Genetic Interactions in Various Environmental Conditions in Caenorhabditis elegans
Abstract
:1. Introduction
2. Materials and Methods
2.1. Query Genes
2.2. C. elegans Strains
2.3. RNA Interference and Bacteria Preparation
2.4. Worm Preparation
2.5. Fitness Assay
2.6. Assay Quality Control, Data Standardization, and Data Quality Control
2.7. S-Score Calculation
2.8. S-Score Significance
2.9. S-Score Distribution Comparison
2.10. Differential Epistasis Calculation
2.11. Sign Epistasis and Reciprocal Sign Epistasis
2.12. Data Availability
3. Results
3.1. Direction of Epistasis
3.2. Differential Epistasis
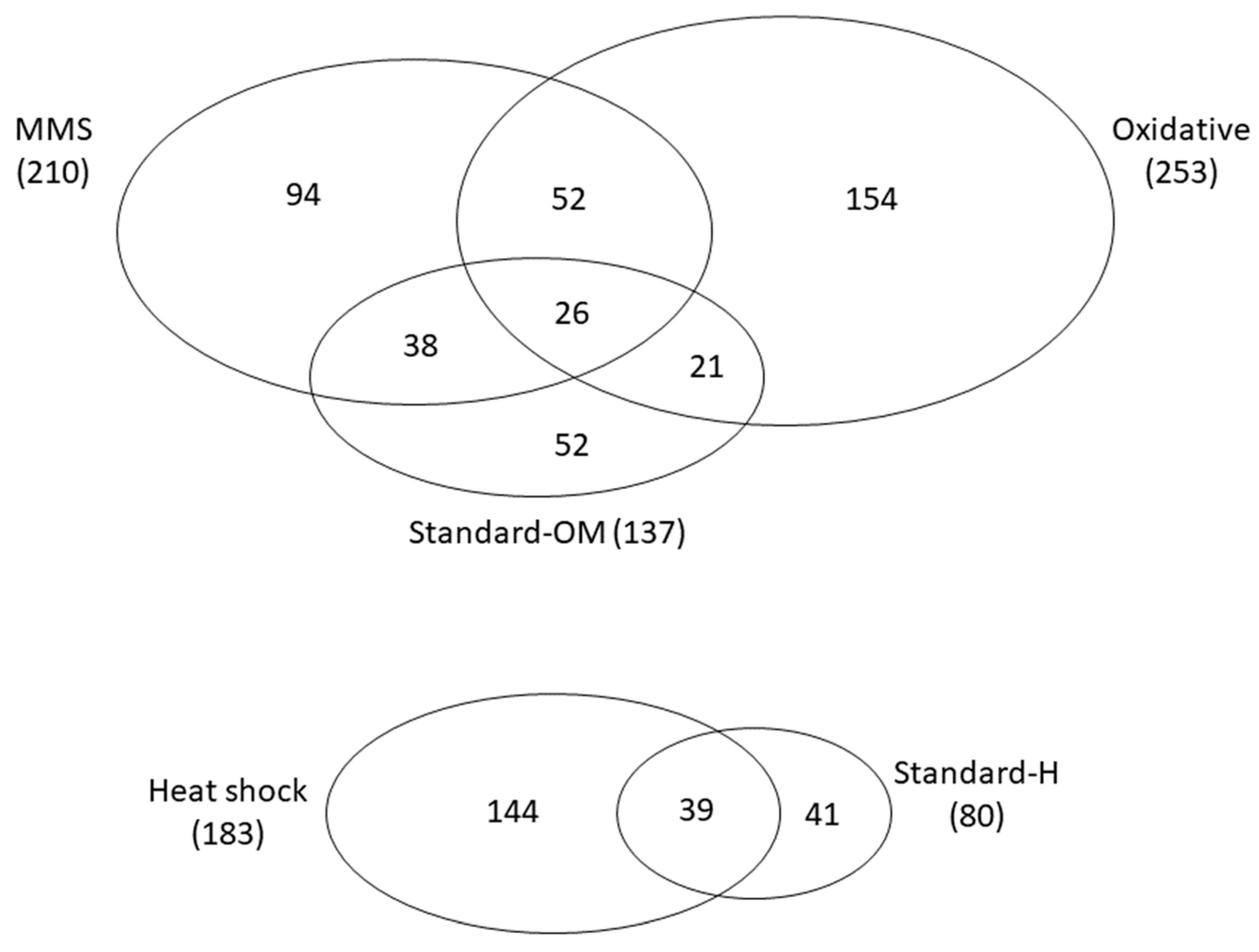
3.3. Persistence of Epistasis across Environments
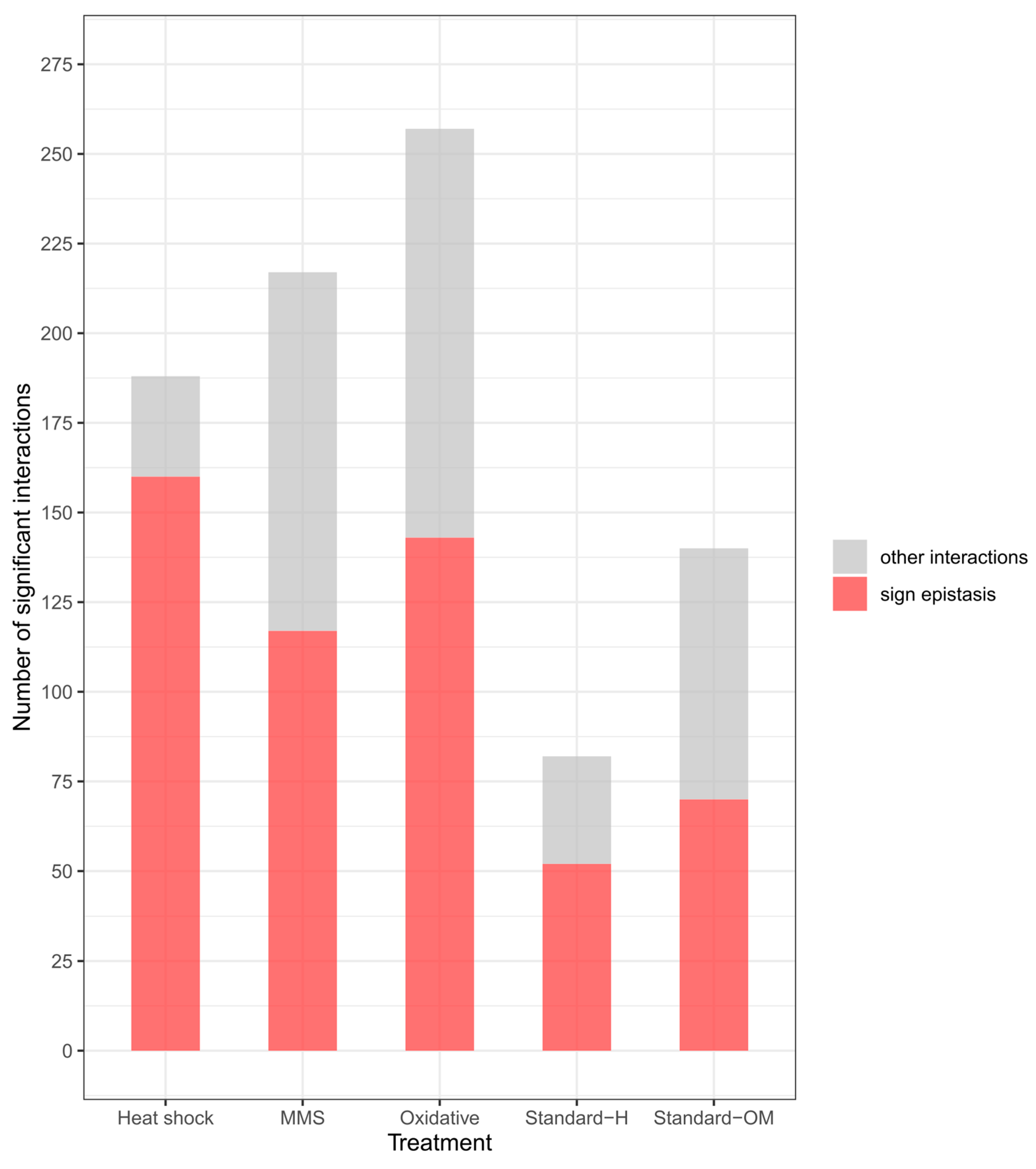
3.4. Sign and Reciprocal Sign Epistasis
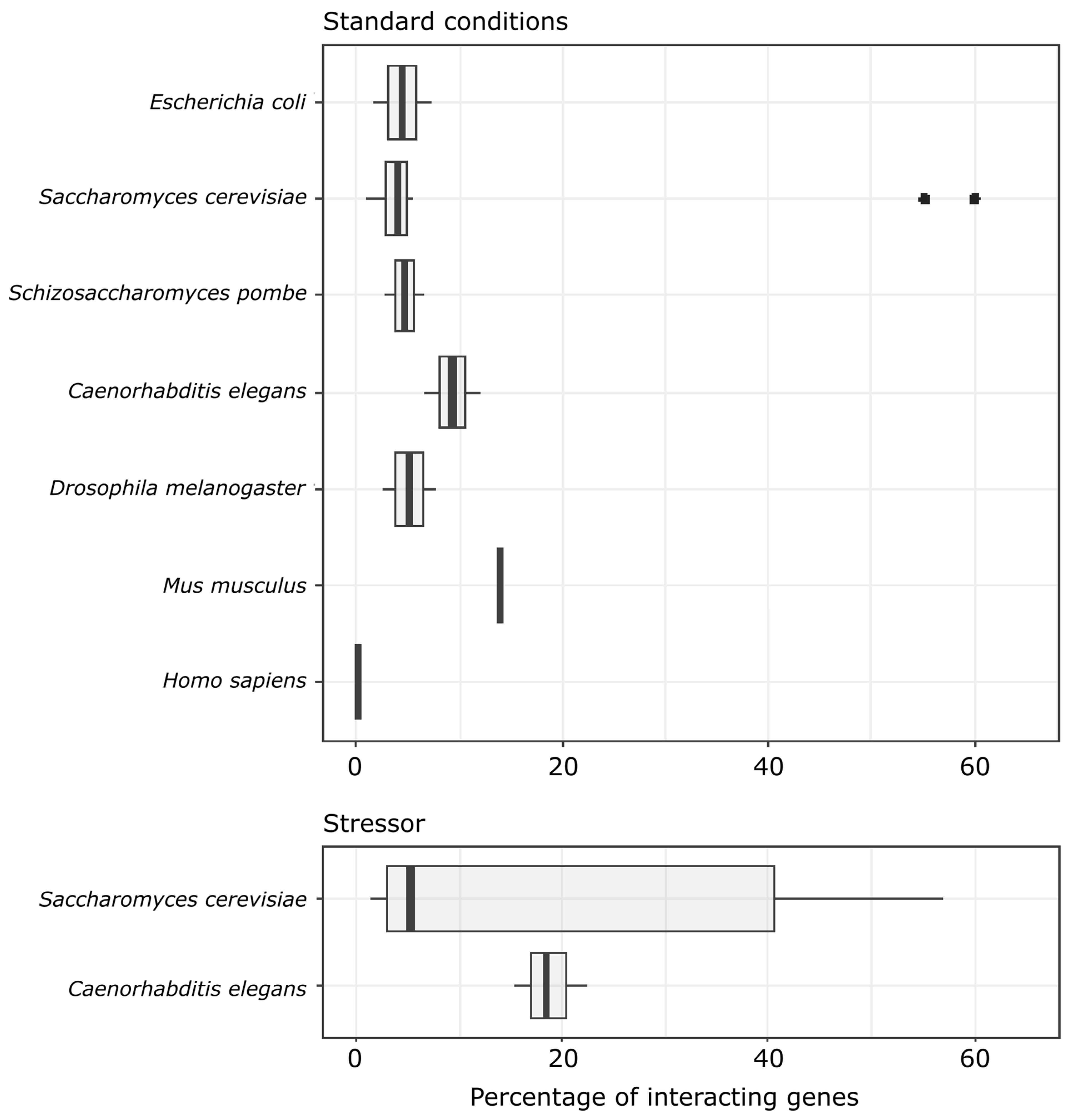
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Presgraves, D.C. Speciation Genetics: Epistasis, Conflict and the Origin of Species. Curr. Biol. 2007, 17, 125–127. [Google Scholar] [CrossRef] [PubMed]
- Butcher, D. Muller’s ratchet, epistasis and mutation effects. Genetics 1995, 141, 431–437. [Google Scholar] [CrossRef] [PubMed]
- Carter, A.J.R.; Hermisson, J.; Hansen, T.F. The role of epistatic gene interactions in the response to selection and the evolution of evolvability. Theor. Popul. Biol. 2005, 68, 179–196. [Google Scholar] [CrossRef] [PubMed]
- Jones, A.G.; Bürger, R.; Arnold, S.J. Epistasis and natural selection shape the mutational architecture of complex traits. Nat. Commun. 2014, 5, 3709. [Google Scholar] [CrossRef]
- Whitlock, M.C.; Philips, P.C. Multiple Fitness Peaks And Epistasis. Annu. Rev. Ecol. Evol. Syst. 1995, 26, 601–629. [Google Scholar] [CrossRef]
- Saltz, J.B.; Bell, A.M.; Flint, J.; Gomulkiewicz, R.; Hughes, K.A.; Keagy, J. Why does the magnitude of genotype-by-environment interaction vary? Ecol. Evol. 2018, 8, 6342–6353. [Google Scholar] [CrossRef]
- Onge, R.P.S.; Mani, R.; Oh, J.; Proctor, M.; Fung, E.; Davis, R.W.; Nislow, C.; Roth, F.P.; Giaever, G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007, 39, 199–206. [Google Scholar] [CrossRef]
