The New Mitochondrial Genome of Hemiculterella wui (Cypriniformes, Xenocyprididae): Sequence, Structure, and Phylogenetic Analyses
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection, DNA Extraction, and Illumina Sequencing
2.2. Mitochondrial Genome Assembly, Annotation, and Sequence Analysis
2.3. Phylogenetic Analysis
3. Results and Discussion
3.1. Mitochondrial Genome Organization of H. wui
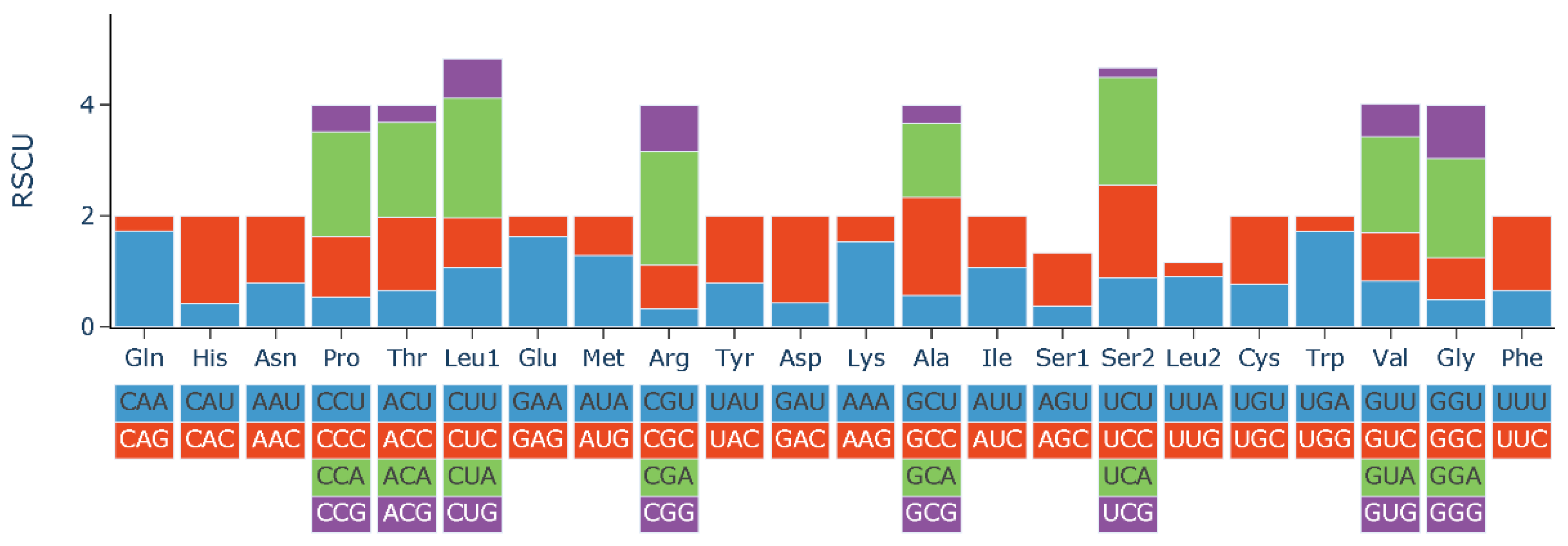
3.2. Protein Coding Genes and Codon Usage
3.3. Ribosomal and Transfer RNA Genes
3.4. Control Region
3.5. Selection Analysis
3.6. Phylogenetic Analysis
4. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Saccone, C.; Gissi, C.; Lanave, C.; Larizza, A.; Pesole, G.; Reyes, A. Evolution of the mitochondrial genetic system: An overview. Gene 2000, 261, 153–159. [Google Scholar] [CrossRef]
- Zhang, H.; Lu, C.C.; Liu, Q.; Zou, T.M.; Qiao, G.X.; Huang, X.L. Insights into the evolution of Aphid mitogenome features from new data and comparative analysis. Animals 2022, 12, 1970. [Google Scholar] [CrossRef]
- Satoh, T.P.; Miya, M.; Mabuchi, K.; Nishida, M. Structure and variation of the mitochondrial genome of fishes. BMC Genom. 2016, 17, 719. [Google Scholar] [CrossRef] [PubMed]
- Brown, K.H. Fish mitochondrial genomics: Sequence, inheritance and functional variation. J. Fish Biol. 2008, 72, 355–374. [Google Scholar] [CrossRef]
- Zhou, C.; Ma, A.; Wang, X.; Yang, C.; Li, Z.; Zhang, C.; Zhu, G.; Gu, Q. Progress on fish mitochondrial genome. J. Henan Normal Univer. 2019, 47, 74–82. [Google Scholar] [CrossRef]
- Sharma, A.; Siva, C.; Ali, S.; Sahoo, P.K.; Nath, R.; Laskar, M.A.; Sarma, D. The complete mitochondrial genome of the medicinal fish, Cyprinion semiplotum: Insight into its structural features and phylogenetic implications. Int. J. Biol. Macromol. 2020, 164, 939–948. [Google Scholar] [CrossRef] [PubMed]
- Fricke, R.; Eschmeyer, W.N.; Van der Laan, R.; Eschmeyer’s Catalog of Fishes: Genera, Species, References. Electronic Version. Available online: http://researcharchive.calacademy.org/research/ichthyology/catalog/fishcatmain.asp (accessed on 14 October 2023).
