Comparative Transcriptome Analysis of Galeruca daurica Reveals Cold Tolerance Mechanisms
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collecting of G. daurica Larvae and Indoor and Outdoor Rearing
2.2. Indoor and Outdoor Temperature and Humidity Data Collection
2.3. RNA Extraction, cDNA Library Construction and Transcriptome Sequencing
2.4. Quantitative qRT-PCR for Verification
2.5. dsRNA Synthesis
2.6. Microinjection of RNAi
2.7. Relative Expression of HSP70A1, Hsp70-2 and Hsp70-3 after RNAi
2.8. Determination of Super-Cooling Point and Freezing Point
2.9. Gene Functional Annotation
2.10. Normalization of Gene Expression Levels and Analysis of DEGs
2.11. Enrichment and Analysis of DEGs by GO and KEGG
3. Results
3.1. Gene Annotation Results and Bioinformatics Analysis
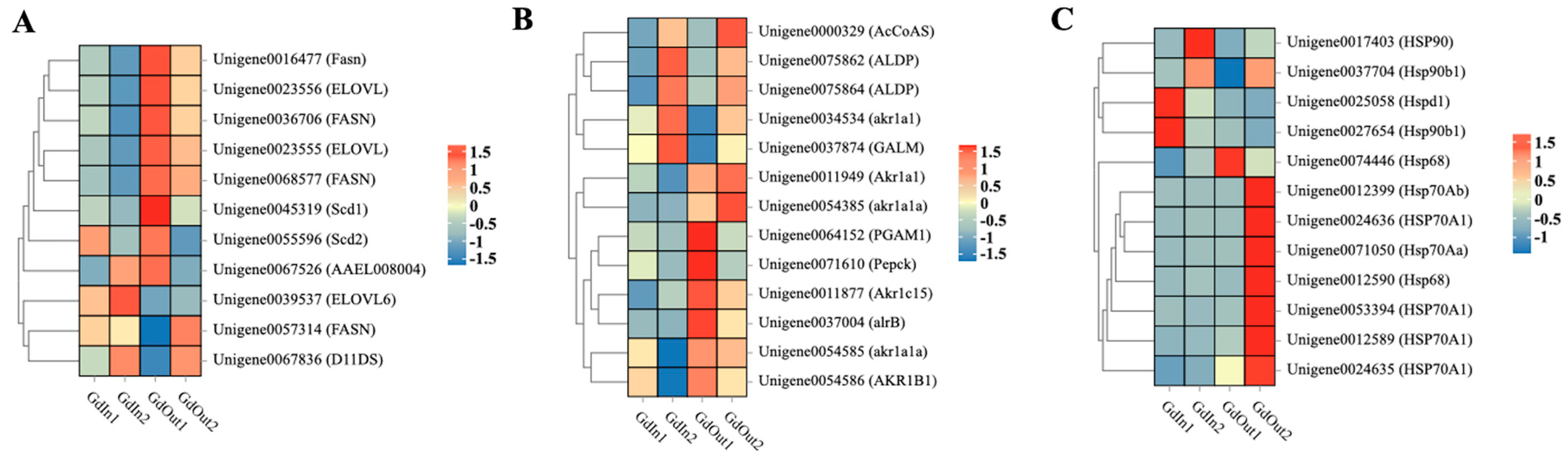
3.2. Analysis of the Expression of Biosynthesis-Related and HSP-Related Genes in G. daurica
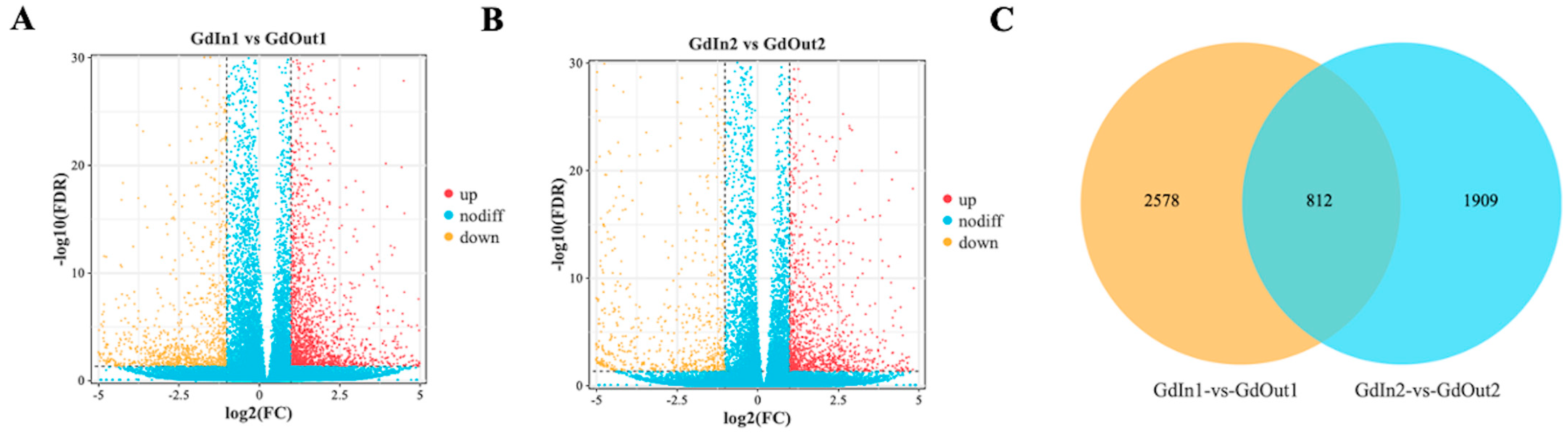
3.3. Identification and Analysis of DEGs
3.4. GO Functional Enrichment of DEGs
3.5. KOG Annotation Analysis of DEGs
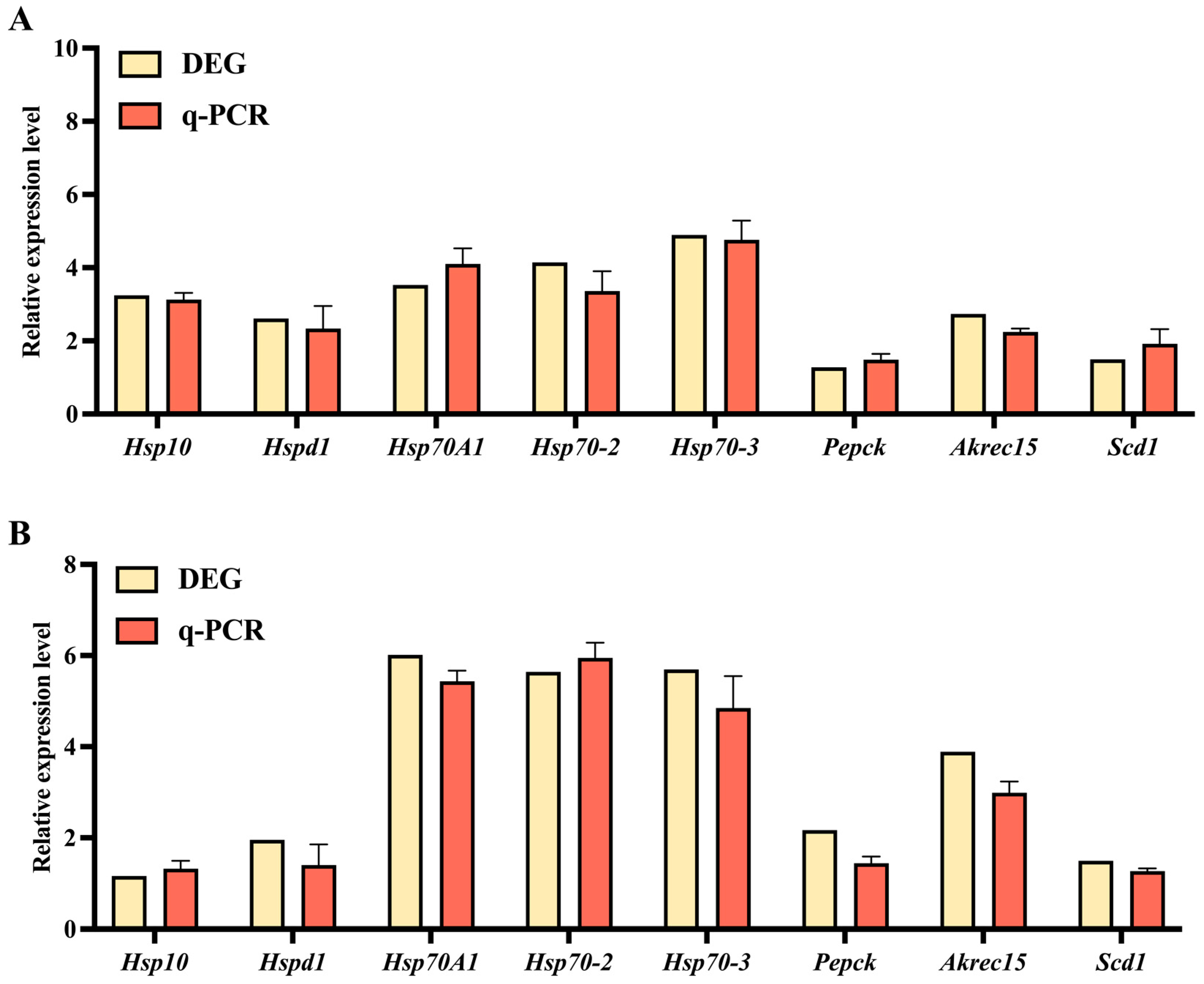
3.6. The qPCR Validation of DEGs
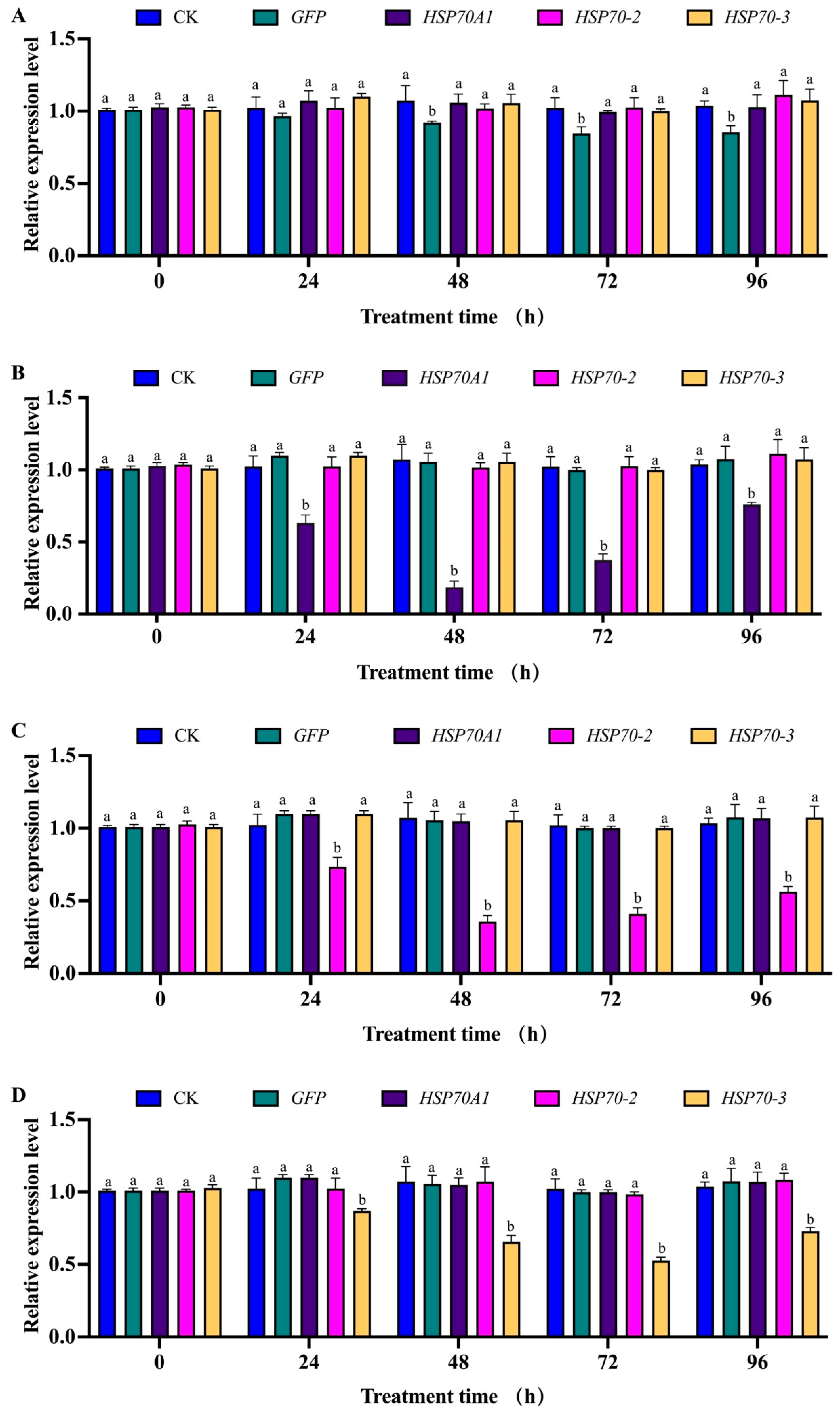
3.7. Expression Levels of Hsp70A1, Hsp70-2 and Hsp70-3 in Second-Instar Larvae of G. daurica after RNAi Using Microinjection Method
3.8. Super-Cooling Point and Body Fluid Freezing Point of G. daurica Larvae after RNAi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Enriquez, T.; Colinet, H. Cold acclimation triggers major transcriptional changes in Drosophila suzukii. BMC Genom. 2019, 20, 413. [Google Scholar] [CrossRef]
- Khurshid, A.; Ali, S.; Inayat, R.; Tamkeen, A.; Tahir, M.B.; Niaz, Y.; Haq, I.; Ghramh, H.A.; Boamah, S.; Zhang, K.; et al. Effect of short-term heat stress on life table parameters of green peach aphid [Myzus persicae (Sulzer) (Hemiptera: Aphididae)]. J. King Saud. Univ. Sci. I 2022, 34, 102342. [Google Scholar] [CrossRef]
- Mutamiswa, R.; Machekano, H.; Chidawanyika, F.; Nyamukondiwa, C. Life-stage related responses to. combined effects of acclimation temperature and humidity on the thermal tolerance of Chilo partellus (Swinhoe) (Lepidoptera: Crambidae). J. Therm. Biol. 2019, 79, 85–94. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.W.; Zhang, X.X.; Lu, M.X.; Gong, W.R.; Du, Y.Z. Transcriptome analysis of Liriomyza trifolii (Diptera: Agromyzidae) in response to temperature stress. Comp. Biochem. Phys. D 2020, 34, 100677. [Google Scholar] [CrossRef]
