Statistical Dissection of the Genetic Determinants of Phenotypic Heterogeneity in Genes with Multiple Associated Rare Diseases
Abstract
1. Introduction
2. Materials and Methods
2.1. Data Collection
2.2. Gene-Set Enrichment Analysis
2.3. Phenotypic Similarity Analysis
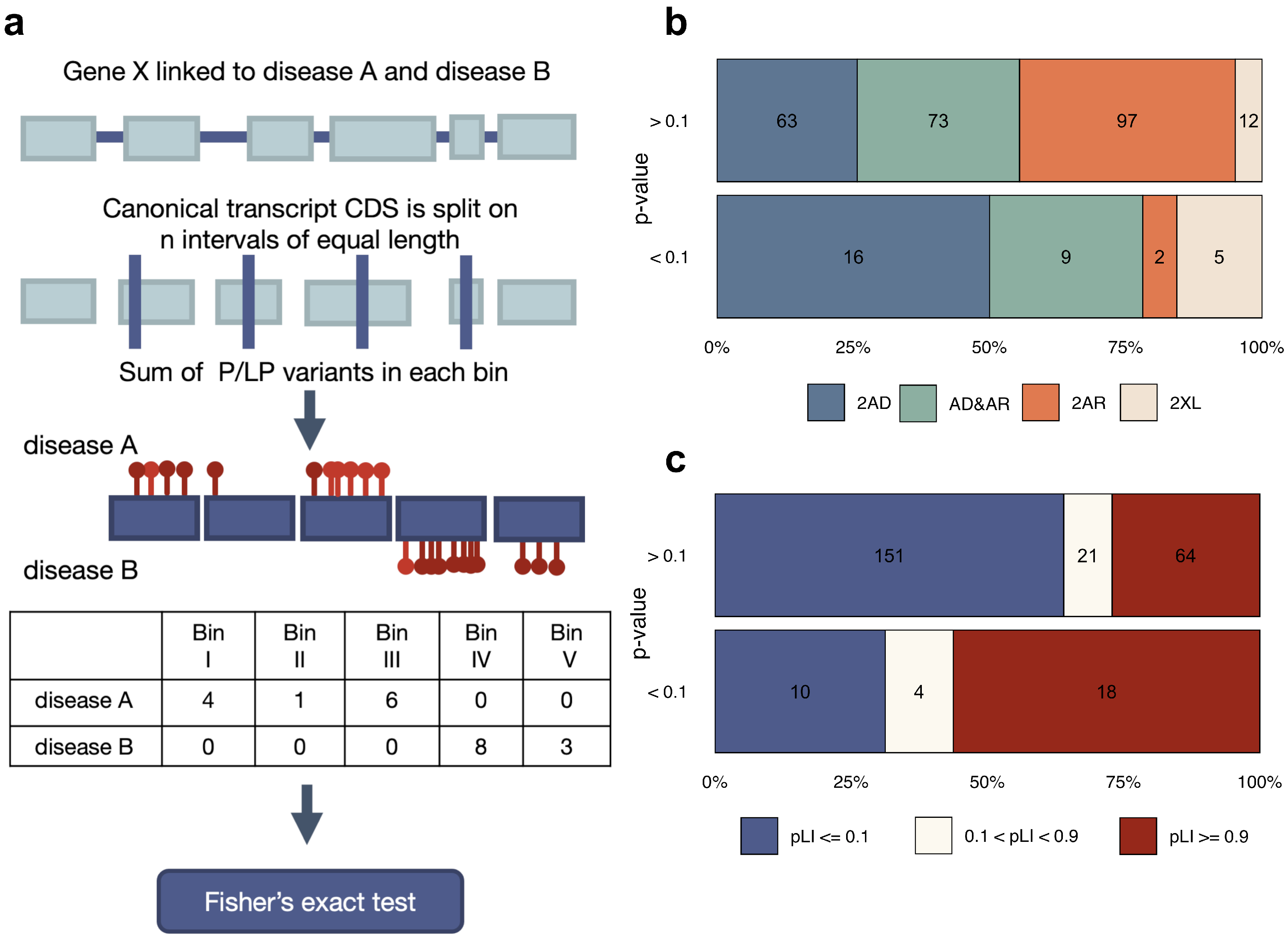
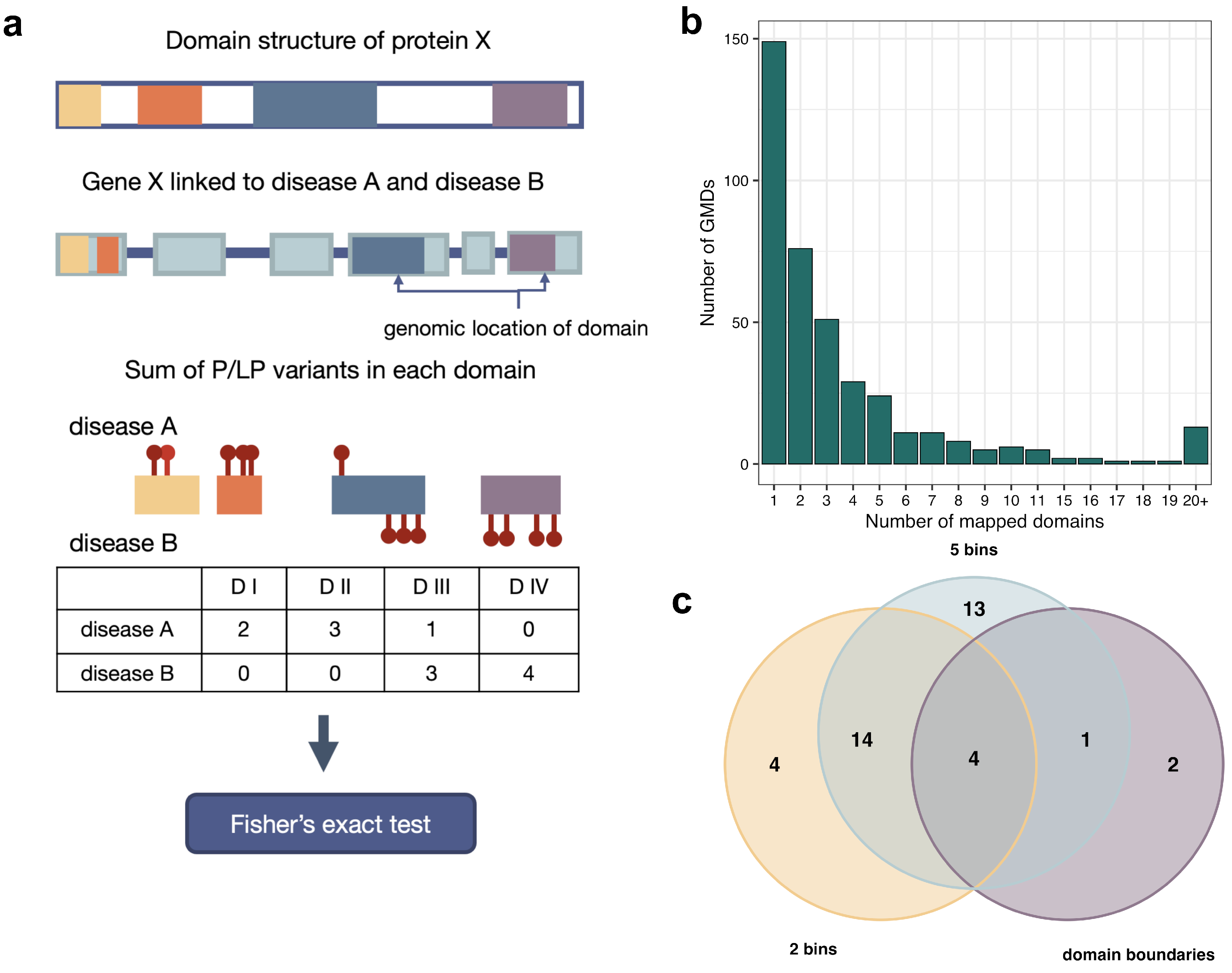
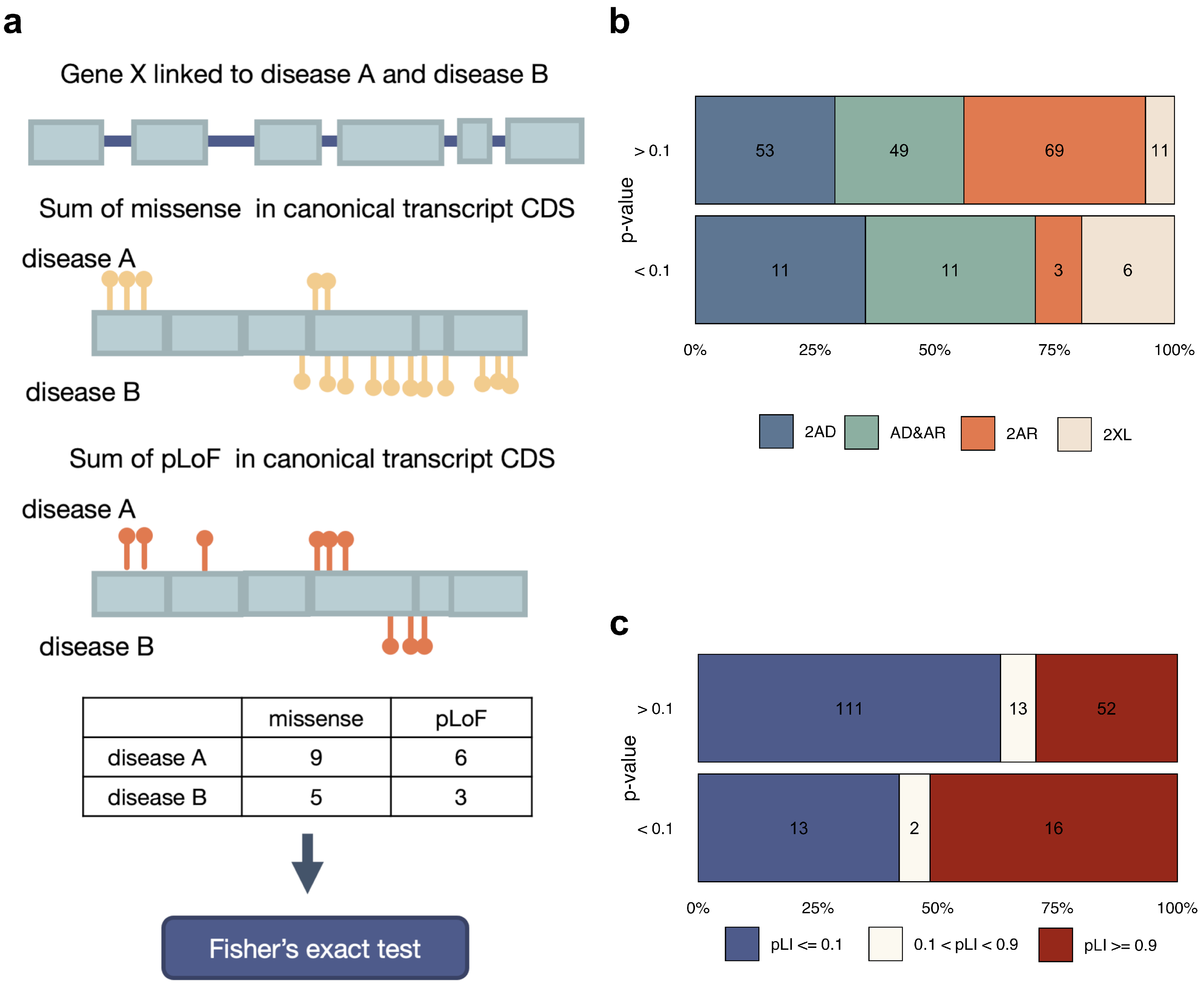
2.4. Statistical Analysis of Within-Gene Distribution and Type of Variants
2.5. Data and Code Availability
3. Results
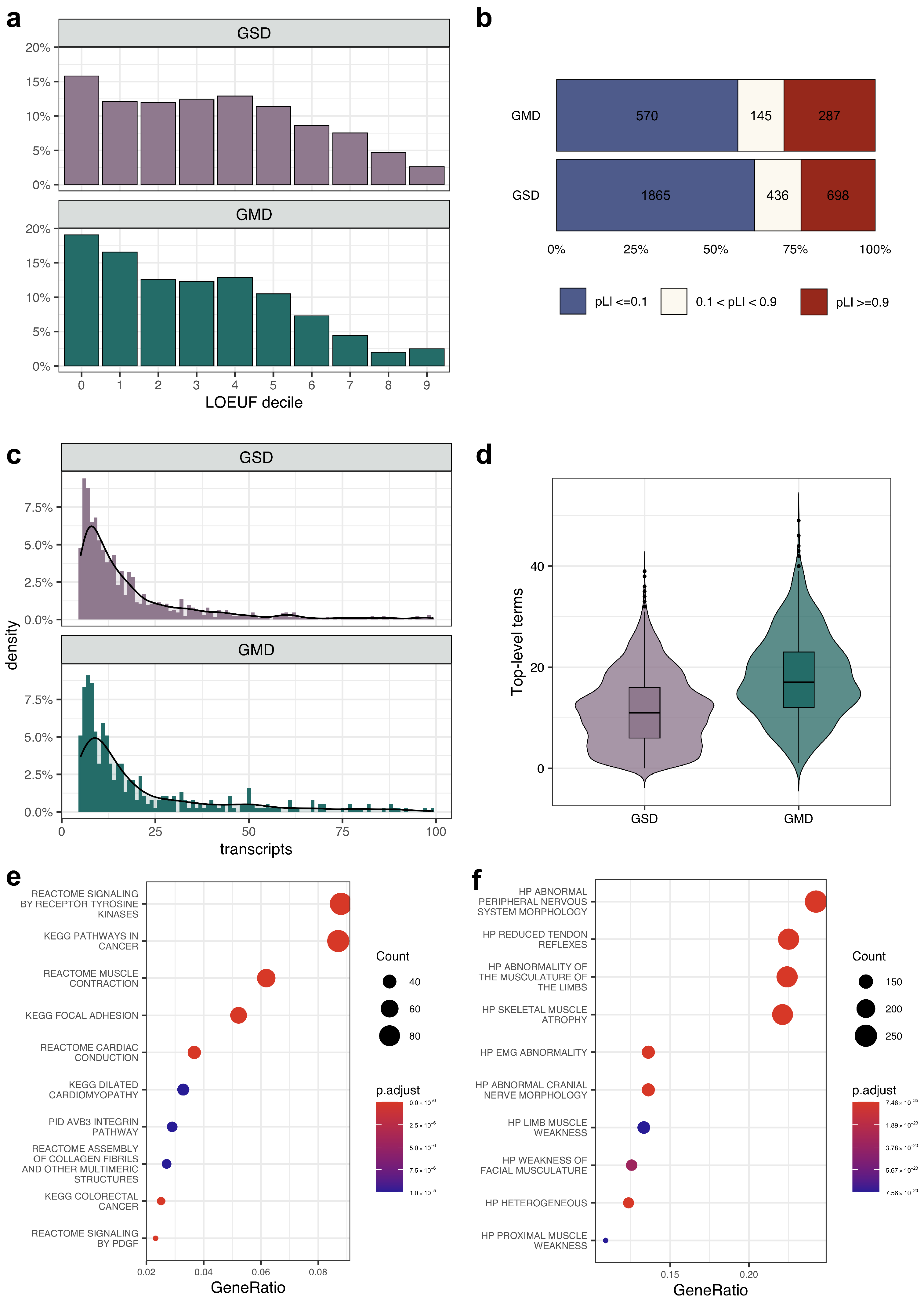
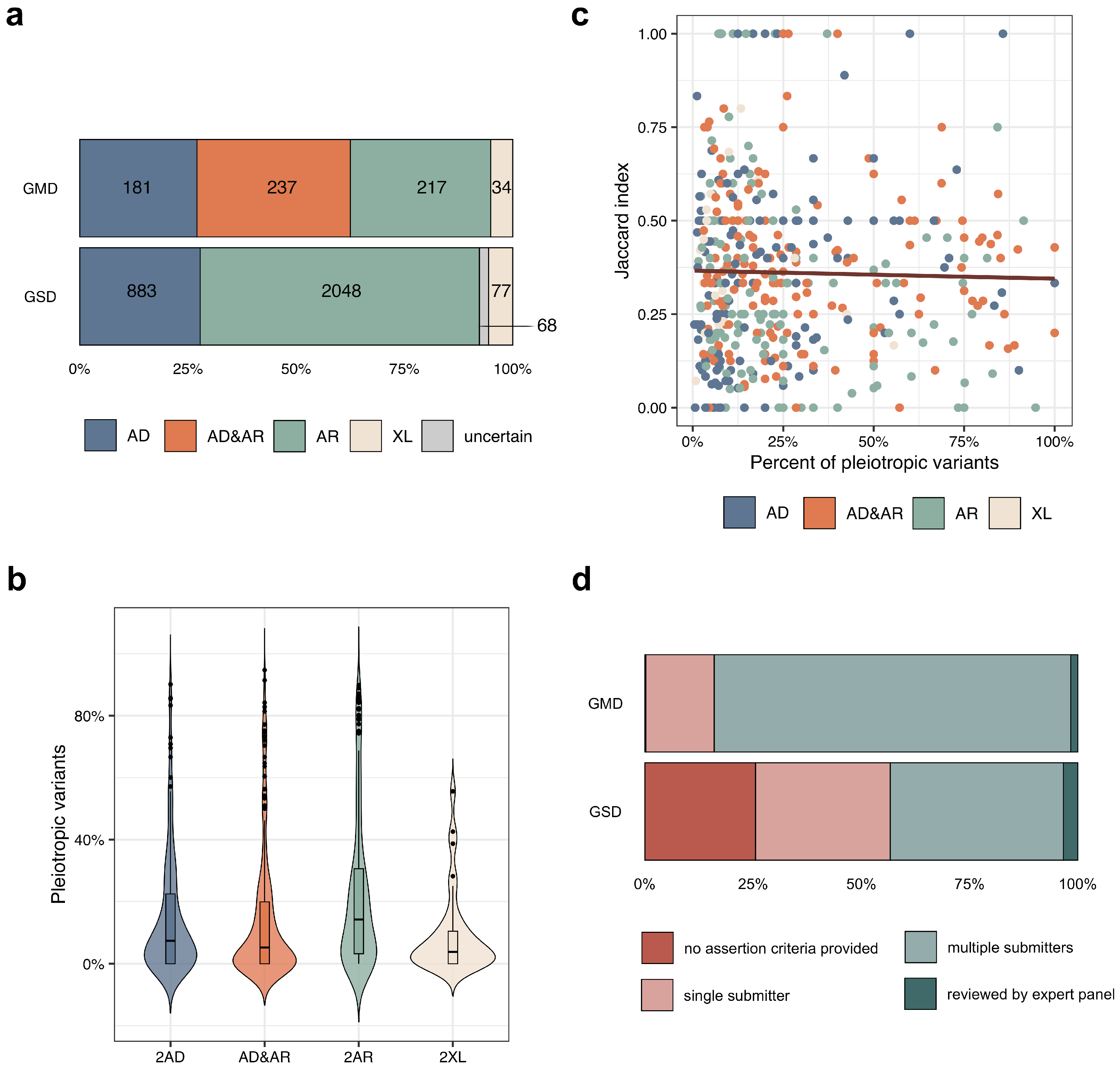
3.1. Common Properties of Genes Linked to Multiple Rare Diseases
3.2. The Effects of Variant Localization and Type on the Phenotypic Outcome
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| HPO | Human Phenotype Ontology |
| NGS | next-generation sequencing |
| OMIM | Online Mendelian Inheritance in Man |
| P/LP | pathogenic/likely pathogenic variants |
| pLoF | predicted loss-of-function variant |
| WES | whole-exome sequencing |
| WGS | whole-genome sequencing |
References
- Wright, M.; Menon, V.; Taylor, L.; Shashidharan, M.; Westercamp, T.; Ternent, C.A. Factors predicting reclassification of variants of unknown significance. Am. J. Surg. 2018, 216, 1148–1154. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef]
- Nykamp, K.; Anderson, M.; Powers, M.; Garcia, J.; Herrera, B.; Ho, Y.Y.; Kobayashi, Y.; Patil, N.; Thusberg, J.; Westbrook, M.; et al. Sherloc: A comprehensive refinement of the ACMG-AMP variant classification criteria. Genet. Med. 2017, 19, 1105–1117. [Google Scholar] [CrossRef]
