Cloning, Identification, and Functional Analysis of the Foxl2 Gene in Procambarus clarkii
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Materials
2.2. Primer Design and Synthesis
2.3. Total RNA Extraction and cDNA Synthesis
2.4. PcFoxl2 Gene Cloning
2.5. Bioinformatics Analysis
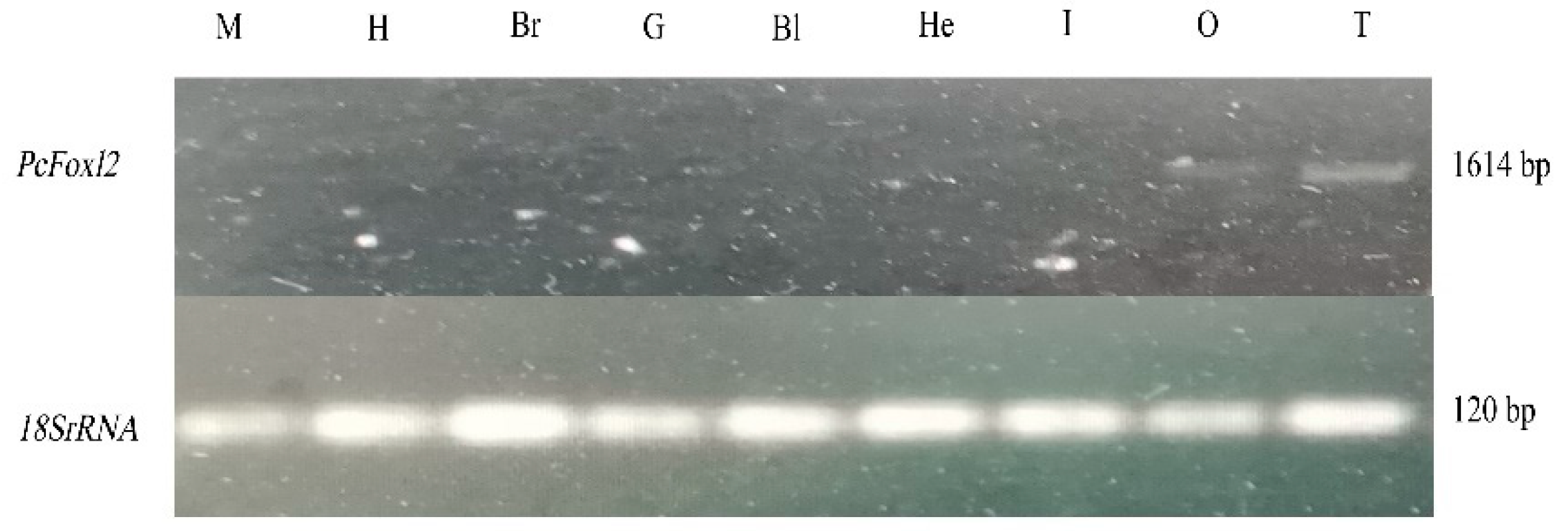
2.6. Expression of the PcFoxl2 Gene in Different Tissues of P. clarkii
2.7. In Situ Hybridization
2.8. Exploration of the Function of PcFoxl2 in Gonadal Development
3. Results
3.1. PcFoxl2 Gene Sequence Characteristics
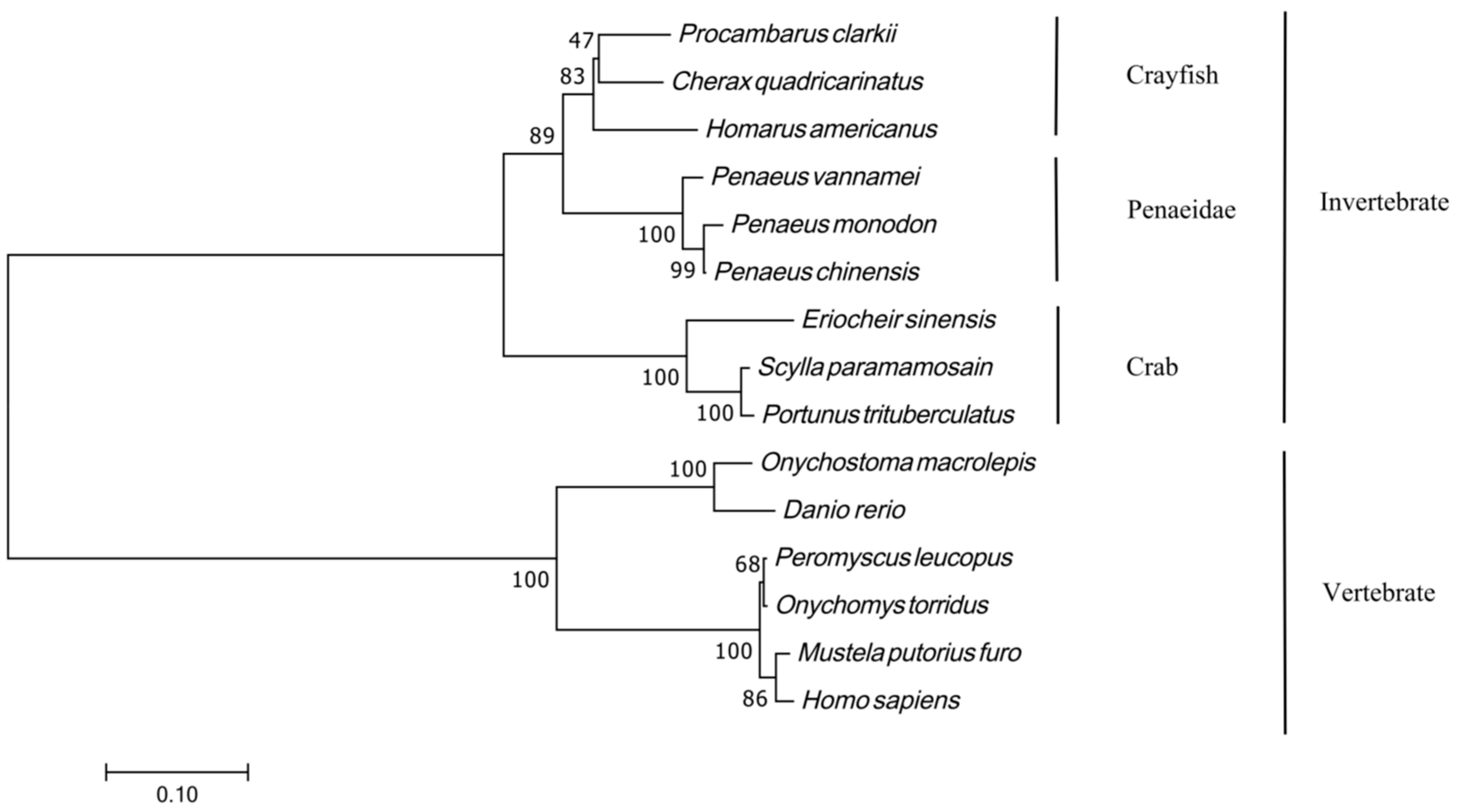
3.2. PcFoxl2 Amino Acid Homology Comparison and Phylogenetic Tree Construction
3.3. Expression Level of PcFoxl2 in the Gonadal Tissue
3.4. Expression Pattern of PcFoxl2 during Ovarian Development
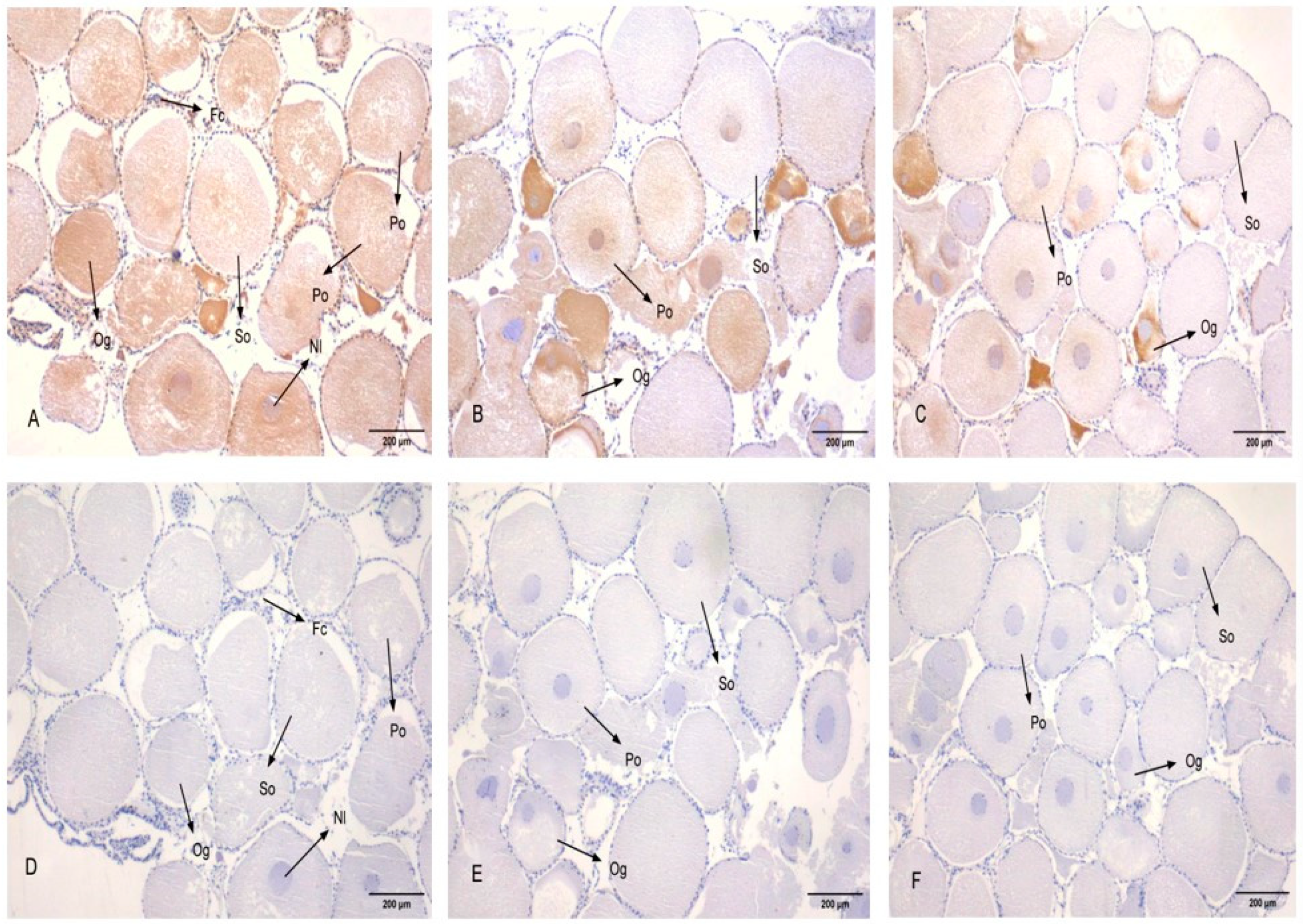
3.5. Subcellular Localization of PcFoxl2 in Gonadal Tissue
3.6. Expression Effects of PcFoxl2 Gene after Interference
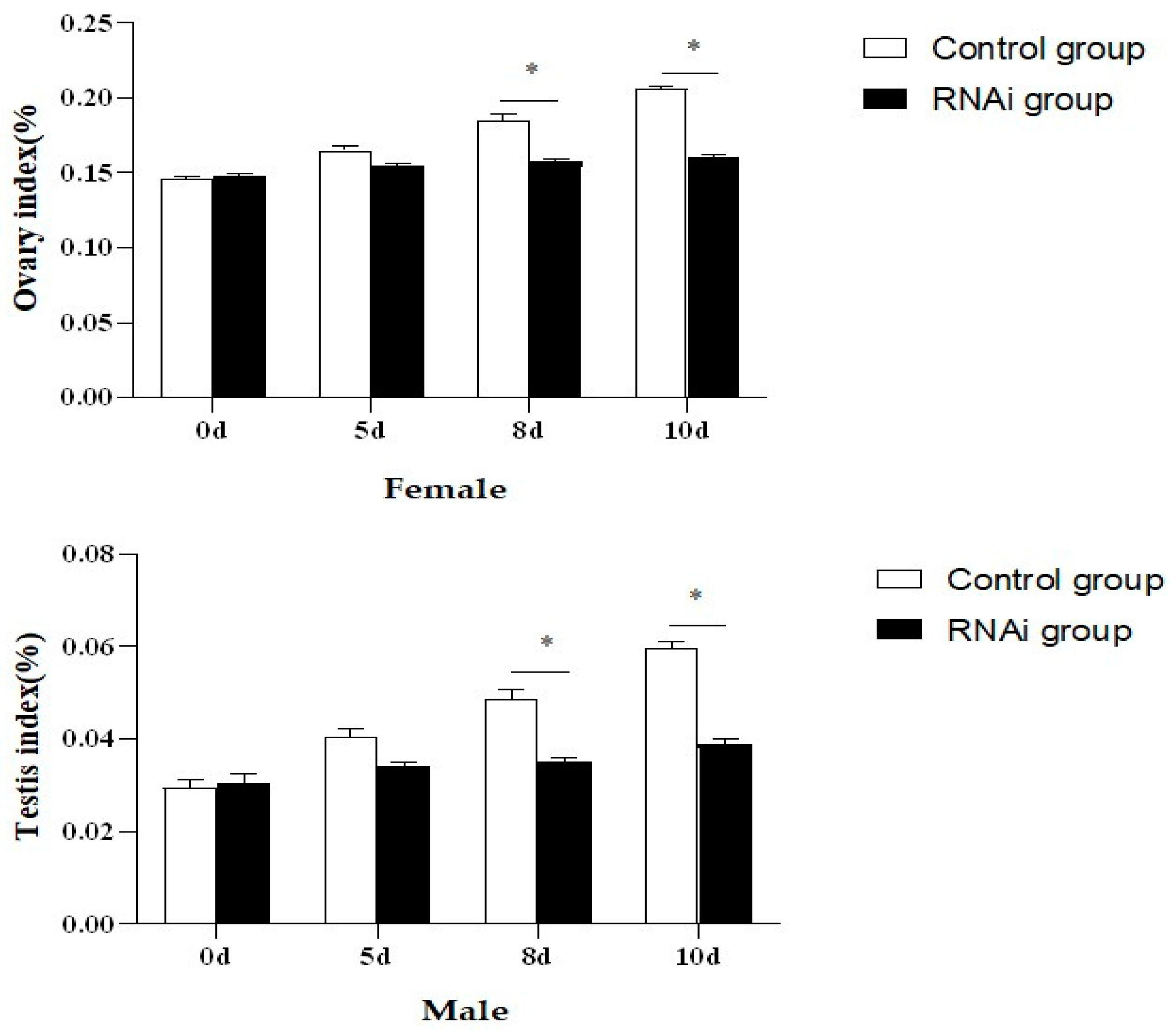
3.7. Expression Characteristics of Genes Related to Gonadal Development in P. clarkii after PcFoxl2 Interference
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Yue, G.H.; Li, J.; Bai, Z.Y.; Wang, C.M.; Feng, F. Genetic diversity and population structure of the invasive alien red swamp crayfish. Biol. Invasions 2010, 12, 2697–2706. [Google Scholar] [CrossRef]
- Azra, M.N.; Wong, L.L.; Aouissi, H.A.; Zekker, I.; Amin, M.A.; Adnan, W.N.W.; Abdullah, M.F.; Abd Latif, Z.; Noor, M.I.M.; Lananan, F.; et al. Crayfish Research: A Global Scientometric Analysis Using Cite Space. Animals 2023, 13, 1240. [Google Scholar] [CrossRef] [PubMed]
- Peng, B.; Tan, Y.F.; Peng, G.H.; Xiong, L.J.; Bai, X.F. Path Analysis of Effects of Phenotypic Traits Attributes on Abdomen Meat Weight of Red Swamp Crayfish Procambarus clarkii. Fish. Sci. 2021, 40, 718–725. [Google Scholar]
- Yuan, J.; Liao, C.S.; Zhang, T.L.; Guo, C.B.; Liu, J.S. Advances in Ecology Research on Integrated Rice Field Aquaculture in China. Water 2022, 14, 2333. [Google Scholar] [CrossRef]