- Bale, J.S.; Hayward, S.A. Insect overwintering in a changing climate. J. Exp. Biol. 2010, 213, 980–994. [Google Scholar] [CrossRef] [PubMed]
- Hayward, S.A. Application of functional ‘Omics’ in environmental stress physiology: Insights, limitations, and future challenges. Curr. Opin. Insect Sci. 2014, 4, 35–41. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wu, H.; Xie, J.; Jiang, R.; Deng, C.; Pang, H. Transcriptome responses to heat- and cold-stress in ladybirds (Cryptolaemus montrouzieri Mulasnt) analyzed by deep-sequencing. Biol. Res. 2015, 48, 66. [Google Scholar] [CrossRef] [PubMed]
- Dunning, L.T.; Dennis, A.B.; Park, D.; Sinclair, B.J.; Newcomb, R.D.; Buckley, T.R. Identification of cold-responsive genes in a New Zealand alpine stick insect using RNA-Seq. Comp. Biochem. Phys. D 2013, 8, 24–31. [Google Scholar] [CrossRef]
- Dunning, L.T.; Dennis, A.B.; Sinclair, B.J.; Newcomb, R.D.; Buckley, T.R. Divergent transcriptional responses to low temperature among populations of alpine and lowland species of New Zealand stick insects (Micrarchus). Mol. Ecol. 2014, 23, 2712–2726. [Google Scholar] [CrossRef]
- Tusong, K.; Guo, X.; Meng, S.; Liu, X.; Ma, J. Comparative analysis of the transcriptome of the overwintering desert beetle Microdera punctipennis. Cryobiology 2017, 78, 80–89. [Google Scholar] [CrossRef]
- Govaere, L.; Morin, M.D.; Frigault, J.J.; Boquel, S.; Cohen, A.; Lamarre, S.G. Transcriptome and proteome analyses to investigate the molecular underpinnings of cold response in the Colorado potato beetle, Leptinotarsa decemlineata. Cryobiology 2019, 88, 54–63. [Google Scholar] [CrossRef] [PubMed]
- Vatanparast, M.; Park, Y. Comparative RNA-Seq analyses of Solenopsis japonica (Hymenoptera: Formicidae) reveal gene in response to cold stress. Genes 2021, 12, 1610. [Google Scholar] [CrossRef] [PubMed]
- Ashraf, H.J.; Ramos, A.L.C.; Ahmed, S.; Haq, I.U.; Ali, H.; Ilyas, M.; Gu, S.; Wang, L. Comparative transcriptome analysis of Tamarixia radiata (Hymenoptera: Eulophidae) reveals differentially expressed genes upon heat shock. Comp. Biochem. Phys. Part D 2022, 41, 100940. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Shi, F.; Li, Y.; Zong, S.; Tao, J. Genome-wide identification and expression analysis of the Hsp gene superfamily in Asian long-horned beetle (Anoplophora glabripennis). Int. J. Biol. Macromol. 2022, 200, 583–592. [Google Scholar] [CrossRef]
- Luo, D.; Liu, Q.; Wang, J.; Jashenko, R.; Ji, R. Transcriptome analysis of the differentially expressed heat-resistant genes between Calliptamus italicus and Gomphocerus sibiricus. Environ. Entomol. 2023, 52, 129–137. [Google Scholar] [CrossRef]