- Biesecker, L.G.; Harrison, S. Clinical Genome Resource Sequence Variant Interpretation Working Group. Available online: https://clinicalgenome.org/working-groups/sequence-variant-interpretation/ (accessed on 29 May 2023).
- Weatherall, D.J. Phenotype—Genotype relationships in monogenic disease: Lessons from the thalassaemias. Nat. Rev. Genet. 2001, 2, 245–255. [Google Scholar] [CrossRef] [PubMed]
- Mestroni, L. Phenotypic Heterogeneity of Sarcomeric Gene Mutations. J. Am. Coll. Cardiol. 2009, 54, 343–345. [Google Scholar] [CrossRef][Green Version]
- Brennan, M.L.; Schrijver, I. Cystic Fibrosis: A Review of Associated Phenotypes, Use of Molecular Diagnostic Approaches, Genetic Characteristics, Progress, and Dilemmas. J. Mol. Diagn. 2016, 18, 3–14. [Google Scholar] [CrossRef]
- Martínez-Campelo, L.; Cruz, R.; Blanco-Verea, A.; Moscoso, I.; Ramos-Luis, E.; Lage, R.; Álvarez-Barredo, M.; Sabater-Molina, M.; Peñafiel-Verdú, P.; Jiménez-Jáimez, J.; et al. Searching for genetic modulators of the phenotypic heterogeneity in Brugada syndrome. PLoS ONE 2022, 17, e0263469. [Google Scholar] [CrossRef] [PubMed]
- Dubik, N.; Mai, S. Lamin A/C: Function in normal and tumor cells. Cancers 2020, 12, 3688. [Google Scholar] [CrossRef]
- Thaxton, C.; Goldstein, J.; DiStefano, M.; Wallace, K.; Witmer, P.D.; Haendel, M.A.; Hamosh, A.; Rehm, H.L.; Berg, J.S. Lumping versus splitting: How to approach defining a disease to enable accurate genomic curation. Cell Genom. 2022, 2, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Zschocke, J.; Byers, P.H.; Wilkie, A.O.M. nature reviews genetics Mendelian inheritance revisited: Dominance and recessiveness in medical genetics. Nat. Rev. Genet. 2023, 24, 442–463. [Google Scholar] [CrossRef]