- Luo, Y.; Chen, R. Cultrinae. In Fauna Sinica, Osteichthyes, Cypriniformes (II); Chen, Y., Ed.; Science Press: Beijing, China, 1998; pp. 232–378. [Google Scholar]
- Zhang, R.; Zhu, T.; Li, H.; Deng, L. The Mitochondrial Genome of Linichthys laticeps (Cypriniformes: Cyprinidae): Characterization and Phylogeny. Genes 2023, 14, 1938. [Google Scholar] [CrossRef]
- Jin, J.J.; Yu, W.B.; Yang, J.B.; Song, Y.; dePamphilis, C.W.; Yi, T.S.; Li, D.Z. GetOrganelle: A fast and versatile toolkit for accurate de novo assembly of organelle genomes. Genome Biol. 2020, 21, 241. [Google Scholar] [CrossRef]
- Zhu, T.; Sato, Y.; Sado, T.; Miya, M.; Iwasaki, W. MitoFish, MitoAnnotator, and MiFish Pipeline: Updates in ten years. Mol. Biol. Evol. 2023, 40, msad035. [Google Scholar] [CrossRef]
- Bernt, M.; Donath, A.; Juhling, F.; Externbrink, F.; Florentz, C.; Fritzsch, G.; Putz, J.; Middendorf, M.; Stadler, P.F. MITOS: Improved de novo metazoan mitochondrial genome annotation. Mol. Phylogenet. Evol. 2013, 69, 313–319. [Google Scholar] [CrossRef]
- Perna, N.T.; Kocher, T.D. Patterns of nucleotide composition at fourfold degenerate sites of animal mitochondrial genomes. J. Mol. Evol. 1995, 41, 353–358. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Gao, F.; Jakovlić, I.; Zou, H.; Zhang, J.; Li, W.X.; Wang, G.T. PhyloSuite: An integrated and scalable desktop platform for streamlined molecular sequence data management and evolutionary phylogenetics studies. Mol. Ecol. Resour. 2020, 20, 348–355. [Google Scholar] [CrossRef] [PubMed]
- Rozas, J.; Ferrer-Mata, A.; Sanchez-DelBarrio, J.C.; Guirao-Rico, S.; Librado, P.; Ramos-Onsins, S.E.; Sanchez-Gracia, A. DnaSP 6: DNA sequence polymorphism analysis of large data sets. Mol. Biol. Evol. 2017, 34, 3299–3302. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Lanfear, R.; Frandsen, P.B.; Wright, A.M.; Senfeld, T.; Calcott, B. PartitionFinder 2: New methods for selecting partitioned models of evolution for molecular and morphological phylogenetic analyses. Mol. Biol. Evol. 2017, 34, 772–773. [Google Scholar] [CrossRef] [PubMed]
- Ronquist, F.; Teslenko, M.; van der Mark, P.; Ayres, D.L.; Darling, A.; Hohna, S.; Larget, B.; Liu, L.; Suchard, M.A.; Huelsenbeck, J.P. MrBayes 3.2: Efficient Bayesian phylogenetic inference and model choice across a large model space. Syst. Biol. 2012, 61, 539–542. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, L.T.; Schmidt, H.A.; von Haeseler, A.; Minh, B.Q. IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 2015, 32, 268–274. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Shen, Y. Characterization of the complete mitochondrial genome of Rhinogobius leavelli (Perciformes: Gobiidae: Gobionellinae) and its phylogenetic analysis for Gobionellinae. Biologia 2019, 74, 493–499. [Google Scholar] [CrossRef]