- Iqbal, J.; Zhang, X.X.; Chang, Y.W.; Du, Y.Z. Comparative transcriptome analysis of Liriomyza trifolii (Burgess) and Liriomyza sativae (Blanchard) (Diptera: Agromyzidae) in response to rapid cold hardening. PLoS ONE 2022, 17, e0279254. [Google Scholar] [CrossRef]
- Zhang, C.; Wang, H.; Zhuang, G.; Zheng, H.; Zhang, X. Comparative transcriptome analysis of Callosobruchus chinensis (L.) (Coleoptera: Chrysomelidae-Bruchinae) after heat and cold stress exposure. J. Therm. Biol. 2023, 112, 103479. [Google Scholar] [CrossRef]
- Wang, L.; Etebari, K.; Walter, G.H.; Furlong, M.J. Sex dependent transcriptome responses of the diamondback moth, Plutella xylostella L. to cold stress. Comp. Biochem. Phys. D 2023, 45, 101053. [Google Scholar] [CrossRef]
- Noer, N.K.; Nielsen, K.L.; Sverrisdóttir, E.; Kristensen, T.N.; Bahrndorff, S. Temporal regulation of temperature tolerances and gene expression in an arctic insect. J. Exp. Biol. 2023, 226, jeb245097. [Google Scholar] [CrossRef]
- Yang, X.K.; Huang, D.C.; Ge, S.Q.; Bai, M.; Zhang, R.Z. One million mu of meadow in Inner Mongolia suffer from the harm of breaking out of Galeruca daurica (Joannis). Chin. Bull. Entomol. 2010, 47, 812. (In Chinese) [Google Scholar]
- Zhou, X.R.; Shan, Y.M.; Tan, Y.; Zhang, Z.R.; Pang, B.P. Comparative analysis of transcriptome responses to cold stress in Galeruca daurica (Coleoptera: Chrysomelidae). J. Insect Sci. 2019, 19, 8. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Zhou, X.R.; Pang, B.P.; Zhang, Z.R.; Ma, C.Y. Effects of host plants on feeding amount, growth and development of Galeruca daurica Joannis larvae (Coleoptera: Chrysomelidae). Acta Agrestia Sin. 2014, 22, 854–858. (In Chinese) [Google Scholar]
- Wang, H.C.; Li, L.; Li, Y.Y.; Tan, Y.; Pang, B.P. Evaluation of reference genes for miRNA expression analysis in Galeruca daurica (Coleoptera: Chrysomelidae) using qRT-PCR. Entomol. Res. 2021, 51, 393–402. [Google Scholar] [CrossRef]
- Zhou, X.R.; Gao, J.C.; Pang, B.P. Effects of temperature on the termination of egg diapause and post-diapause embryonic development of Galeruca daurica (Coleoptera: Chrysomelidae). Environ. Entomol. 2016, 45, 1076–1080. [Google Scholar] [CrossRef] [PubMed]
- Hao, X.; Zhou, X.R.; Pang, B.P.; Zhang, Z.R.; Bao, X. Morphological and biological characteristics of Galeruca daurica Joannis (Coleoptera: Chrysomelidae). Acta Agrestia Sin. 2015, 23, 1106–1108. (In Chinese) [Google Scholar]