- Kingdom, R.; Wright, C.F. Incomplete Penetrance and Variable Expressivity: From Clinical Studies to Population Cohorts. Front. Genet. 2022, 13, 920390. [Google Scholar] [CrossRef] [PubMed]
- Martinelli, S.; Krumbach, O.H.; Pantaleoni, F.; Coppola, S.; Amin, E.; Pannone, L.; Nouri, K.; Farina, L.; Dvorsky, R.; Lepri, F.; et al. Functional Dysregulation of CDC42 Causes Diverse Developmental Phenotypes. Am. J. Hum. Genet. 2018, 102, 309–320. [Google Scholar] [CrossRef]
- Leen, W.G.; Klepper, J.; Verbeek, M.M.; Leferink, M.; Hofste, T.; van Engelen, B.G.; Wevers, R.A.; Arthur, T.; Bahi-Buisson, N.; Ballhausen, D.; et al. Glucose transporter-1 deficiency syndrome: The expanding clinical and genetic spectrum of a treatable disorder. Brain 2010, 39, 133–655. [Google Scholar] [CrossRef] [PubMed]
- Köhler, S.; Gargano, M.; Matentzoglu, N.; Carmody, L.C.; Lewis-Smith, D.; Vasilevsky, N.A.; Danis, D.; Balagura, G.; Baynam, G.; Brower, A.M.; et al. The Human Phenotype Ontology in 2021. Nucleic Acids Res. 2020, 49, D1207–D1217. [Google Scholar] [CrossRef]
- Karczewski, K.J.; Francioli, L.C.; Tiao, G.; Cummings, B.B.; Alföldi, J.; Wang, Q.; Collins, R.L.; Laricchia, K.M.; Ganna, A.; Birnbaum, D.P.; et al. The mutational constraint spectrum quantified from variation in 141,456 humans, Genome Aggregation Database Consortium. Nature 2020, 581, 19. [Google Scholar] [CrossRef] [PubMed]
- Lonsdale, J.; Thomas, J.; Salvatore, M.; Phillips, R.; Lo, E.; Shad, S.; Hasz, R.; Walters, G.; Garcia, F.; Young, N.; et al. The genotype-tissue expression (GTEx) project. Nat. Genet. 2013, 45, 580–585. [Google Scholar] [CrossRef]
- Landrum, M.J.; Lee, J.M.; Benson, M.; Brown, G.R.; Chao, C.; Chitipiralla, S.; Gu, B.; Hart, J.; Hoffman, D.; Jang, W.; et al. ClinVar: Improving access to variant interpretations and supporting evidence. Nucleic Acids Res. 2018, 46, D1062–D1067. [Google Scholar] [CrossRef]