- Xu, H.; Zhao, Y.Y.; Sun, D.Y.; Ke, Y.Y.; Zhang, L.L.; Chen, X.; Wei, F.Z.; Li, J.C. Progress in Integrated Rice-crayfish Farming System. J. Agric. Sci. Technol. 2022, 24, 160–168. [Google Scholar]
- Jayasankar, V.; Tomy, S.; Wilder, M.N. Insights on Molecular Mechanisms of Ovarian Development in Decapod Crustacea: Focus on Vitellogenesis-Stimulating Factors and Pathways. Front. Endocrinol. 2020, 11, 577925. [Google Scholar] [CrossRef]
- Huang, X.; Tang, Z.; Liu, J.; Zhang, H.Y.; Zhong, Y.Z.; Lu, Z.F.; Hou, S.J.; Wang, D.P.; Lu, Z.L. Genetic diversity microsatellite analysis of Procambarus clarkii populations in different regions of Guangxi. J. South. Agric. 2020, 51, 437–444. [Google Scholar]
- Yu, J.; Xiong, M.; Ye, S.; Li, W.; Xiong, F.; Liu, J.S.; Zhang, T.L. Effects of stocking density and artificial macrophyte shelter on survival, growth and molting of juvenile red swamp crayfish (Procambarus clarkii) under experimental conditions. Aquaculture 2020, 521, 735001. [Google Scholar] [CrossRef]
- Song, G.T.; Jiang, H.; Wang, F.; Jiang, Y.L.; Chen, Z.; Wang, J.J.; Wu, C.C. Effect of Greenhouse and Broodstock Size on Breeding Performance of Red Swamp Crayfish Procambarus clarkii. Fish. Sci. 2022, 41, 130–136. [Google Scholar]
- Wan, H.F.; Zhong, J.Y.; Zhang, Z.P.; Xie, Y.C.; Wang, Y.L. Characterization of the foxl2 gene involved in the vtg expression in mud crab (Scylla paramamosain). Gene 2021, 798, 145807. [Google Scholar] [CrossRef]
- Liu, J.; Han, L.; Li, R.F.; Xu, P.; Lu, Y.; Yang, M.; Wang, S.H. Eukaryotic expression and purification of FOXL2 gene in leghorn. J. Northwest A F Univ. Nat. Sci. Ed. 2016, 44, 10–15. [Google Scholar]
- Wu, J.; Wang, D.; Miao, C.L.; Han, Y.Y. Current Research and the Regulation Mechanism of FOXL2. Chin. J. Cell Biol. 2018, 40, 602–607. [Google Scholar]
- Schmidt, D.; Ovitt, C.E.; Anlag, K.; Fehsenfeld, S.; Gredsted, L.; Treier, A.C.; Treier, M. The murine winged-helix transcription factor Foxl2 is required for granulosa cell differentiation and ovary maintenance. Development 2004, 131, 933–942. [Google Scholar] [CrossRef] [PubMed]