- Gao, J.C.; Zhou, X.R.; Pang, B.P.; Bao, X.; Luo, J.P. Erdengqimuge. Effects of low temperature on the survivorship and development of overwintering eggs of Galeruca daurica (Coleoptera: Chrysomelidae). Acta Entomol. Sin. 2015, 58, 881–886. (In Chinese) [Google Scholar]
- Li, H.; Zhou, X.R.; Pang, B.P.; Chang, J. Supercooling capacity and cold hardiness of Galeruca daurica (Coleoptera: Chrysomelidae). Acta Entomol. Sin. 2014, 57, 212–217. (In Chinese) [Google Scholar]
- Li, H.; Zhou, X.R.; Pang, B.P.; Zhang, Z.R.; Chang, J.; Shan, Y.M. Effects of low-temperature stress on the supercooling capacity and development of Galeruca daurica (Joannis) larvae (Coleoptera: Chrysomelidae). Chin. J. Appl. Entomol. 2015, 52, 434–439. (In Chinese) [Google Scholar]
- Tan, Y.; Zhou, X.R.; Pang, B.P. Reference gene selection and evaluation for expression analysis using qRT-PCR in Galeruca daurica (Joannis). Bull. Entomol. Res. 2017, 107, 359–368. [Google Scholar] [CrossRef]
- Tan, Y.; Zhang, Y.; Huo, Z.J.; Zhou, X.R.; Pang, B.P. Molecular cloning of heat shock protein 10 (Hsp10) and 60 (Hsp60) cDNAs from Galeruca daurica (Coleoptera: Chrysomelidae) and their expression analysis. Bullet. Entomol. Res. 2018, 108, 510–522. [Google Scholar] [CrossRef]
- Zhang, H.L.; Ren, H.; Li, K.X.; Tian, Y.; Zhang, H.; Li, Y.Y.; Li, L.; Pang, B.P.; Tan, Y. Effects of RNAi-mediated silencing of GdHsp60 and GdHsp70 genes on the cold hardiness in Galeruca daurica (Coleoptera: Chrysomelidae). Acta Entomol. Sin. 2022, 65, 807–817. (In Chinese) [Google Scholar]
- Zhang, H.L.; Ren, H.; Tian, Y.; Shan, Y.M.; Wang, Y.; Li, L.; Li, Y.Y.; Pang, B.P.; Tan, Y. Cloning, identification and expression profiling analysis of GdHsp70-2, GdHsp70-3 genes in Galeruca daurica (Coleoptera: Chrysomelidae). Acta Agrestia Sin. 2023, 31, 61–72. (In Chinese) [Google Scholar]
- Gurusamy, D.; Mogilicherla, K.; Palli, S.R. Chitosan nanoparticles help double-stranded RNA escape from endosomes and improve RNA interference in the fall armyworm, Spodoptera frugiperda. Arch. Insect Biochem. Physiol. 2020, 104, e21677. [Google Scholar] [CrossRef] [PubMed]
- You, L.; Zhang, F.; Huang, S.; Merchant, A.; Zhou, X.; Li, Z. Over-expression of RNA interference (RNAi) core machinery improves susceptibility to RNAi in silkworm larvae. Insect Mol. Biol. 2020, 29, 353–362. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.H.; Hu, J.R.; Li, C.R.; Zhang, Y.J. Research progress in the cold tolerance mechanism of insects under environmental stress. Plant Prot. 2016, 42, 21–28. [Google Scholar]