- Minh, B.Q.; Nguyen, M.A.; von Haeseler, A. Ultrafast approximation for phylogenetic bootstrap. Mol. Biol. Evol. 2013, 30, 1188–1195. [Google Scholar] [CrossRef]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Gao, X.; Li, Y.; Zhang, R.; Lv, Y.; Wang, Y.; Shi, J.; Xie, J.; Kong, C.; Li, K. The Complete Mitochondrial Genome of a Rare Cavefish (Sinocyclocheilus cyphotergous) and Comparative Genomic Analyses in Sinocyclocheilus. Pak. J. Zool. 2023, 55, 1–10. [Google Scholar] [CrossRef]




| No. | Species | GenBank Accession No. | Size (bp) | A% | T% | G% | C% | A + T% | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|---|
| 1 | Anabarilius brevianalis Zhou & Cui, 1992 | MK757491.1 | 16,610 | 28.3 | 24.6 | 18.7 | 28.4 | 52.9 | 0.070 | −0.205 |
| 2 | Anabarilius duoyiheensis Li, Mao & Lu, 2002 | NC_068241.1 | 16,614 | 28.9 | 24.9 | 18.4 | 27.9 | 53.8 | 0.074 | −0.205 |
| 3 | Anabarilius grahami (Regan, 1908) | MF370204.1 | 16,612 | 28.9 | 24.8 | 18.3 | 27.9 | 53.7 | 0.077 | −0.208 |
| 4 | Anabarilius liui (Chang, 1944) | MG702493.1 | 16,608 | 28.5 | 24.4 | 18.5 | 28.5 | 52.9 | 0.077 | −0.212 |
| 5 | Ancherythroculter kurematsui (Kimura, 1934) | NC_029707.1 | 16,621 | 31.2 | 24.9 | 16.2 | 27.7 | 56.1 | 0.112 | −0.263 |
| 6 | Ancherythroculter lini Luo, 1994 | NC_027741.1 | 16,616 | 30.8 | 24.7 | 16.5 | 28.0 | 55.5 | 0.109 | −0.259 |
| 7 | Ancherythroculter nigrocauda Yih & Wu, 1964 | NC_021414.1 | 16,623 | 31.2 | 24.8 | 16.2 | 27.8 | 56.0 | 0.114 | −0.262 |
| 8 | Ancherythroculter wangi (Tchang, 1932) | NC_037405.1 | 16,622 | 31.2 | 24.8 | 16.2 | 27.8 | 56.0 | 0.113 | −0.264 |
| 9 | Aphyocypris arcus (Lin, 1931) | NC_015540.1 | 16,617 | 31.4 | 27.1 | 15.7 | 25.7 | 58.5 | 0.074 | −0.241 |
| 10 | Aphyocypris chinensis Günther, 1868 | NC_008650.1 | 16,606 | 30.7 | 27.7 | 16.5 | 25.1 | 58.4 | 0.053 | −0.207 |
| 11 | Aphyocypris kikuchii (Oshima, 1919) | NC_019620.1 | 16,601 | 30.8 | 27.7 | 16.5 | 25.0 | 58.5 | 0.052 | −0.206 |
| 12 | Aphyocypris lini (Weitzman & Chan, 1966) | NC_062821.1 | 16,613 | 30.4 | 26.9 | 16.6 | 26.0 | 57.3 | 0.061 | −0.221 |
| 13 | Aphyocypris moltrechti (Regan, 1908) | NC_019621.1 | 16,617 | 31.2 | 27.4 | 16.0 | 25.5 | 58.6 | 0.066 | −0.229 |
| 14 | Aphyocypris normalis Nichols & Pope, 1927 | NC_015538.1 | 16,619 | 31.2 | 27.0 | 15.9 | 25.8 | 58.2 | 0.073 | −0.237 |
| 15 | Aphyocypris pulchrilineata Zhu, Zhao & Huang, 2013 | MK387702.1 | 16,610 | 30.5 | 26.6 | 16.6 | 26.2 | 57.1 | 0.069 | −0.224 |
| 16 | Candidia barbata (Regan, 1908) | NC_037156.1 | 16,608 | 30.3 | 26.9 | 16.7 | 26.1 | 57.2 | 0.059 | −0.219 |
| 17 | Candidia pingtungensis Chen, Wu & Hsu, 2008 | NC_028596.1 | 16,612 | 29.9 | 26.9 | 17.1 | 26.2 | 56.8 | 0.053 | −0.211 |