- Nakamoto, M.; Matsuda, M.; Wang, D.S.; Nagahama, Y.; Shibata, N. Molecular cloning and analysis of gonadal expression of Foxl2 in the medaka, Oryzias latipes. Biochem. Biophys. Res. Commun. 2006, 344, 353–361. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.P.; Zhu, C.H.; Sun, J.; Wang, W.D.; Wu, T.L.; Chen, H.P.; Shi, S.L.; Li, G.L. Foxl2 of the Hong Kong catfish (Clarias fuscus): cDNA cloning, tissue distribution and changes in gene expression towards methyltestosterone, estradiol and letrozole exposure of the fries during gonadal differentiation. Genes Genom. 2015, 37, 669–677. [Google Scholar] [CrossRef]
- Hu, Q.; Guo, W.; Gao, Y.; Tang, R.; Li, D.P. Molecular cloning and analysis of gonadal expression of Foxl2 in the rice-field eel Monopterus albus. Sci. Rep. 2014, 4, 6884. [Google Scholar] [CrossRef] [PubMed]
- Yang, Y.J.; Wang, Y.; Li, Z.; Zhou, L.; Gui, J.F. Sequential, Divergent, and Cooperative Requirements of Foxl2a and Foxl2b in Ovary Development and Maintenance of Zebrafish. Genetics 2017, 205, 1551–1572. [Google Scholar] [CrossRef]
- Yarmohammadi, M.; Pourkazemi, M.; Kazemi, R. Differential expression of foxl2 and cyp19a1a mRNA during gonad developmental stages in great sturgeon Huso huso. J. Fish Biol. 2017, 90, 1104–1111. [Google Scholar] [CrossRef]
- Yarmohammadi, M.; Pourkazemi, M.; Kazemi, R. Expression pattern of foxl2 and dmrt1 in gonad of Amur sturgeon Acipenser schrenckii in relation to sex differentiation. Aquaculture 2017, 479, 712–720. [Google Scholar]
- Liu, H.F.; Mu, X.J.; Gui, L.; Su, M.L.; Li, H.; Zhang, G.; Liu, Z.H.; Zhang, J.B. Characterization and gonadal expression of FOXL2 relative to Cyp19a genes in spotted scat Scatophagus argus. Gene 2015, 561, 6–14. [Google Scholar] [CrossRef]
- He, J.Q.; Zhang, H.; She, Z.C.; Zhen, W.Q.; You, S.Y.; Wang, P.L.; Xu, Y.H.; Zhu, P. Cloning and Expression Pattern Investigation of Crassostrea hongkongensis Foxl2 Gene. Genom. Appl. Biol. 2020, 39, 1519–1528. [Google Scholar]