- Hunter, S.; Apweiler, R.; Attwood, T.K.; Bairoch, A.; Bateman, A. InterPro: The integrative protein signature database. Nucleic Acids Res. 2009, 37, D211–D215. [Google Scholar] [CrossRef] [PubMed]
- Aparicio, G.; Gotz, S.; Conesa, A.; Segrelles, D.; Blanquer, I.; García, J.M.; Hernandez, V.; Robles, M.; Talon, M. Blast2GO goes. grid: Developing a grid-enabled prototype for functional genomics analysis. Stud. Health Technol. Inform. 2006, 120, 194–204. [Google Scholar] [PubMed]
- Deng, Y.Y.; Li, J.Q.; Wu, S.F.; Zhu, Y.P.; Chen, Y.W.; He, F.C. Integrated nr database in protein annotation system and its localization. Comput. Eng. 2006, 32, 71–74. [Google Scholar]
- Apweiler, R.; Bairoch, A.; Wu, C.H.; Barker, W.C.; Boeckmann, B.; Ferro, S.; Gasteiger, E.; Huang, H.; Lopez, R.; Magrane, M.; et al. UniProt: The universal protein knowledge base. Nucleic Acids Res. 2004, 32, D115–D119. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef]
- Kanehisa, M.; Goto, S.; Kawashima, S.; Okuno, Y.; Hattori, M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004, 32, D277–D280. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Dong, F.; Qian, K.; Miao, L.; Yang, X.; Ge, H.; Wu, Z.; Wang, J. Transcriptome analysis reveals global gene expression changes of Chilo suppressalis in response to sublethal dose of chlorantraniliprole. Chemosphere 2019, 234, 648–657. [Google Scholar] [CrossRef] [PubMed]
- Benjamini, Y.; Yekutieli, D. The control of the false discovery rate in multiple testing under dependency. Ann. Stat. 2001, 29, 1165–1188. [Google Scholar] [CrossRef]
- Xue, B.; Li, F.; Hu, J.; Tian, J.; Li, J.; Cheng, X.; Hu, J.; Li, B. Differentially expressed genes in the silk gland of silkworm (Bombyx mori) treated with TiO2 NPs. Gene 2017, 611, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T.; et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008, 36, 480–484. [Google Scholar] [CrossRef] [PubMed]
- Mao, X.; Cai, T.; Olyarchuk, J.G.; Wei, L. Automated genome annotation and pathway identification. using the KEGG orthology (KO) as a controlled vocabulary. Bioinformatics 2005, 21, 3787–3793. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, H.; Li, X.; Meng, Q.; Shu, R.; Wang, M.; Zhou, G.; Wang, H.; Miao, L.; Zhang, J.; et al. Cold adaptation mechanisms in the ghost moth Hepialus xiaojinensis: Metabolic regulation and thermal compensation. J. Insect Physiol. 2016, 85, 76–85. [Google Scholar] [CrossRef]