- Jasnos, L.; Tomala, K.; Paczesniak, D.; Korona, R. Interactions between stressful environment and gene deletions alleviate the expected average loss of fitness in yeast. Genetics 2008, 178, 2105–2111. [Google Scholar] [CrossRef]
- Bandyopadhyay, S.; Mehta, M.; Kuo, D.; Sung, M.-K.; Chuang, R.; Jaehnig, E.J.; Bodenmiller, B.; Licon, K.; Copeland, W.; Shales, M.; et al. Rewiring of genetic networks in response to DNA damage. Science 2010, 330, 1385–1389. [Google Scholar] [CrossRef]
- Babu, M.; Díaz-Mejía, J.J.; Vlasblom, J.; Gagarinova, A.; Phanse, S.; Graham, C.; Yousif, F.; Ding, H.; Xiong, X.; Nazarians-Armavil, A.; et al. Genetic interaction maps in Escherichia coli reveal functional crosstalk among cell envelope biogenesis pathways. PLoS Genet. 2011, 7, e1002377. [Google Scholar] [CrossRef]
- Cheverud, J.M.; Routman, E.J. Epistasis as a source of increased additive genetic variance at population bottlenecks. Evolution 1996, 50, 1042–1051. [Google Scholar] [CrossRef] [PubMed]
- Weinreich, D.M. The rank ordering of genotypic fitness values predicts genetic constraint on natural selection on landscapes lacking sign epistasis. Genetics 2005, 171, 1397–1405. [Google Scholar] [CrossRef] [PubMed]
- Yukilevich, R.; Lachance, J.; Aoki, F.; True, J.R. Long-term adaptation of epistatic genetic networks. Evolution 2008, 62, 2215–2235. [Google Scholar] [CrossRef] [PubMed]
- Hansen, T.F. Why epistasis is important for selection and adaptation. Evolution 2013, 67, 3501–3511. [Google Scholar] [CrossRef] [PubMed]
- Barker, B.; Xu, L.; Gu, Z. Dynamic epistasis under varying environmental perturbations. PLoS ONE 2015, 10, e0114911. [Google Scholar] [CrossRef] [PubMed]
- Fiedler, D.; Braberg, H.; Mehta, M.; Chechik, G.; Cagney, G.; Mukherjee, P.; Silva, A.C.; Shales, M.; Collins, S.R.; van Wageningen, S.; et al. Functional Organization of the S. cerevisiae Phosphorylation Network. Cell 2009, 136, 952–963. [Google Scholar] [CrossRef]
- Collins, S.R.; Miller, K.M.; Maas, N.L.; Roguev, A.; Fillingham, J.; Chu, C.S.; Schuldiner, M.; Gebbia, M.; Recht, J.; Shales, M.; et al. Functional dissection of protein complexes involved in yeast chromosome biology using a genetic interaction map. Nature 2007, 446, 806–810. [Google Scholar] [CrossRef]