| 18 | Chanodichthys dabryi (Bleeker, 1871) | NC_021418.1 | 16,622 | 31.5 | 25.0 | 15.9 | 27.6 | 56.5 | 0.116 | −0.268 |
| 19 | Chanodichthys erythropterus (Basilewsky, 1855) | MN105126.1 | 16,623 | 31.0 | 25.1 | 16.3 | 27.6 | 56.1 | 0.106 | −0.255 |
| 20 | Chanodichthys mongolicus (Basilewsky, 1855) | NC_008683.1 | 16,622 | 31.2 | 24.9 | 16.2 | 27.8 | 56.1 | 0.113 | −0.264 |
| 21 | Chanodichthys recurviceps (Richardson, 1846) | NC_024277.1 | 16,622 | 31.4 | 24.8 | 16.1 | 27.8 | 56.2 | 0.117 | −0.266 |
| 22 | Ctenopharyngodon idella (Valenciennes, 1844) | NC_010288.1 | 16,609 | 31.9 | 26.2 | 15.6 | 26.3 | 58.1 | 0.098 | −0.253 |
| 23 | Culter alburnus Basilewsky, 1855 | NC_013616.1 | 16,622 | 31.2 | 24.8 | 16.2 | 27.8 | 56.0 | 0.114 | −0.262 |
| 24 | Culter compressocorpus (Yih & Chu, 1959) | NC_024183.1 | 16,623 | 31.1 | 25.1 | 16.3 | 27.5 | 56.2 | 0.105 | −0.254 |
| 25 | Culter oxycephaloides Kreyenberg & Pappenheim, 1908 | KY404014.1 | 16,619 | 31.3 | 24.8 | 16.1 | 27.9 | 56.1 | 0.117 | −0.269 |
| 26 | C. carpio Linnaeus, 1758 | NC_018035.1 | 16581 | 31.9 | 24.8 | 15.7 | 27.5 | 56.7 | 0.125 | −0.272 |
| 27 | Distoechodon compressus (Nichols, 1925) | NC_067894.1 | 16,621 | 31.4 | 25.4 | 16.0 | 27.2 | 56.8 | 0.107 | −0.260 |
| 28 | Distoechodon tumirostris Peters, 1881 | NC_011208.1 | 16,620 | 31.4 | 25.4 | 16.0 | 27.2 | 56.8 | 0.106 | −0.261 |
| 29 | Elopichthys bambusa (Richardson, 1845) | NC_024834.1 | 16,619 | 30.1 | 27.2 | 16.9 | 25.8 | 57.3 | 0.049 | −0.209 |
| 30 | G. rarus Ye & Fu, 1983 | NC_018099.1 | 16,601 | 29.5 | 27.6 | 17.2 | 25.7 | 57.1 | 0.034 | −0.198 |
| 31 | Hemiculter bleekeri Warpachowski, 1888 | NC_029831.1 | 16,615 | 29.3 | 25.6 | 17.8 | 27.3 | 54.9 | 0.067 | −0.211 |
| 32 | Hemiculter leucisculus (Basilewsky, 1855) | NC_022929.1 | 16,617 | 30.4 | 25.4 | 17.1 | 27.2 | 55.8 | 0.090 | −0.228 |
| 33 | Hemiculter nigromarginis Fang, 1942 | NC_036740.1 | 16,621 | 30.4 | 25.4 | 17.1 | 27.1 | 55.8 | 0.089 | −0.228 |
| 34 | Hemiculterella sauvagei Warpachowski, 1888 | NC_026693.1 | 16,618 | 29.9 | 25.6 | 17.4 | 27.0 | 55.5 | 0.078 | −0.216 |
| 35 | H. wui (Wang, 1935) | OR574832 | 16,619 | 29.9 | 25.3 | 17.5 | 27.4 | 55.2 | 0.084 | −0.22 |
| 36 | Hemigrammocypris neglecta (Stieler, 1907) | NC_015548.1 | 16,615 | 31.2 | 26.3 | 16.7 | 25.8 | 57.5 | 0.085 | −0.215 |
| 37 | Hypophthalmichthys molitrix (Valenciennes, 1844) | NC_010156.1 | 16,620 | 31.8 | 25.6 | 15.8 | 26.9 | 57.4 | 0.109 | −0.261 |
| 38 | Hypophthalmichthys nobilis (Richardson, 1845) | NC_010194.1 | 16,621 | 31.6 | 25.3 | 15.9 | 27.1 | 56.9 | 0.111 | −0.259 |
| 39 | Ischikauia steenackeri (Sauvage, 1883) | NC_008667.1 | 16,620 | 31.4 | 25.1 | 16.1 | 27.4 | 56.5 | 0.111 | −0.261 |