- Zhu, W.; Zhang, H.; Meng, Q.; Wang, M.; Zhou, G.; Li, X.; Wang, H.; Miao, L.; Qin, Q.; Zhang, J. Metabolic insights into the cold survival strategy and overwintering of the common cutworm, Spodoptera litura (Fabricius) (Lepidoptera: Noctuidae). J. Insect Physiol. 2017, 100, 53–64. [Google Scholar] [CrossRef]
- Chen, J.; Wu, Y.C.; Chen, J.K.; Zhu, X.J.; Merkler, D.; Liao, C.H.; Han, Q. Elongases of Long-Chain Fatty Acids ELO2 and ELO9 Are Involved in Cuticle Formation and Function in Fecundity in the Yellow Fever Mosquito, Aedes aegypti. Insects 2023, 14, 189. [Google Scholar] [CrossRef]
- Duell, M.E.; Gray, M.T.; Roe, A.D.; MacQuarrie, C.J.K.; Sinclair, B.J. Plasticity drives extreme cold tolerance of emerald ash borer (Agrilus planipennis) during a polar vortex. Curr. Res. Insect Sci. 2022, 3, 100031. [Google Scholar] [CrossRef]
- Muluvhahothe, M.M.; Joubert, E.; Foord, S.H. Thermal tolerance responses of the two-spotted. stink bug, Bathycoelia distincta (Hemiptera: Pentatomidae), vary with life stage and the sex of adults. J. Therm. Biol. 2023, 111, 103395. [Google Scholar] [CrossRef] [PubMed]
- Kuo, Y.; Lu, Y.H.; Lin, Y.H.; Lin, Y.C.; Wu, Y.L. Elevated temperature affects energy. metabolism and behavior of bumblebees. Insect Biochem. Mol. Biol. 2023, 155, 103932. [Google Scholar] [CrossRef] [PubMed]
- Lebenzon, J.E.; Overgaard, J.; Jørgensen, L.B. Chilled, starved or frozen: Insect mitochondrial. adaptations to overcome the cold. Curr. Opin. Insect Sci. 2023, 58, 101076. [Google Scholar] [CrossRef] [PubMed]
- Wang, N.; Ji, A.; Masoudi, A.; Li, S.; Hu, Y.; Zhang, Y.; Yu, Z.; Wang, H.; Wang, H.; Liu, J. Protein regulation mechanism of cold tolerance in Haemaphysalis longicornis. Insect Sci. 2023, 30, 725–740. [Google Scholar] [CrossRef]
- Mikucki, E.E.; Lockwood, B.L. Local thermal environment and warming influence. supercooling and drive widespread shifts in the metabolome of diapausing Pieris rapae butterflies. J. Exp. Biol. 2021, 224, jeb243118. [Google Scholar] [CrossRef] [PubMed]
- Kojić, D.; Popović, Ž.D.; Orčić, D.; Purać, J.; Orčić, S.; Vukašinović, E.L.; Nikolić, T.V.; Blagojević, D.P. The influence of low temperature and diapause phase on sugar and polyol content in the European corn borer Ostrinia nubilalis (Hbn.). J. Insect Physiol. 2018, 109, 107–113. [Google Scholar] [CrossRef] [PubMed]
- Enriquez, T.; Teets, N.M. Lipid Metabolism in Response to Cold. 2023. Available online: https://ecoevorxiv.org/repository/view/5092/ (accessed on 13 April 2023).