- Costanzo, M.; Baryshnikova, A.; Bellay, J.; Kim, Y.; Spear, E.D.; Sevier, C.S.; Ding, H.; Koh, J.L.Y.; Toufighi, K.; Mostafavi, S.; et al. The genetic landscape of a cell. Science 2010, 327, 425–431. [Google Scholar] [CrossRef]
- Horn, T.; Sandmann, T.; Fischer, B.; Axelsson, E.; Huber, W.; Boutros, M. Mapping of signaling networks through synthetic genetic interaction analysis by RNAi. Nat. Methods 2011, 8, 341–346. [Google Scholar] [CrossRef]
- Roguev, A.; Talbot, D.; Negri, G.L.; Shales, M.; Cagney, G.; Bandyopadhyay, S.; Panning, B.; Krogan, N.J. Quantitative genetic-interaction mapping in mammalian cells. Nat. Methods 2013, 10, 432–437. [Google Scholar] [CrossRef]
- Roguev, A.; Bandyopadhyay, S.; Zofall, M.; Zhang, K.; Fischer, T.; Collins, S.R.; Qu, H.; Shales, M.; Park, H.O.; Hayles, J.; et al. Conservation and rewiring of functional modules revealed by an epistasis map in fission yeast. Science 2008, 322, 405–410. [Google Scholar] [CrossRef] [PubMed]
- Kishony, R.; Leibler, S. Environmental stresses can alleviate the average deleterious effect of mutations. J. Biol. 2003, 2, 14. [Google Scholar] [CrossRef] [PubMed]
- Guénolé, A.; Srivas, R.; Vreeken, K.; Wang, Z.Z.; Wang, S.; Krogan, N.J.; Ideker, T.; van Attikum, H. Dissection of DNA Damage Responses Using Multiconditional Genetic Interaction Maps. Mol. Cell 2013, 49, 346–358. [Google Scholar] [CrossRef] [PubMed]
- Szafraniec, K.; Borts, R.H.; Korona, R. Environmental stress and mutational load in diploid strains of the yeast Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2001, 98, 1107–1112. [Google Scholar] [CrossRef] [PubMed]
- Rzezniczak, T.Z.; Merritt, T.J.S. Interactions of NADP-reducing enzymes across varying environmental conditions: A model of biological complexity. G3 Genes Genomes Genet. 2012, 2, 1613–1623. [Google Scholar] [CrossRef]
- Stiernagle, T. Maintenance of C. elegans; University of Minnesota: Minneapolis, MN, USA, 2006; pp. 1–11. [Google Scholar] [CrossRef]
- Fire, A.; Xu, S.; Montgomery, M.K.; Kostas, S.A.; Driver, S.E.; Mello, C.C. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 1998, 391, 806–811. [Google Scholar] [CrossRef]