| 40 | Macrochirichthys macrochirus (Valenciennes, 1844) | NC_015551.1 | 16857 | 32.5 | 26.1 | 15.2 | 26.2 | 58.6 | 0.108 | −0.266 |
| 41 | Megalobrama amblycephala Yih, 1955 | NC_010341.1 | 16,623 | 31.2 | 24.7 | 16.2 | 27.9 | 55.9 | 0.117 | −0.265 |
| 42 | Megalobrama skolkovii (Basilewsky, 1855) | NC_024422.1 | 16,620 | 31.2 | 24.7 | 16.2 | 27.9 | 55.9 | 0.116 | −0.266 |
| 43 | Megalobrama terminalis (Richardson, 1846) | NC_018816.1 | 16,623 | 31.1 | 24.8 | 16.3 | 27.8 | 55.9 | 0.113 | −0.26 |
| 44 | Metzia formosae (Oshima, 1920) | NC_022458.1 | 16,614 | 31.4 | 25.6 | 16.2 | 26.8 | 57.0 | 0.102 | −0.246 |
| 45 | Metzia lineata (Pellegrin, 1907) | NC_031541.1 | 16,614 | 32.1 | 26.9 | 15.6 | 25.4 | 59.0 | 0.089 | −0.241 |
| 46 | Metzia longinasus Gan, Lan & Zhang, 2009 | NC_024729.1 | 16,614 | 31.9 | 26.2 | 15.7 | 26.2 | 58.1 | 0.098 | −0.251 |
| 47 | Metzia mesembrinum (Jordan & Evermann, 1902) | NC_023797.1 | 16,611 | 32.0 | 26.8 | 15.7 | 25.5 | 58.8 | 0.088 | −0.238 |
| 48 | Mylopharyngodon piceus (Richardson, 1846) | NC_011141.1 | 16,609 | 32.0 | 24.5 | 15.7 | 27.8 | 56.5 | 0.132 | −0.278 |
| 49 | Nipponocypris koreanus (Kim, Oh & Hosoya, 2005) | NC_025286.1 | 16,615 | 30.0 | 26.8 | 17.1 | 26.1 | 56.8 | 0.056 | −0.209 |
| 50 | Nipponocypris sieboldii (Temminck & Schlegel, 1846) | NC_008653.1 | 16,616 | 30.1 | 25.8 | 16.9 | 27.2 | 55.9 | 0.078 | −0.233 |
| 51 | Nipponocypris temminckii (Temminck & Schlegel, 1846) | NC_027664.1 | 16,615 | 30.3 | 26.5 | 16.8 | 26.3 | 56.8 | 0.067 | −0.221 |
| 52 | Ochetobius elongatus (Kner, 1867) | NC_025646.1 | 16,613 | 31.0 | 25.4 | 16.3 | 27.4 | 56.4 | 0.099 | −0.255 |
| 53 | Opsariichthys acutipinnis (Bleeker, 1871) | NC_028595.1 | 16,615 | 28.2 | 26.6 | 18.0 | 27.2 | 54.8 | 0.030 | −0.204 |
| 54 | Opsariichthys bidens Günther, 1873 | NC_008744.1 | 16,611 | 27.2 | 26.7 | 19.1 | 27.1 | 53.9 | 0.010 | −0.174 |
| 55 | Opsariichthys evolans (Jordan & Evermann, 1902) | NC_033948.1 | 16,616 | 28.2 | 26.6 | 18.0 | 27.2 | 54.8 | 0.029 | −0.203 |
| 56 | Opsariichthys pachycephalus Günther, 1868 | NC_033949.1 | 16,606 | 27.9 | 26.8 | 18.3 | 27.1 | 54.7 | 0.020 | −0.194 |
| 57 | Opsariichthys uncirostris (Temminck & Schlegel, 1846) | NC_008652.1 | 16,613 | 27.2 | 26.7 | 18.9 | 27.2 | 53.9 | 0.008 | −0.181 |
| 58 | Parabramis pekinensis (Basilewsky, 1855) | NC_022678.1 | 16,622 | 31.1 | 24.8 | 16.3 | 27.8 | 55.9 | 0.113 | −0.261 |
| 59 | Parazacco spilurus (Günther, 1868) | NC_023786.1 | 16,612 | 30.4 | 26.9 | 16.6 | 26.0 | 57.3 | 0.062 | −0.220 |
| 60 | Plagiognathops microlepis (Bleeker, 1871) | NC_022711.1 | 16,623 | 30.6 | 25.2 | 16.9 | 27.3 | 55.8 | 0.097 | −0.236 |
| 61 | Pseudobrama simoni (Bleeker, 1864) | NC_022852.1 | 16,618 | 31.6 | 27.2 | 15.7 | 25.5 | 58.8 | 0.075 | −0.238 |