- Li, Y.J.; Ma, C.S.; Yan, Y.; Renault, D.; Colinet, H. The interspecific variations in molecular responses to various doses of heat and cold stress: The case of cereal aphids. J. Insect Physiol. 2023, 147, 104520. [Google Scholar] [CrossRef]
- Tonione, M.A.; Bi, K.; Tsutsui, N.D. Transcriptomic signatures of cold adaptation and heat. stress in the winter ant (Prenolepis imparis). PLoS ONE 2020, 15, e0239558. [Google Scholar] [CrossRef]
- Liu, X.; Fu, Z.X.; Kang, Z.W.; Li, H.; Liu, T.X.; Wang, D. Identification and characterization of antioxidant enzyme genes in parasitoid Aphelinus asychis (Hymenoptera: Aphelinidae) and expression profiling analysis under temperature stress. Insects 2022, 13, 447. [Google Scholar] [CrossRef]
- Yuan, J.W.; Song, H.X.; Chang, Y.W.; Yang, F.; Xie, H.F.; Gong, W.R.; Du, Y.Z. Identification, expression analysis and functional verification of two genes encoding small heat shock proteins in the western flower thrips, Frankliniella occidentalis (Pergande). Int. J. Biol. Macromol. 2022, 211, 74–84. [Google Scholar] [CrossRef]
- Ashraf, H.; Qamar, A. A review on heat stress altering the insect life history strategies and underlying mechanisms: Special reference to an economically important Lepidoptera, Bombyx mori (Linnaeus, 1758) (Lepidoptera: Bombycidae). Shilap-Rev. Lepidopt. 2023, 51, 339–357. [Google Scholar] [CrossRef]
- Yang, C.L.; Meng, J.Y.; Zhou, L.; Yao, M.S.; Zhang, C.Y. Identification of five small heat. Shock protein genes in Spodoptera frugiperda and expression analysis in response to different environmental stressors. Cell Stress Chaperones 2021, 26, 527–539. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Miano, F.N.; Jiang, T.; Peng, Y.; Zhang, W.; Xiao, H. Characterization of Three Heat Shock Protein Genes in Pieris melete and Their Expression Patterns in Response to Temperature Stress and Pupal Diapause. Insects 2022, 13, 430. [Google Scholar] [CrossRef] [PubMed]
- Ruan, H.Y.; Meng, J.Y.; Yang, C.L.; Zhou, L.; Zhang, C.Y. Identification of six small heat shock protein genes in Ostrinia furnacalis (Lepidoptera: Pyralidae) and analysis of their expression patterns in response to environmental stressors. J. Insect Sci. 2022, 22, 7. [Google Scholar] [CrossRef] [PubMed]
- Jin, J.; Liu, Y.; Liang, X.; Pei, Y.; Wan, F.; Guo, J. Regulatory mechanism of transcription. factor AhHsf modulates AhHsp70 transcriptional expression enhancing heat tolerance in Agasicles hygrophila (Coleoptera: Chrysomelidae). Int. J. Mol. Sci. 2022, 23, 3210. [Google Scholar] [CrossRef] [PubMed]
- Yi, J.; Wu, H.; Liu, J.; Lai, X.; Guo, J.; Li, D.; Zhang, G. Molecular characterization and expression of six heat shock protein genes in relation to development and temperature in Trichogramma chilonis. PLoS ONE 2018, 13, e0203904. [Google Scholar] [CrossRef] [PubMed]
- Seddigh, S. Proteomics analysis of two heat shock proteins in insects. J. Biomol. Struct. Dyn. 2019, 37, 2652–2668. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Li, X.J.; Lü, J.H.; Huo, M.F. The effect of acclimation on heat tolerance of Lasioderma serricorne (Fabricius) (Coleoptera: Anobiidae). J. Therm. Biol. 2018, 71, 153–157. [Google Scholar] [CrossRef]