- Elvin, M.; Snoek, L.B.; Frejno, M.; Klemstein, U.; Kammenga, J.E.; Poulin, G.B. A fitness assay for comparing RNAi effects across multiple C. elegans genotypes. BMC Genom. 2011, 12, 510. [Google Scholar] [CrossRef]
- Zhang, X.D.; Ferrer, M.; Espeseth, A.S.; Marine, S.D.; Stec, E.M.; Crackower, M.A.; Holder, D.J.; Heyse, J.F.; Strulovici, B. The use of strictly standardized mean difference for hit selection in primary RNA interference high-throughput screening experiments. J. Biomol. Screen. 2007, 12, 497–509. [Google Scholar] [CrossRef]
- Labocha, M.K.; Yuan, W.; Aleman-Meza, B.; Zhong, W. A strategy to apply quantitative epistasis analysis on developmental traits. BMC Genet. 2017, 18, 42. [Google Scholar] [CrossRef]
- Zhang, X.D. A method for effectively comparing gene effects in multiple conditions in RNAi and expression-profiling research. Pharmacogenomics 2009, 10, 345–358. [Google Scholar] [CrossRef]
- Pastore, M. Overlapping: A R package for Estimating Overlapping in Empirical Distributions. J. Open Source Softw. 2018, 3, 1023. [Google Scholar] [CrossRef]
- Sanjuán, R.; Elena, S.F. Epistasis correlates to genomic complexity. Proc. Natl. Acad. Sci. USA 2006, 103, 14402–14405. [Google Scholar] [CrossRef]
- Gaertner, B.E.; Parmenter, M.D.; Rockman, M.V.; Kruglyak, L.; Phillips, P.C. More than the sum of its parts: A complex epistatic network underlies natural variation in thermal preference behavior in Caenorhabditis elegans. Genetics 2012, 192, 1533–1542. [Google Scholar] [CrossRef] [PubMed]
- Matsui, T.; Mullis, M.N.; Roy, K.R.; Hale, J.J.; Schell, R.; Levy, S.F.; Ehrenreich, I.M. The interplay of additivity, dominance, and epistasis on fitness in a diploid yeast cross. Nat. Commun. 2022, 13, 1463. [Google Scholar] [CrossRef] [PubMed]
- Jaffe, M.; Dziulko, A.; Smith, J.D.; St. Onge, R.P.; Levy, S.F.; Sherlock, G. Improved discovery of genetic interactions using CRISPRiSeq across multiple environments. Genome Res. 2019, 29, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Martin, H.; Shales, M.; Fernandez-Piñar, P.; Wei, P.; Molina, M.; Fiedler, D.; Shokat, K.M.; Beltrao, P.; Lim, W.; Krogan, N.J. Differential genetic interactions of yeast stress response MAPK pathways. Mol. Syst. Biol. 2015, 11, 800. [Google Scholar] [CrossRef]