| 62 | P. dispar (Peters, 1881) | NC_020435.1 | 16,620 | 30.3 | 25.5 | 17.1 | 27.1 | 55.8 | 0.086 | −0.224 |
| 63 | Pseudohemiculter hainanensis (Boulenger, 1900) | NC_065693.1 | 16,647 | 29.7 | 24.8 | 17.5 | 28.0 | 54.5 | 0.089 | −0.230 |
| 64 | Pseudolaubuca engraulis (Nichols, 1925) | NC_020462.1 | 16,612 | 27.1 | 25.9 | 19.5 | 27.5 | 53.0 | 0.023 | −0.171 |
| 65 | Pseudolaubuca sinensis Bleeker, 1864 | NC_026712.1 | 16,617 | 29.6 | 25.8 | 17.6 | 26.9 | 55.4 | 0.069 | −0.209 |
| 66 | Sinibrama macrops (Günther, 1868) | NC_020013.1 | 16,626 | 30.7 | 24.7 | 16.6 | 27.9 | 55.4 | 0.110 | −0.253 |
| 67 | Sinibrama melrosei (Nichols & Pope, 1927) | NC_063731.1 | 16,619 | 30.7 | 25.5 | 16.6 | 27.2 | 56.2 | 0.092 | −0.242 |
| 68 | Sinibrama taeniatus (Nichols, 1941) | NC_026119.1 | 16,623 | 31.3 | 25.8 | 16.1 | 26.8 | 57.1 | 0.097 | −0.248 |
| 69 | Sinibrama wui (Rendahl, 1933) | NC_068747.1 | 16,626 | 30.7 | 24.6 | 16.7 | 28.0 | 55.3 | 0.110 | −0.253 |
| 70 | Squaliobarbus curriculus (Richardson, 1846) | NC_019652.1 | 16,619 | 31.2 | 25.0 | 16.1 | 27.7 | 56.2 | 0.111 | −0.264 |
| 71 | Toxabramis houdemeri Pellegrin, 1932 | NC_029348.1 | 16,618 | 30.8 | 25.3 | 16.6 | 27.3 | 56.1 | 0.098 | −0.243 |
| 72 | Toxabramis swinhonis Günther, 1873 | NC_029249.1 | 16,622 | 31.2 | 26.5 | 16.1 | 26.1 | 57.7 | 0.081 | −0.237 |
| 73 | Xenocypris davidi Bleeker, 1871 | NC_013072.1 | 16,630 | 31.3 | 25.4 | 16.2 | 27.2 | 56.7 | 0.104 | −0.254 |
| 74 | Xenocypris fangi Tchang, 1930 | NC_056130.1 | 16,619 | 31.2 | 25.3 | 16.2 | 27.3 | 56.5 | 0.105 | −0.255 |
| 75 | Xenocypris yunnanensis Nichols, 1925 | NC_035954.1 | 16,630 | 31.3 | 25.4 | 16.1 | 27.2 | 56.7 | 0.104 | −0.255 |
| 76 | Zacco acanthogenys (Bleeker, 1871) | NC_028546.1 | 16,611 | 29.1 | 27.2 | 17.6 | 26.0 | 56.3 | 0.033 | −0.192 |
| 77 | Zacco platypus (Temminck & Schlegel, 1846) | NC_023105.1 | 16,611 | 29.0 | 27.2 | 17.8 | 26.1 | 56.2 | 0.032 | −0.190 |
| Gene | Strand | Location (bp) | Size (bp) | Intergenic Nucleotide | Anticodon | Start/Stop Codons |
|---|---|---|---|---|---|---|
| tRNAPhe | H | 1–69 | 69 | 0 | GAA | |
| 12S rRNA | H | 70–1033 | 964 | 0 | ||
| tRNAVal | H | 1034–1105 | 72 | 0 | TAC | |
| 16S rRNA | H | 1106–2795 | 1690 | 0 | ||
| tRNALeu (UUR) | H | 2796–2871 | 76 | 1 | TAA | |
| ND1 | H | 2873–3847 | 975 | 4 | ATG/TAA | |
| tRNAIle | H | 3852–3923 | 72 | −2 | GAT | |
| tRNAGln | L | 3922–3992 | 71 | 1 | TTG | |
| tRNAMet | H | 3994–4062 | 69 | 0 | CAT | |
| ND2 | H | 4063–5107 | 1045 | 0 | ATG/T | |
| tRNATrp | H | 5108–5178 | 71 | 1 | TCA | |
| tRNAAla | L | 5180–5248 | 69 | 1 | TGC | |
| tRNAAsn | L | 5250–5322 | 73 | 32 | GTT | |
| tRNACys | L | 5355–5422 | 68 | 1 | GCA | |
| tRNATyr | L | 5424–5494 | 71 | 1 | GTA | |