- Liu, X.L.; Li, Y.; Liu, J.G.; Cui, L.B.; Zhang, Z.F. Gonadogenesis in scallop Chlamys farreri and Cf-foxl2 expression pattern during gonadal sex differentiation. Aquac. Res. 2016, 47, 1605–1611. [Google Scholar] [CrossRef]
- Fan, S.T.; Li, X.X.; Lin, S.Y.; Li, Y.P.; Ma, H.X.; Zhang, Z.F.; Qin, Z.K. Screening and Identification of Transcription Factors Potentially Regulating Foxl2 Expression in Chlamys farreri Ovary. Biology 2022, 11, 113. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.L.; Liu, P.; Jia, F.L.; Li, J.; Gao, B.Q. De novo Transcriptome Analysis of Portunus trituberculatus Ovary and Testis by RNA-Seq: Identification of Genes Involved in Gonadal Development. PLoS ONE 2015, 10, e0128659. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Xie, J.; He, L.; Wang, Y.L.; Yang, H.D.; Duan, Z.L.; Wang, Q. FOXL2 down-regulates vitellogenin expression at mature stage in Eriocheir sinensis. Biosci. Rep. 2015, 35, e00278. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.K.; Chen, H.L.; Zhang, Q.F.; Tan, K.; Liao, S.L.; Wang, W.M. Molecular cloning and expression patterns of a sex-biased transcriptional factor Foxl2 in the giant freshwater prawn (Macrobrachium rosenbergii). Mol. Biol. Rep. 2023, 50, 3581–3591. [Google Scholar] [CrossRef]
- Zhong, Y.Z.; Zhao, W.B.; Tang, Z.S.; Huang, L.M.; Zhu, X.X.; Liang, X.; Yan, A.; Lu, Z.; Yu, Y.L.; Tang, D.S.; et al. Comparative transcriptomic analysis of the different developmental stages of ovary in red swamp crayfish Procambarus clarkii. BMC Genom. 2021, 22, 199. [Google Scholar] [CrossRef]
- Dai, Y.; Gong, X.J.; Li, B.; Wang, Y.F.; Huang, W.G. Reproductive Period of Procambarus clarkii in Wuhan Area. Chin. J. Zool. 2008, 43, 21–27. [Google Scholar]
- Jiang, H.C.; Xing, Z.J.; Lu, W.; Qian, Z.J.; Yu, H.W.; Li, J. Transcriptome Analysis of Red Swamp Crawfish Procambarus clarkii Reveals Genes Involved in Gonadal Development. PLoS ONE 2014, 9, e105122. [Google Scholar] [CrossRef]