- Pang, Y.L.J.; Abo, R.; Levine, S.S.; Dedon, P.C. Diverse cell stresses induce unique patterns of tRNA up- and down-regulation: tRNA-seq for quantifying changes in tRNA copy number. Nucleic Acids Res. 2014, 42, e170. [Google Scholar] [CrossRef]
- Feder, M.E.; Hofmann, G.E. Heat-shock proteins, molecular chaperones, and the stress response: Evolutionary and ecological physiology. Annu. Rev. Physiol. 1999, 61, 243–282. [Google Scholar] [CrossRef]
- Hillenmeyer, M.E.; Fung, E.; Wildenhain, J. The chemical genomic portrait of yeast: Uncovering a phenotype for all genes. Chemtracts 2008, 20, 503–504. [Google Scholar] [CrossRef]
- Mäki-Tanila, A.; Hill, W.G. Influence of gene interaction on complex trait variation with multilocus models. Genetics 2014, 198, 355–367. [Google Scholar] [CrossRef]
- Barton, N.H. How does epistasis influence the response to selection? Heredity 2017, 118, 96–109. [Google Scholar] [CrossRef] [PubMed]
- Fraisse, C.; Welch, J.J. The distribution of epistasis on simple fitness landscapes. bioRxiv 2018, 490573. [Google Scholar] [CrossRef] [PubMed]
- Paixão, T.; Barton, N.H. The effect of gene interactions on the long-term response to selection. Proc. Natl. Acad. Sci. USA 2016, 113, 4422–4427. [Google Scholar] [CrossRef] [PubMed]
- Forneris, N.S.; Vitezica, Z.G.; Legarra, A.; Pérez-Enciso, M. Influence of epistasis on response to genomic selection using complete sequence data. Genet. Sel. Evol. 2017, 49, 66. [Google Scholar] [CrossRef]
- Dagilis, A.J.; Kirkpatrick, M.; Bolnick, D.I. The evolution of hybrid fitness during speciation. PLoS Genet. 2019, 15, e1008125. [Google Scholar] [CrossRef]
- Guerrero, R.F.; Muir, C.D.; Josway, S.; Moyle, L.C. Pervasive antagonistic interactions among hybrid incompatibility loci. PLoS Genet. 2017, 13, e1006817. [Google Scholar] [CrossRef]
- Johnson, N.A. Speciation: Dobzhansky–Muller incompatibilities, dominance and gene interactions. Trends Ecol. Evol. 2000, 15, 480–482. [Google Scholar] [CrossRef]