- Yu, G.; Wang, L.G.; Han, Y.; He, Q.Y. ClusterProfiler: An R package for comparing biological themes among gene clusters. OMICS J. Integr. Biol. 2012, 16, 284–287. [Google Scholar] [CrossRef]
- Cunningham, F.; Allen, J.E.; Allen, J.; Alvarez-Jarreta, J.; Amode, M.R.; Armean, I.M.; Austine-Orimoloye, O.; Azov, A.G.; Barnes, I.; Bennett, R.; et al. Ensembl 2022. Nucleic Acids Res. 2022, 50, D988–D995. [Google Scholar] [CrossRef]
- Kent, W.J.; Zweig, A.S.; Barber, G.; Hinrichs, A.S.; Karolchik, D. BigWig and BigBed: Enabling browsing of large distributed datasets. Bioinformatics 2010, 26, 2204–2207. [Google Scholar] [CrossRef]
- Miller, M.P.; Parker, J.D.; Rissing, S.W.; Kumar, S. Quantifying the Intragenic Distribution of Human Disease Mutations. Ann. Hum. Genet. 2003, 67, 567–579. [Google Scholar] [CrossRef] [PubMed]
- Turner, T.N.; Douville, C.; Kim, D.; Stenson, P.D.; Cooper, D.N.; Chakravarti, A.; Karchin, R. Proteins linked to autosomal dominant and autosomal recessive disorders harbor characteristic rare missense mutation distribution patterns. Hum. Mol. Genet. 2015, 24, 5995–6002. [Google Scholar] [CrossRef] [PubMed]
- Cerrone, M.; Remme, C.A.; Tadros, R.; Bezzina, C.R.; Delmar, M. Beyond the one gene-one disease paradigm complex genetics and pleiotropy in inheritable cardiac disorders. Circulation 2019, 140, 595–610. [Google Scholar] [CrossRef]
- Magrinelli, F.; Balint, B.; Bhatia, K.P. Challenges in Clinicogenetic Correlations: One Gene—Many Phenotypes. Mov. Disord. Clin. Pract. 2021, 8, 299–310. [Google Scholar] [CrossRef] [PubMed]