- Wang, G.L.; Dong, S.S.; Guo, P.F.; Cui, X.Y.; Duan, S.H.; Li, J. Identification of Foxl2 in freshwater mussel Hyriopsis cumingii and its involvement in sex differentiation. Gene 2020, 754, 144853. [Google Scholar] [CrossRef]
- Ning, J.H.; Cao, W.A.; Lu, X.; Chen, M.; Liu, B.; Wang, C.D. Identification and functional analysis of a sex-biased transcriptional factor Foxl2 in the bay scallop Argopecten irradians irradians. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2021, 256, 110638. [Google Scholar] [CrossRef]
- Ashida, H.; Ueyama, N.; Kinoshita, M.; Kobayashi, T. Molecular identification and expression of FOXL2 and DMRT1 genes from willow minnow Gnathopogon caerulescens. Reprod. Biol. 2013, 13, 317–324. [Google Scholar] [CrossRef]
- Baron, D.; Cocquet, J.; Xia, X.H.; Fellous, M.; Guiguen, Y.; Veitia, R.A. An evolutionary and functional analysis of FoxL2 in rainbow trout gonad differentiation. J. Mol. Endocrinol. 2004, 33, 705–715. [Google Scholar] [CrossRef]
- Wang, D.S.; Kobayashi, T.; Zhou, L.Y.; Nagahama, Y. Molecular cloning and gene expression of Foxl2 in the Nile tilapia, Oreochromis niloticus. Biochem. Biophys. Res. Commun. 2004, 320, 83–89. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.D.; Zhang, G.R.; Wei, K.J.; Ji, W.; Gardner, J.P.A.; Yang, R.B.; Chen, K.C. Molecular identification and expression of the Foxl2 gene during gonadal sex differentiation in northern snakehead Channa argus. Fish Physiol. Biochem. 2015, 41, 1419–1433. [Google Scholar] [CrossRef]
- Bhat, I.A.; Rather, M.A.; Dar, J.Y.; Sharma, R. Molecular cloning, computational analysis and expression pattern of forkhead box l2 (Foxl2) gene in catfish. Comput. Biol. Chem. 2016, 64, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.L.; Liang, S.Q.; Zhong, Y.F.; Wang, Y.R.; Lao, J.T.; Li, G.L.; Huang, H.; Chen, H.P. Molecular Cloning and Tissue Expression Patterns of foxl2 Gene in Bettas plendens. Genom. Appl. Biol. 2021, 40, 109–116. [Google Scholar]