- Poelwijk, F.J.; Tǎnase-Nicola, S.; Kiviet, D.J.; Tans, S.J. Reciprocal sign epistasis is a necessary condition for multi-peaked fitness landscapes. J. Theor. Biol. 2011, 272, 141–144. [Google Scholar] [CrossRef]
- Silva, R.F.; MendonçaSí, S.C.M.; CarvalhoLuí, L.M.; Reis, A.M.; Gordo, I.; Trindade, S.; Dionisio, F. Pervasive sign epistasis between conjugative plasmids and Drug-Resistance chromosomal mutations. PLoS Genet. 2011, 7, e1002181. [Google Scholar] [CrossRef]
- Lali, J.; Elena, S.F. Magnitude and sign epistasis among deleterious mutations in a positive-sense plant RNA virus. Heredity 2012, 109, 71–77. [Google Scholar] [CrossRef]
- Ono, J.; Gerstein, A.C.; Otto, S.P. Widespread Genetic Incompatibilities between First-Step Mutations during Parallel Adaptation of Saccharomyces cerevisiae to a Common Environment. PLoS Biol. 2017, 23, e1002591. [Google Scholar] [CrossRef] [PubMed]
- Bendixsen, D.P.; Pollock, T.B.; Peri, G.; Hayden, E.J. Experimental Resurrection of Ancestral Mammalian CPEB3 Ribozymes Reveals Deep Functional Conservation. Mol. Biol. Evol. 2021, 38, 2843–2853. [Google Scholar] [CrossRef] [PubMed]
- Butland, G.; Babu, M.; Díaz-Mejía, J.J.; Bohdana, F.; Phanse, S.; Gold, B.; Yang, W.; Li, J.; Gagarinova, A.G.; Pogoutse, O.; et al. eSGA: E. coli synthetic genetic array analysis. Nat. Methods 2008, 5, 789–795. [Google Scholar] [CrossRef] [PubMed]
- Schuldiner, M.; Collins, S.R.; Thompson, N.J.; Denic, V.; Bhamidipati, A.; Punna, T.; Ihmels, J.; Andrews, B.; Boone, C.; Greenblatt, J.F.; et al. Exploration of the Function and Organization of the Yeast Early Secretory Pathway through an Epistatic Miniarray Profile. Cell 2005, 123, 507–519. [Google Scholar] [CrossRef]
- Szappanos, B.; Kovács, K.; Szamecz, B.; Honti, F.; Costanzo, M.; Baryshnikova, A.; Gelius-Dietrich, G.; Lercher, M.J.; Jelasity, M.; Myers, C.L.; et al. An integrated approach to characterize genetic interaction networks in yeast metabolism. Nat. Genet. 2011, 43, 656–662. [Google Scholar] [CrossRef]