- Gosu, V.; Won, K.; Oh, J.D.; Shin, D. Conformational changes induced by S34Y and R98C variants in the death domain of Myd88. Front. Mol. Biosci. 2020, 7, 27. [Google Scholar] [CrossRef]
- Quinodoz, M.; Peter, V.G.; Cisarova, K.; Royer-Bertrand, B.; Stenson, P.D.; Cooper, D.N.; Unger, S.; Superti-Furga, A.; Rivolta, C. Analysis of missense variants in the human genome reveals widespread gene-specific clustering and improves prediction of pathogenicity. Am. J. Hum. Genet. 2022, 109, 457–470. [Google Scholar] [CrossRef]
- Turkunova, M.E.; Barbitoff, Y.A.; Serebryakova, E.A.; Polev, D.E.; Berseneva, O.S.; Bashnina, E.B.; Baranov, V.S.; Glotov, O.S.; Glotov, A.S. Molecular Genetics and Pathogenesis of the Floating Harbor Syndrome: Case Report of Long-Term Growth Hormone Treatment and a Literature Review. Front. Genet. 2022, 13, 846101. [Google Scholar] [CrossRef]
- Rots, D.; Chater-Diehl, E.; Dingemans, A.J.; Goodman, S.J.; Siu, M.T.; Cytrynbaum, C.; Choufani, S.; Hoang, N.; Walker, S.; Awamleh, Z.; et al. Truncating SRCAP variants outside the Floating-Harbor syndrome locus cause a distinct neurodevelopmental disorder with a specific DNA methylation signature. Am. J. Hum. Genet. 2021, 108, 1053–1068. [Google Scholar] [CrossRef]
- Arboleda, V.A.; Lee, H.; Parnaik, R.; Fleming, A.; Banerjee, A.; de Souza, B.F.; Délot, E.C.; Rodriguez-Fernandez, I.A.; Braslavsky, D.; Bergadá, I.; et al. Mutations in the PCNA-binding domain of CDKN1C cause IMAGe syndrome. Nat. Genet. 2012, 44, 788–792. [Google Scholar] [CrossRef]
- Peterson, T.A.; Park, D.; Kann, M.G. A protein domain-centric approach for the comparative analysis of human and yeast phenotypically relevant mutations. BMC Genom. 2013, 14, S5. [Google Scholar] [CrossRef]