| COI | H | 5496–7046 | 1551 | 0 | GTG/TAA | |
| tRNASer (UCN) | L | 7047–7117 | 71 | 3 | TGA | |
| tRNAAsp | H | 7121–7194 | 74 | 13 | GTC | |
| COII | H | 7208–7898 | 691 | 0 | ATG/T | |
| tRNALys | H | 7899–7974 | 76 | 1 | TTT | |
| ATPase 8 | H | 7976–8140 | 165 | −7 | GTG/TAG | |
| ATPase 6 | H | 8134–8816 | 683 | 0 | ATG/TA | |
| COIII | H | 8817–9601 | 785 | 0 | ATG/TA | |
| tRNAGly | H | 9602–9673 | 72 | 0 | TCC | |
| ND3 | H | 9674–10,022 | 349 | 0 | ATG/T | |
| tRNAArg | H | 10,023–10,092 | 70 | 0 | TCG | |
| ND4L | H | 10,093–10,389 | 297 | −7 | ATG/TAA | |
| ND4 | H | 10,383–11,764 | 1382 | 0 | ATG/TA | |
| tRNAHis | H | 11,765–11,833 | 69 | 0 | GTG | |
| tRNASer (AGY) | H | 11,834–11,902 | 69 | 1 | GCT | |
| tRNALeu (CUN) | H | 11,904–11,976 | 73 | 0 | TAG | |
| ND5 | H | 11,977–13,812 | 1836 | −4 | ATG/TAA | |
| ND6 | L | 13,809–14,330 | 522 | 0 | ATG/TAA | |
| tRNAGlu | L | 14,331–14,399 | 69 | 4 | TTC | |
| Cyt b | H | 14,404–15,544 | 1141 | 0 | ATG/T | |
| tRNAThr | H | 15,545–15,616 | 72 | −1 | TGT | |
| tRNAPro | L | 15,616–15,685 | 70 | 0 | TGG | |
| Control region | H | 15,686–16,619 | 934 | 0 |
| Regions | Size (bp) | A% | T% | G% | C% | A + T% | G + C% | AT Skew | GC Skew |
|---|---|---|---|---|---|---|---|---|---|
| Full genome | 16,619 | 29.9 | 25.3 | 17.5 | 27.4 | 55.2 | 44.9 | 0.084 | −0.22 |
| PCGs | 11,422 | 27.6 | 27.0 | 17.1 | 28.3 | 54.6 | 45.4 | 0.011 | −0.248 |
| 1st codon position | 3804 | 26.5 | 21.0 | 26.4 | 26.1 | 47.5 | 52.5 | 0.116 | 0.005 |
| 2nd codon position | 3804 | 18.5 | 40.5 | 13.7 | 27.3 | 59.0 | 41.0 | −0.374 | −0.333 |
| 3rd codon position | 3804 | 37.9 | 19.4 | 11.1 | 31.6 | 57.3 | 42.7 | 0.322 | −0.478 |
| ATPase 6 | 683 | 27.1 | 29.6 | 14.5 | 28.8 | 56.7 | 43.3 | −0.044 | −0.331 |
| ATPase 8 | 165 | 35.8 | 23.6 | 12.1 | 28.5 | 59.4 | 40.6 | 0.204 | −0.403 |
| COI | 1551 | 26.6 | 28.2 | 18.6 | 26.6 | 54.8 | 45.2 | −0.028 | −0.178 |
| COII | 691 | 29.8 | 26.0 | 16.6 | 27.5 | 55.8 | 44.1 | 0.067 | −0.246 |
| COIII | 785 | 27.3 | 27.6 | 17.1 | 28.0 | 54.9 | 45.1 | −0.007 | −0.243 |
| Cyt b | 1141 | 28.0 | 26.2 | 15.9 | 30.0 | 54.2 | 45.9 | 0.032 | −0.308 |
| ND1 | 975 | 26.8 | 24.9 | 17.5 | 30.8 | 51.7 | 48.3 | 0.036 | −0.274 |
| ND2 | 1045 | 28.3 | 22.9 | 17.2 | 31.6 | 51.2 | 48.8 | 0.107 | −0.294 |
| ND3 | 349 | 25.8 | 28.7 | 17.2 | 28.4 | 54.5 | 45.6 | −0.053 | −0.245 |
| ND4L | 1382 | 29.1 | 27.0 | 15.9 | 28.0 | 56.1 | 43.9 | 0.037 | −0.275 |
| ND4 | 297 | 25.9 | 25.3 | 16.5 | 32.3 | 51.2 | 48.8 | 0.013 | −0.324 |
| ND5 | 1836 | 30.4 | 26.0 | 14.4 | 29.2 | 56.4 | 43.6 | 0.077 | −0.34 |
| ND6 | 522 | 14.0 | 39.1 | 32.0 | 14.9 | 53.1 | 46.9 | −0.473 | 0.363 |
| rRNAs | 2654 | 33.4 | 20.3 | 22.1 | 24.2 | 53.7 | 46.3 | 0.245 | −0.046 |