- Gutin, J.; Sadeh, A.; Rahat, A.; Aharoni, A.; Friedman, N. Condition-specific genetic interaction maps reveal crosstalk between the cAMP / PKA and the HOG MAPK pathways in the activation of the general stress response. Mol. Syst. Biol. 2015, 11, 829. [Google Scholar] [CrossRef]
- Ryan, C.J.; Roguev, A.; Patrick, K.; Xu, J.; Jahari, H.; Tong, Z.; Beltrao, P.; Shales, M.; Qu, H.; Collins, S.R.; et al. Hierarchical Modularity and the Evolution of Genetic Interactomes across Species. Mol. Cell 2012, 46, 691–704. [Google Scholar] [CrossRef]
- Bakal, C.; Linding, R.; Llense, F.; Heffern, E.; Martin-Blanco, E.; Pawson, T.; Perrimon, N. Phosphorylation Networks Regulating JNK Activity in Diverse Genetic Backgrounds. Science 2008, 322, 453–456. [Google Scholar] [CrossRef]
- Fischer, B.; Sandmann, T.; Horn, T.; Billmann, M.; Chaudhary, V.; Huber, W.; Boutros, M. A map of directional genetic interactions in a metazoan cell. eLife 2015, 4, e05464. [Google Scholar] [CrossRef]
- Laufer, C.; Fischer, B.; Billmann, M.; Huber, W.; Boutros, M. Mapping genetic interactions in human cancer cells with RNAi and multiparametric phenotyping. Nat. Methods 2013, 10, 427–431. [Google Scholar] [CrossRef]
- Billmann, M.; Chaudhary, V.; ElMaghraby, M.F.; Fischer, B.; Boutros, M. Widespread Rewiring of Genetic Networks upon Cancer Signaling Pathway Activation. Cell Syst. 2017, 6, 52–64.e4. [Google Scholar] [CrossRef] [PubMed]



Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Toch, K.; Buczek, M.; Labocha, M.K. Genetic Interactions in Various Environmental Conditions in Caenorhabditis elegans. Genes 2023, 14, 2080. https://doi.org/10.3390/genes14112080
Toch K, Buczek M, Labocha MK. Genetic Interactions in Various Environmental Conditions in Caenorhabditis elegans. Genes. 2023; 14(11):2080. https://doi.org/10.3390/genes14112080
Chicago/Turabian StyleToch, Katarzyna, Mateusz Buczek, and Marta K. Labocha. 2023. "Genetic Interactions in Various Environmental Conditions in Caenorhabditis elegans" Genes 14, no. 11: 2080. https://doi.org/10.3390/genes14112080
APA StyleToch, K., Buczek, M., & Labocha, M. K. (2023). Genetic Interactions in Various Environmental Conditions in Caenorhabditis elegans. Genes, 14(11), 2080. https://doi.org/10.3390/genes14112080