| Gene | Associated Disease | OMIM Identifier | CDS Quintiles with Causal Variants Prevalence † |
|---|---|---|---|
| CDKN1C | Beckwith–Wiedemann syndrome | OMIM:130650 | II, IV, V |
| IMAGE syndrome | OMIM:614732 | I | |
| CREBBP | Rubinstein–Taybi syndrome 1 | OMIM:180849 | II, III, V |
| Menke–Hennekam syndrome 1 | OMIM:618332 | II | |
| DEAF1 ‡ | Neurodevelopmental disorder with hypotonia, impaired expressive language, and with or without seizures | OMIM:617171 | II, IV |
| Vulto–van Silfout–de Vries syndrome | OMIM:615828 | III | |
| FLT4 ‡ | Lymphatic malformation 1 | OMIM:153100 | I, II |
| Congenital heart defects, multiple types, 7 | OMIM:618780 | I, IV | |
| FN1 ‡ | Glomerulopathy with fibronectin deposits 2 | OMIM:601894 | II |
| Spondylometaphyseal dysplasia, corner fracture type | OMIM:184255 | V | |
| F8 ‡ | Hemophilia A | OMIM:306700 | I, V |
| Thrombophilia 13, X-linked, due to factor VIII defect | OMIM:301071 | I | |
| KMT2D ‡ | Branchial arch abnormalities, choanal atresia, athelia, hearing loss, and hypothyroidism syndrome | OMIM:620186 | II |
| Kabuki syndrome 1 | OMIM:147920 | I, II, IV | |
| MAF ‡ | Ayme-Gripp syndrome | OMIM:601088 | II, V |
| Cataract 21, multiple types | OMIM:610202 | II | |
| NFIX ‡ | Marshall-Smith syndrome | OMIM:602535 | I, IV |
| Malan syndrome | OMIM:614753 | I, II | |
| OCRL | Dent disease 2 | OMIM:300555 | I, III |
| Lowe syndrome | OMIM:309000 | II, III | |
| SETBP1 ‡ | Intellectual developmental disorder, autosomal dominant 29 | OMIM:616078 | II |
| Schinzel-Giedion midface retraction syndrome | OMIM:269150 | III | |
| SPTB | Spherocytosis type 2 | OMIM:616649 | II, IV, V |
| Elliptocytosis-3 | OMIM:617948 | I | |
| SRCAP ‡ | Developmental delay, hypotonia, musculoskeletal defects, and behavioral abnormalities | OMIM:619595 | - |
