidopsis, but not rice, affects trichome and root hair formation. Sci. Rep. 2016, 6, 19254. [Google Scholar] [CrossRef] [PubMed]
- Arteaga, N.; Mendez-Vigo, B.; Fuster-Pons, A.; Savic, M.; Murillo-Sanchez, A.; Pico, F.X.; Alonso-Blanco, C. Differential environmental and genomic architectures shape the natural diversity for trichome patterning and morphology in different Arabidopsis organs. Plant Cell Environ. 2022, 45, 3018–3035. [Google Scholar] [CrossRef]
- Marian, C.O.; Bordoli, S.J.; Goltz, M.; Santarella, R.A.; Jackson, L.P.; Danilevskaya, O.; Beckstette, M.; Meeley, R.; Bass, H.W. The maize Single myb histone 1 gene, Smh1, belongs to a novel gene family and encodes a protein that binds telomere DNA repeats in vitro. Plant Physiol. 2003, 133, 1336–1350. [Google Scholar] [CrossRef]
- Byun, M.Y.; Hong, J.-P.; Kim, W.T. Identification and characterization of three telomere repeat-binding factors in rice. Biochem. Biophys. Res. Commun. 2008, 372, 85–90. [Google Scholar] [CrossRef]
- Hwang, M.G.; Chung, I.K.; Kang, B.G.; Cho, M.H. Sequence-specific binding property of Arabidopsis thaliana telomeric DNA binding protein 1 (AtTBP1). FEBS Lett. 2001, 503, 35–40. [Google Scholar] [CrossRef]
- Schrumpfova, P.; Kuchar, M.; Mikova, G.; Skrisovska, L.; Kubicarova, T.; Fajkus, J. Characterization of two Arabidopsis thaliana myb-like proteins showing affinity to telomeric DNA sequence. Genome 2004, 47, 316–324. [Google Scholar] [CrossRef]
- Karamysheva, Z.N.; Surovtseva, Y.V.; Vespa, L.; Shakirov, E.V.; Shippen, D.E. A C-terminal Myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J. Biol. Chem. 2004, 279, 47799–47807. [Google Scholar] [CrossRef]
- Kuchar, M. Plant telomere-binding proteins. Biol. Plant. 2006, 50, 1–7. [Google Scholar] [CrossRef]
- Yang, S.W.; Kim, S.K.; Kim, W.T. Perturbation of NgTRF1 expression induces apoptosis-like cell death in tobacco BY-2 cells and implicates NgTRF1 in the control of telomere length and stability. Plant Cell 2004, 16, 3370–3385. [Google Scholar] [CrossRef] [PubMed]
- Hong, J.P.; Byun, M.Y.; Koo, D.H.; An, K.; Bang, J.W.; Chung, I.K.; An, G.; Kim, W.T. Suppression of RICE TELOMERE BINDING PROTEIN 1 results in severe and gradual developmental defects accompanied by genome instability in rice. Plant Cell 2007, 19, 1770–1781. [Google Scholar] [CrossRef] [PubMed]
- Hamaguchi, A.; Yamashino, T.; Koizumi, N.; Kiba, T.; Kojima, M.; Sakakibara, H.; Mizuno, T. A small subfamily of Arabidopsis RADIALIS-LIKE SANT/MYB genes: A link to HOOKLESS1-mediated signal transduction during early morphogenesis. Biosci. Biotechnol. Biochem. 2008, 72, 2687–2696. [Google Scholar] [CrossRef] [PubMed]
- Raz, V.; Ecker, J.R. Regulation of differential growth in the apical hook of Arabidopsis. Development 1999, 126, 3661–3668. [Google Scholar] [CrossRef]
- Lehman, A.; Black, R.; Ecker, J.R. HOOKLESS1, an Ethylene Response Gene, Is Required for Differential Cell Elongation in the Arabidopsis Hypocotyl. Cell 1996, 85, 183–194. [Google Scholar] [CrossRef]
- Yang, B.; Song, Z.; Li, C.; Jiang, J.; Zhou, Y.; Wang, R.; Wang, Q.; Ni, C.; Liang, Q.; Chen, H.; et al. RSM1, an Arabidopsis MYB protein, interacts with HY5/HYH to modulate seed germination and seedling development in response to abscisic acid and salinity. PLoS Genet. 2018, 14, e1007839. [Google Scholar] [CrossRef]
- Wu, P.; Peng, M.; Li, Z.; Yuan, N.; Hu, Q.; Foster, C.E.; Saski, C.; Wu, G.; Sun, D.; Luo, H. DRMY1, a Myb-Like Protein, Regulates Cell Expansion and Seed Production in Arabidopsis thaliana. Plant Cell Physiol. 2019, 60, 285–302. [Google Scholar] [CrossRef]
- Zhu, M.; Chen, W.; Mirabet, V.; Hong, L.; Bovio, S.; Strauss, S.; Schwarz, E.M.; Tsugawa, S.; Wang, Z.; Smith, R.S.; et al. Robust organ size requires robust timing of initiation orchestrated by focused auxin and cytokinin signalling. Nat. Plants 2020, 6, 686–698. [Google Scholar] [CrossRef]
- Lee, W.J.; Truong, H.A.; Trinh, C.S.; Kim, J.H.; Lee, S.; Hong, S.W.; Lee, H. NITROGEN RESPONSE DEFICIENCY 1-mediated CHL1 induction contributes to optimized growth performance during altered nitrate availability in Arabidopsis. Plant J. 2020, 104, 1382–1398. [Google Scholar] [CrossRef] [PubMed]
- Peng, H.; Phung, J.; Zhai, Y.; Neff, M.M. Self-transcriptional repression of the Arabidopsis NAC transcription factor ATAF2 and its genetic interaction with phytochrome A in modulating seedling photomorphogenesis. Planta 2020, 252, 48. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, P.; Indoliya, Y.; Chauhan, A.S.; Pande, V.; Chakrabarty, D. Over-expression of rice R1-type MYB transcription factor confers different abiotic stress tolerance in transgenic Arabidopsis. Ecotoxicol. Environ. Saf. 2020, 206, 111361. [Google Scholar] [CrossRef] [PubMed]

Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, G.; Cao, A.; Wen, Y.; Bao, W.; She, F.; Wu, W.; Zheng, S.; Yang, N. Characteristics and Functions of MYB (v-Myb avivan myoblastsis virus oncogene homolog)-Related Genes in Arabidopsis thaliana. Genes 2023, 14, 2026. https://doi.org/10.3390/genes14112026
Wu G, Cao A, Wen Y, Bao W, She F, Wu W, Zheng S, Yang N. Characteristics and Functions of MYB (v-Myb avivan myoblastsis virus oncogene homolog)-Related Genes in Arabidopsis thaliana. Genes. 2023; 14(11):2026. https://doi.org/10.3390/genes14112026
Chicago/Turabian StyleWu, Guofan, Aohua Cao, Yuhan Wen, Wencheng Bao, Fawen She, Wangze Wu, Sheng Zheng, and Ning Yang. 2023. "Characteristics and Functions of MYB (v-Myb avivan myoblastsis virus oncogene homolog)-Related Genes in Arabidopsis thaliana" Genes 14, no. 11: 2026. https://doi.org/10.3390/genes14112026
APA StyleWu, G., Cao, A., Wen, Y., Bao, W., She, F., Wu, W., Zheng, S., & Yang, N. (2023). Characteristics and Functions of MYB (v-Myb avivan myoblastsis virus oncogene homolog)-Related Genes in Arabidopsis thaliana. Genes, 14(11), 2026. https://doi.org/10.3390/genes14112026