- Shu, C.; Wang, L.J.; Zou, C.C.; Tan, X.G.; Zou, Y.X.; Kong, L.M.; Wu, Z.H.; Wu, Q.W.; Wang, L.; Wang, G.Y.; et al. Function of Foxl2 and Dmrt1 proteins during gonadal differentiation in the olive flounder Paralichthys olivaceus. Int. J. Biol. Macromol. 2022, 215, 141–154. [Google Scholar] [CrossRef] [PubMed]
- Jin, S.B.; Fu, H.; Jiang, S.F.; Xiong, Y.W.; Sun, S.M.; Qiao, H.; Zhang, W.Y.; Gong, Y.S.; Wu, Y. Molecular Cloning, Expression, and In Situ Hybridization Analysis of Forkhead Box Protein L2 during Development in Macrobrachium nipponense. J. World Aquac. Soc. 2018, 49, 429–440. [Google Scholar] [CrossRef]
- Monné, R.J.M.; Frisk, A.L.; Kreutzer, R.; Lemarchand, T.; Lezmi, S.; Saravanan, C.; Stierstorfer, B.; Thuilliez, C.; Vezzali, E.; Wieczorek, G.; et al. European Society of Toxicologic Pathology (Pathology 2.0 Molecular Pathology Special Interest Group): Review of In Situ Hybridization Techniques for Drug Research and Development. Toxicol. Pathol. 2023, 51, 92–111. [Google Scholar] [CrossRef]
- Wei, H.L.; Li, W.; Liu, T.; Li, Y.J.; Liu, L.J.; Shu, Y.; Zhang, L.J.; Xing, Q.; Zhang, L.L.; Bao, Z.M. Sexual Development of the Hermaphroditic Scallop Argopecten irradians Revealed by Morphological, Endocrine and Molecular Analysis. Front. Cell Dev. Biol. 2021, 9, 646754. [Google Scholar] [CrossRef] [PubMed]
- Tan, K.; Li, Y.H.; Zhou, M.; Wang, W.M. siRNA knockdown of MrIR induces sex reversal in Macrobrachium rosenbergii. Aquaculture 2020, 523, 735172. [Google Scholar] [CrossRef]
- Sun, D.F.; Yu, H.; Li, Q. Examination of the roles of Foxl2 and Dmrt1 in sex differentiation and gonadal development of oysters by using RNA interference. Aquaculture 2022, 548, 737732. [Google Scholar] [CrossRef]
- Zhou, Y.Y.; Li, Y.Q.; Liao, Q.L.; Gongpengyang, S.; Qin, Y.P.; Zhang, Y.H.; Ma, H.T.; Li, J.; Yu, Z.N. Examination of the role of the forkhead box L2 (Foxl2) in gonadal and embryonic development in the boring giant clam Tridacna crocea. Aquaculture 2022, 560, 738554. [Google Scholar] [CrossRef]
- Shen, S.; Hu, J.X.; Shen, Q.; Chen, H.; Gao, H.; Lai, X.F. Cloning of notch1 and its role in the growth and development of Exopalaemon carinicauda. Aquac. Rep. 2023, 30, 101537. [Google Scholar] [CrossRef]