| Floating-Harbor syndrome | OMIM:136140 | IV |
| Gene | Associated Disease | OMIM Identifier | Causal Variant Count | |
|---|---|---|---|---|
| Missense | pLoF | |||
| CAPN3 | Muscular dystrophy, limb-girdle, autosomal recessive 1 | OMIM:253600 | 67 | 109 |
| Muscular dystrophy, limb-girdle, autosomal dominant 4 | OMIM:618129 | 15 | 3 | |
| CDKN1C | Beckwith–Wiedemann syndrome | OMIM:130650 | 5 | 55 |
| IMAGE syndrome | OMIM:614732 | 7 | 1 | |
| COL4A3 | Alport syndrome 3, autosomal dominant | OMIM:104200 | 68 | 23 |
| Alport syndrome 2, autosomal recessive | OMIM:203780 | 51 | 65 | |
| CREBBP | Rubinstein–Taybi syndrome 1 | OMIM:180849 | 8 | 76 |
| Menke–Hennekam syndrome 1 | OMIM:618332 | 5 | 1 | |
| FLT4 | Lymphatic malformation 1 | OMIM:153100 | 13 | 0 |
| Congenital heart defects, multiple types, 7 | OMIM:618780 | 0 | 7 | |
| KCNQ2 | Seizures, benign neonatal, 1 | OMIM:121200 | 69 | 25 |
| Developmental and epileptic encephalopathy 7 | OMIM:613720 | 115 | 13 | |
| KMT2D | Kabuki syndrome 1 | OMIM:147920 | 42 | 221 |
| Branchial arch abnormalities, choanal atresia, athelia, hearing loss, and hypothyroidism syndrome | OMIM:620186 | 6 | 0 | |
| NALCN | Hypotonia, infantile, with psychomotor retardation and characteristic facies 1 | OMIM:615419 | 5 | 11 |
| Congenital contractures of the limbs and face, hypotonia, and developmental delay | OMIM:616266 | 31 | 2 | |
| NOTCH2 | Hajdu-Cheney syndrome | OMIM:102500 | 0 | 15 |
| Alagille syndrome-2 | OMIM:610205 | 5 | 1 | |
| SLC12A6 | Agenesis of the corpus callosum with peripheral neuropathy | OMIM:218000 | 0 | 42 |
| Charcot-Marie-Tooth disease, axonal, type 2 | OMIM:620068 | 5 | 0 | |