- Liu, J.; Liu, Y.; Li, Q.; Lu, Y. Heat shock protein 70 and Cathepsin B genes are involved in the thermal tolerance of Aphis gossypii. Pest. Manag. Sci. 2023, 79, 2075–2086. [Google Scholar] [CrossRef]
- Chen, K.; Tang, T.; Song, Q.; Wang, Z.; He, K.; Liu, X.; Song, J.; Wang, L.; Yang, Y.; Feng, C. Transcription Analysis of the Stress and Immune Response Genes to Temperature Stress in Ostrinia furnacalis. Front. Physiol. 2019, 10, 1289. [Google Scholar] [CrossRef]




| Gene ID | Gene Name | Primer Sequences | Product Size | Purpose |
|---|---|---|---|---|
| NM_010477.4 | Hspd1 | F: CGCTCTGCTTCGGTGCAT | 275 | q-PCR |
| R: CAGCAGCATCCAGTAAGGCA | ||||
| KY460464.1 | Hsp10 | F: GGCAACTACGAAAACGAAAGG | 126 | |
| R: CCGCATTGTCTTCGAGGTTT | ||||
| KY460462.1 | Hsp70A1 | F: ACACTGGCCGTTTAAGGTGG | 276 | |
| R: GCTGCTGCTGTTGGTTCATT | ||||
| MZ853083.1 | Hsp70-2 | F: GGAACCATTGCTGGTCTTGT | 214 | |
| R: CGAAATCTTCACCTCCCAAA | ||||
| KO585088 | Hsp70-3 | TTCAACACTGGCCATTCAAA | 218 | |
| TTTTGTAGCTTGTCTCTGTG | ||||
| NM_001109900.1 | Akr1c15 | F: GGAAGTAGATGCAGGTCGAGT | 259 | |
| R: AACACCTGGCAGCTTGTGAT | ||||
| AY776138.1 | Pepck | F: CGGTTTCTTCGGTGTTGCTC | 243 | |
| R: TCCGCAGGAGTGCAAAATCT | ||||
| ON054187.1 | Scd1 | F: ATGCTGATCCTCACAACGCT | 212 | |
| R: TAAATACATTGGGGCGGCGA | ||||
| KU240575.1 | SDHA | F: GGGAGACCACAATCTCCTCA | 192 | |
| R: AGCTGGTGCTCCTAAGTCCA | ||||
| dsHsp70-1 | F: taatacgactcactatagggGGTTCGACTCGTATTCCCAA | 435 | RNAi | |
| R: taatacgactcactatagggCCTTGGTCATTGGACGTTCT | ||||
| dsHsp70-2 | F: taatacgactcactatagggGGTTCGACTCGTATTCCCAA | 334 | ||
| R: taatacgactcactatagggCCTTGGTCATTGGACGTTCT | ||||
| dsHsp70-3 | F: taatacgactcactatagggAAGGAAGAGGATGAACGCCAGA | 323 | ||
| R: taatacgactcactatagggAATCCACCTGCTTGTTGACC | ||||
| dsGFP | F: taatacgactcactatagggGGAGAAGAACTTTTCACTGG | 560 | ||
| R: taatacgactcactatagggAGTTGAACGGATCCATCTTC |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Sun, F.; Zhang, W.; Gao, X.; Du, L.; Yun, X.; Li, Y.; Li, L.; Pang, B.; Tan, Y. Comparative Transcriptome Analysis of Galeruca daurica Reveals Cold Tolerance Mechanisms. Genes 2023, 14, 2177. https://doi.org/10.3390/genes14122177
Zhang H, Sun F, Zhang W, Gao X, Du L, Yun X, Li Y, Li L, Pang B, Tan Y. Comparative Transcriptome Analysis of Galeruca daurica Reveals Cold Tolerance Mechanisms. Genes. 2023; 14(12):2177. https://doi.org/10.3390/genes14122177
Chicago/Turabian StyleZhang, Hongling, Feilong Sun, Wenbing Zhang, Xia Gao, Lei Du, Xiaopeng Yun, Yanyan Li, Ling Li, Baoping Pang, and Yao Tan. 2023. "Comparative Transcriptome Analysis of Galeruca daurica Reveals Cold Tolerance Mechanisms" Genes 14, no. 12: 2177. https://doi.org/10.3390/genes14122177
APA StyleZhang, H., Sun, F., Zhang, W., Gao, X., Du, L., Yun, X., Li, Y., Li, L., Pang, B., & Tan, Y. (2023). Comparative Transcriptome Analysis of Galeruca daurica Reveals Cold Tolerance Mechanisms. Genes, 14(12), 2177. https://doi.org/10.3390/genes14122177