- Li, Y.Z.; Zhang, S.S.; Liu, X.W.; Zhang, R.; Lin, X.H.; Wang, L.; Yu, M.; Qiao, Z.G.; Jiang, H.X. cDNA cloning and expression of ribosomal protein S3a gene in Macrobrachium nipponense and its function in ovarian development. Freshw. Fish. 2023, 53, 57–68. [Google Scholar]
- Liu, J.Q.; Sun, M.; Wang, L. Cloning of the partial sequences of nanos1,nanos2 and piwi genes and analysing of their expression patterns in the Sinopotamon henanense. Freshw. Fish. 2020, 50, 10–16. [Google Scholar]
- Iatsui, A.; Chotigeat, W.; Deachamag, P. Effects of ribosomal protein S3a (RPS3a) on the survival of WSSV-infected pacific white shrimp (Litopenaeus vannamei). Scienceasia 2020, 46, 43–50. [Google Scholar] [CrossRef]
- Zhu, Y.K.; Chen, H.L.; Wei, Y.F.; Wang, W.M. Effects of Mrfoxl 2—dsRNA on the expression of foxl 2 and reproduction—Related genes in the ovaries of Macrobrachium rosenbergii. Freshw. Fish. 2022, 52, 38–43. [Google Scholar]
- Zhang, C.L.; He, Q.; Cheng, H.T.; Li, L.H.; Ruan, X.H.; Duan, X.Z.; Huang, F.Q.; Yang, H.; Zhang, H.F.; Shi, H.R.; et al. Transcription factors foxl2 and foxl3 regulate cyp19a1a and cyp11b in orange-spotted grouper (Epinephelus coioides). Aquac. Rep. 2022, 25, 101243. [Google Scholar] [CrossRef]
- He, K.; Wu, H.D.; Zhang, S.L.; Liu, Z.Z.; Ruan, J.Z.; Sun, Y.W.; Li, F.G. Cloning, identification and tissue expression analysis of nanos3 gene in Monopterus albus. J. South. Agric. 2022, 53, 2321–2330. [Google Scholar]
- Li, H.j.; Zhang, J.Q.; Wu, S.G.; Wei, L.L.; Liu, Y. Cloning and expression analysis of short type peptidoglycan recognition protein s3 gene from hyriopsis cumingii. Acta Hydrobiol. Sin. 2016, 40, 921–927. [Google Scholar]
- Navakanitworakul, R.; Deachamag, P.; Wonglapsuwan, M.; Chotigeat, W. The roles of ribosomal protein S3a in ovarian development of Fenneropenaeus merguiensis (De Man). Aquaculture 2012, 338, 208–215. [Google Scholar] [CrossRef]


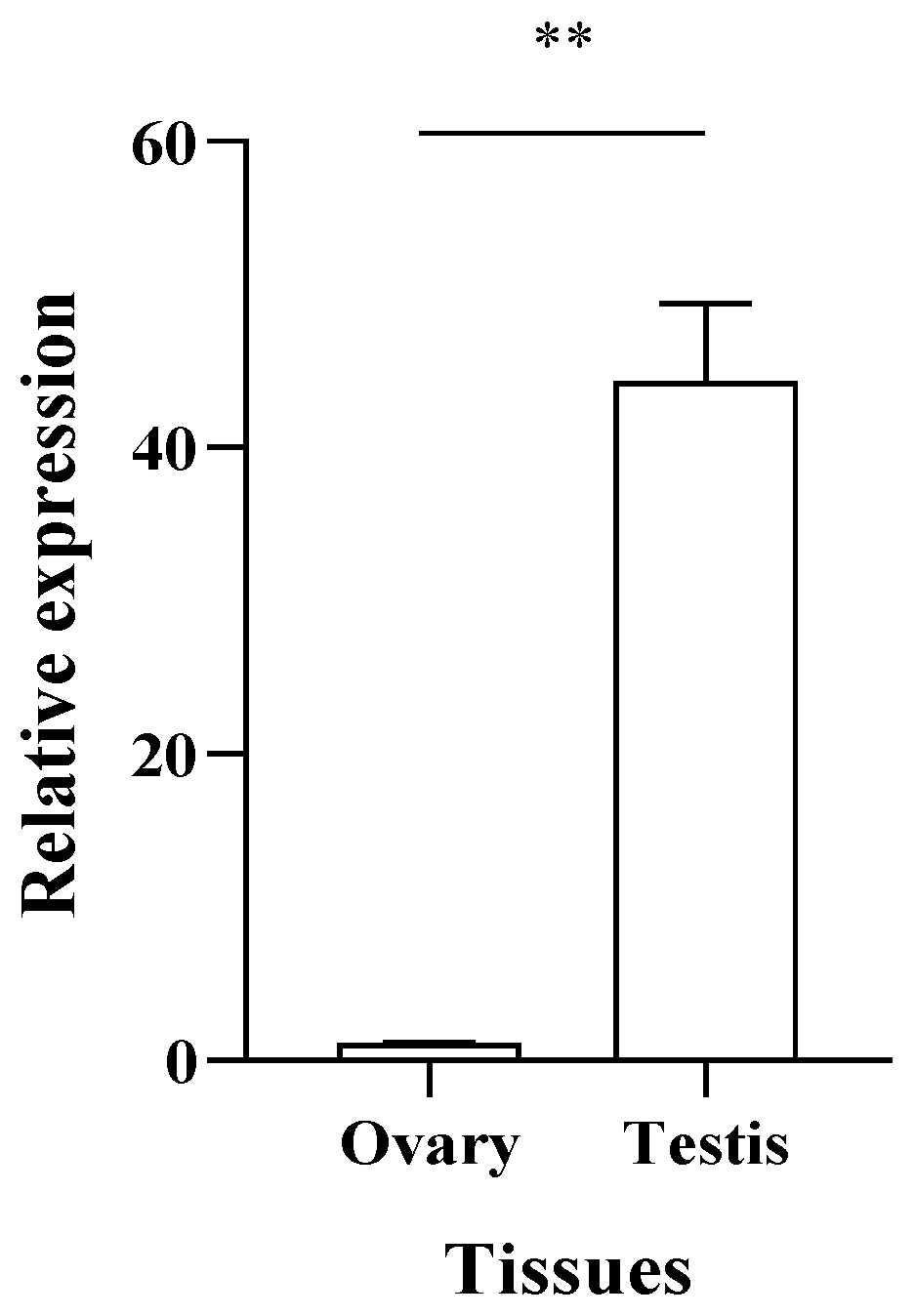





| Primer | Sequence (5′–3′) | Application |