| SLC26A4 | Pendred syndrome | OMIM:274600 | 71 | 49 |
| Deafness, autosomal recessive 4, with enlarged vestibular aqueduct | OMIM:600791 | 96 | 21 | |
| SMARCA4 | Rhabdoid tumor predisposition syndrome-2 | OMIM:613325 | 2 | 38 |
| Coffin-Siris syndrome-4 | OMIM:614609 | 8 | 0 | |
| SMC1A | Cornelia de Lange syndrome 2 | OMIM:300590 | 49 | 21 |
| Developmental and epileptic encephalopathy 85, with or without midline brain defects | OMIM:301044 | 2 | 10 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lazareva, T.E.; Barbitoff, Y.A.; Nasykhova, Y.A.; Pavlova, N.S.; Bogaychuk, P.M.; Glotov, A.S. Statistical Dissection of the Genetic Determinants of Phenotypic Heterogeneity in Genes with Multiple Associated Rare Diseases. Genes 2023, 14, 2100. https://doi.org/10.3390/genes14112100
Lazareva TE, Barbitoff YA, Nasykhova YA, Pavlova NS, Bogaychuk PM, Glotov AS. Statistical Dissection of the Genetic Determinants of Phenotypic Heterogeneity in Genes with Multiple Associated Rare Diseases. Genes. 2023; 14(11):2100. https://doi.org/10.3390/genes14112100
Chicago/Turabian StyleLazareva, Tatyana E., Yury A. Barbitoff, Yulia A. Nasykhova, Nadezhda S. Pavlova, Polina M. Bogaychuk, and Andrey S. Glotov. 2023. "Statistical Dissection of the Genetic Determinants of Phenotypic Heterogeneity in Genes with Multiple Associated Rare Diseases" Genes 14, no. 11: 2100. https://doi.org/10.3390/genes14112100
APA StyleLazareva, T. E., Barbitoff, Y. A., Nasykhova, Y. A., Pavlova, N. S., Bogaychuk, P. M., & Glotov, A. S. (2023). Statistical Dissection of the Genetic Determinants of Phenotypic Heterogeneity in Genes with Multiple Associated Rare Diseases. Genes, 14(11), 2100. https://doi.org/10.3390/genes14112100