| tRNAs | 1566 | 28.7 | 27.1 | 23.3 | 20.8 | 55.8 | 44.1 | 0.029 | 0.056 |
| Control region | 934 | 32.9 | 31.5 | 14.9 | 20.8 | 64.4 | 35.7 | 0.022 | −0.165 |
| Amino Acid | Codon | Count | RSCU | Amino Acid | Codon | Count | RSCU |
|---|---|---|---|---|---|---|---|
| Phe (F) | UUU | 74 | 0.66 | Tyr (Y) | UAU | 45 | 0.79 |
| UUC | 151 | 1.34 | UAC | 69 | 1.21 | ||
| Leu (L) | UUA | 94 | 0.91 | stop codon | UAA | 5 | 3.33 |
| UUG | 26 | 0.25 | UAG | 1 | 0.67 | ||
| CUU | 111 | 1.07 | His (H) | CAU | 22 | 0.42 | |
| CUC | 92 | 0.89 | CAC | 82 | 1.58 | ||
| CUA | 224 | 2.16 | Gln (Q) | CAA | 84 | 1.73 | |
| CUG | 74 | 0.71 | CAG | 13 | 0.27 | ||
| Ile (I) | AUU | 149 | 1.06 | Asn (N) | AAU | 48 | 0.79 |
| AUC | 132 | 0.94 | AAC | 73 | 1.21 | ||
| Met (M) | AUA | 112 | 1.29 | Lys (K) | AAA | 61 | 1.54 |
| AUG | 61 | 0.71 | AAG | 18 | 0.46 | ||
| Val (V) | GUU | 50 | 0.83 | Asp (D) | GAU | 17 | 0.44 |
| GUC | 52 | 0.86 | GAC | 61 | 1.56 | ||
| GUA | 105 | 1.74 | Glu (E) | GAA | 84 | 1.63 | |
| GUG | 35 | 0.58 | GAG | 19 | 0.37 | ||
| Ser (S) | UCU | 35 | 0.88 | Cys (C) | UGU | 10 | 0.77 |
| UCC | 67 | 1.68 | UGC | 16 | 1.23 | ||
| UCA | 77 | 1.93 | Trp (W) | UGA | 104 | 1.72 | |
| UCG | 7 | 0.18 | UGG | 17 | 0.28 | ||
| Pro (P) | CCU | 29 | 0.54 | Arg (R) | CGU | 6 | 0.32 |
| CCC | 59 | 1.09 | CGC | 15 | 0.79 | ||
| CCA | 102 | 1.89 | CGA | 39 | 2.05 | ||
| CCG | 26 | 0.48 | CGG | 16 | 0.84 | ||
| Thr (T) | ACU | 50 | 0.66 | Ser (S) | AGU | 15 | 0.38 |
| ACC | 99 | 1.32 | AGC | 38 | 0.95 | ||
| ACA | 128 | 1.7 | stop codon | AGA | 0 | 0 | |
| ACG | 24 | 0.32 | AGG | 0 | 0 | ||
| Ala (A) | GCU | 48 | 0.57 | Gly (G) | GGU | 30 | 0.49 |
| GCC | 149 | 1.77 | GGC | 46 | 0.75 | ||
| GCA | 112 | 1.33 | GGA | 109 | 1.79 | ||
| GCG | 28 | 0.33 | GGG | 59 | 0.97 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, R.; Zhu, T.; Yu, F. The New Mitochondrial Genome of Hemiculterella wui (Cypriniformes, Xenocyprididae): Sequence, Structure, and Phylogenetic Analyses. Genes 2023, 14, 2110. https://doi.org/10.3390/genes14122110
Zhang R, Zhu T, Yu F. The New Mitochondrial Genome of Hemiculterella wui (Cypriniformes, Xenocyprididae): Sequence, Structure, and Phylogenetic Analyses. Genes. 2023; 14(12):2110. https://doi.org/10.3390/genes14122110
Chicago/Turabian StyleZhang, Renyi, Tingting Zhu, and Feng Yu. 2023. "The New Mitochondrial Genome of Hemiculterella wui (Cypriniformes, Xenocyprididae): Sequence, Structure, and Phylogenetic Analyses" Genes 14, no. 12: 2110. https://doi.org/10.3390/genes14122110
APA StyleZhang, R., Zhu, T., & Yu, F. (2023). The New Mitochondrial Genome of Hemiculterella wui (Cypriniformes, Xenocyprididae): Sequence, Structure, and Phylogenetic Analyses. Genes, 14(12), 2110. https://doi.org/10.3390/genes14122110