|---|---|---|
| Foxl2-F Foxl2-R | GGATCCATGAAGGACGATTACCTCACT CTCGAGGAGCTTGGAGTCCGGCCAGTA | RT-PCR |
| Foxl2-Pro | TTGTAGGTTTTCTCGTAGCACTTCTCGT | Probe of CISH |
| CCTTCGTGTTCTCGTCCTTTTTACTTC | ||
| CCGTAGGACTTAACCTCATGGCTGTC | ||
| Foxl2-QF Foxl2-QR | TGACTACCTTGGGCTCACCT TGCATCTGACCTTGGAGCTG | qPCR |
| 18SrRNA-F 18SrRNA-R | AATGTCTCGTGGTGGAAAAG TTAACTGTTATCTTTACCTTCC | RT-PCR |
| 18SrRNA-QF 18SrRNA-QR | GGGGAGGTAGTGACGAAAAAT TATACGCTAGTGGAGCTGGAA | qPCR |
| Foxl2-dsF Foxl2-dsR | AGTGGATCGCCACCAAGTTT CAGCGCTCCTGAGGACAAT | dsRNA |
| Foxl2-T7-dsF Foxl2-T7-dsR | GGATCCTAATACGACTCACTATAGGAGTGGATCGCCACCAAGTTT GGATCCTAATACGACTCACTATAGGCAGCGCTCCTGAGGACAAT | dsRNA |
| Group | IW (g) | FW (g) | SR (%) | GSI (%) | |
|---|---|---|---|---|---|
| Female | Male | ||||
| Experimental group | 20.94 ± 3.745 | 21.02 ± 3.744 | 50.833 | 0.16 ± 0.015 | 0.04 ± 0.002 |
| Control group | 20.99 ± 4.257 | 21.08 ± 4.252 | 55.833 | 0.21 ± 0.023 | 0.06 ± 0.003 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Huang, J.; Zhu, W.; Peng, M.; Yang, C.; Chen, X.; Wu, T.; Zeng, D.; Zhao, Y.; Chen, X. Cloning, Identification, and Functional Analysis of the Foxl2 Gene in Procambarus clarkii. Genes 2023, 14, 2190. https://doi.org/10.3390/genes14122190
Huang J, Zhu W, Peng M, Yang C, Chen X, Wu T, Zeng D, Zhao Y, Chen X. Cloning, Identification, and Functional Analysis of the Foxl2 Gene in Procambarus clarkii. Genes. 2023; 14(12):2190. https://doi.org/10.3390/genes14122190
Chicago/Turabian StyleHuang, Jin, Weilin Zhu, Min Peng, Chunling Yang, Xiaohan Chen, Tiejun Wu, Digang Zeng, Yongzhen Zhao, and Xiuli Chen. 2023. "Cloning, Identification, and Functional Analysis of the Foxl2 Gene in Procambarus clarkii" Genes 14, no. 12: 2190. https://doi.org/10.3390/genes14122190
APA StyleHuang, J., Zhu, W., Peng, M., Yang, C., Chen, X., Wu, T., Zeng, D., Zhao, Y., & Chen, X. (2023). Cloning, Identification, and Functional Analysis of the Foxl2 Gene in Procambarus clarkii. Genes, 14(12), 2190. https://doi.org/10.3390/genes14122190





