Polymer Modeling Reveals Interplay between Physical Properties of Chromosomal DNA and the Size and Distribution of Condensin-Based Chromatin Loops
Abstract
:1. Introduction
2. Materials and Methods
2.1. Computation Environment
2.2. Polymer Model of Chromatin
2.3. Persistence Length
2.4. Tethering Resistance
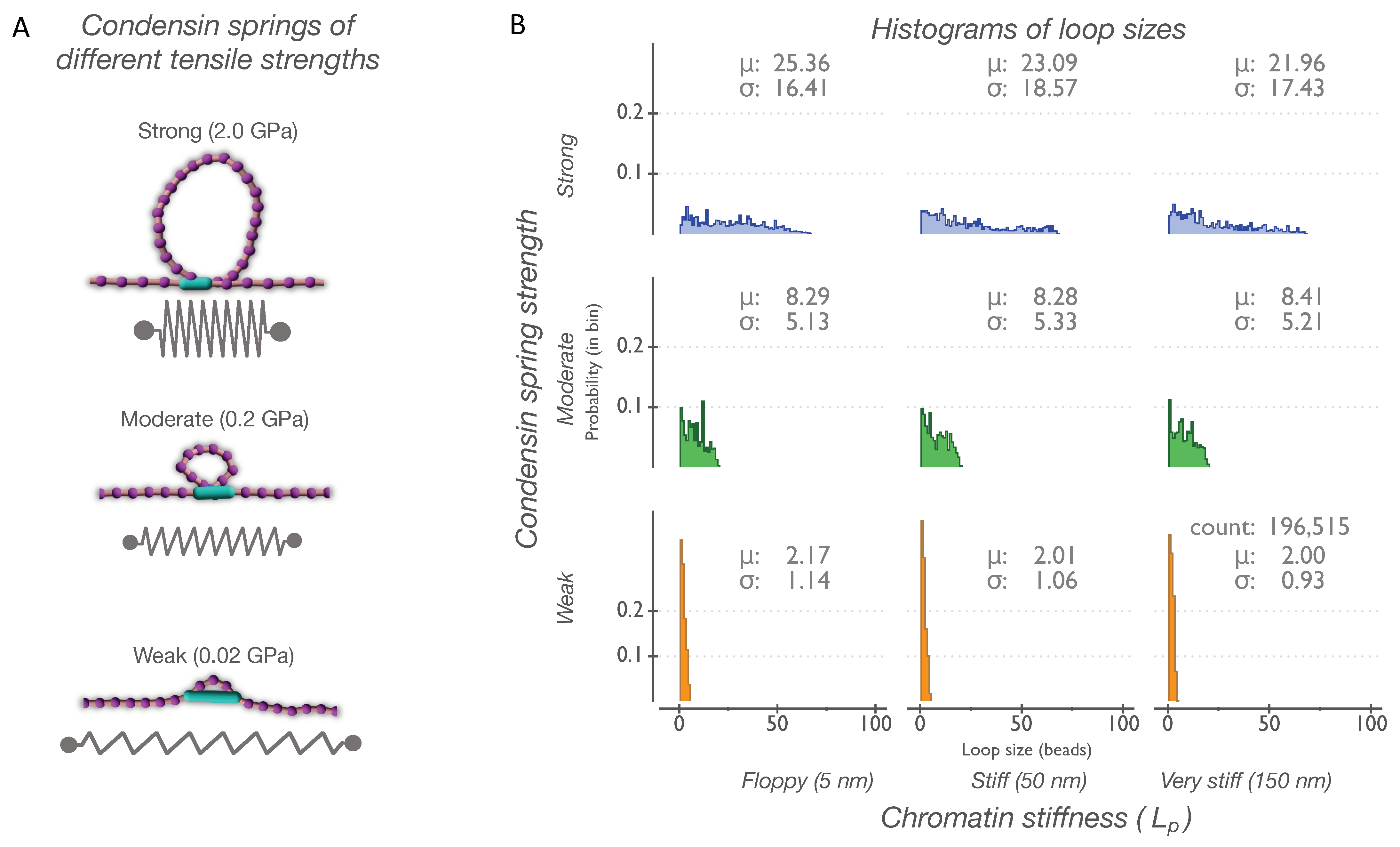
2.5. Condensin Springs
2.6. Recoil Imaging
2.7. Image Analysis
3. Results
3.1. Condensin Spring Strength Dictates Distribution of Loop Sizes on Chains
3.2. Kinetics of Chromatin Compaction in Live Cells
3.3. Stiffness of the DNA Chain Dictates Distribution of Loop Sizes on Unconstrained Chains
3.4. Persistence Length and Condensin Spring Strength Both Determine Loop Sizes in Moderate Tethering Regimes
3.5. Persistence Length of DNA within a Loop Determines the Space It Will Explore
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Narwade, N.; Patel, S.; Alam, A.; Chattopadhyay, S.; Mittal, S.; Kulkarni, A. Mapping of Scaffold/Matrix Attachment Regions in Human Genome: A Data Mining Exercise. Nucleic Acids Res. 2019, 47, 7247–7261. [Google Scholar] [CrossRef]
- Sureka, R.; Avvaru, A.K.; Sowpati, D.T.; Pathak, R.U.; Mishra, R.K. Structural and Developmental Dynamics of Matrix Associated Regions in Drosophila Melanogaster Genome. BMC Genom. 2022, 23, 725. [Google Scholar] [CrossRef]
- Spakman, D.; Clement, T.V.M.; Biebricher, A.S.; King, G.A.; Singh, M.I.; Hickson, I.D.; Peterman, E.J.G.; Wuite, G.J.L. PICH Acts as a Force-Dependent Nucleosome Remodeler. Nat. Commun. 2022, 13, 7277. [Google Scholar] [CrossRef] [PubMed]
- Rajendraprasad, G.; Rodriguez-Calado, S.; Barisic, M. SiR-DNA/SiR-Hoechst-Induced Chromosome Entanglement Generates Severe Anaphase Bridges and DNA Damage. Life Sci. Alliance 2023, 6, e202302260. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, T.; Kinoshita, K.; Hirano, T. Elasticity Control of Entangled Chromosomes: Crosstalk between Condensin Complexes and Nucleosomes. Biophys. J. 2023, 122, 3869–3881. [Google Scholar] [CrossRef] [PubMed]
- Zimmer, C.; Fabre, E. Principles of Chromosomal Organization: Lessons from Yeast. J. Cell Biol. 2011, 192, 723–733. [Google Scholar] [CrossRef] [PubMed]
- Taddei, A.; Gasser, S.M. Structure and Function in the Budding Yeast Nucleus. Genetics 2012, 192, 107–129. [Google Scholar] [CrossRef]
- Verdaasdonk, J.S.; Vasquez, P.A.; Barry, R.M.; Barry, T.; Goodwin, S.; Forest, M.G.; Bloom, K. Centromere Tethering Confines Chromosome Domains. Mol. Cell 2013, 52, 819–831. [Google Scholar] [CrossRef]
- Lawrimore, J.; Barry, T.M.; Barry, R.M.; York, A.C.; Friedman, B.; Cook, D.M.; Akialis, K.; Tyler, J.; Vasquez, P.; Yeh, E.; et al. Microtubule Dynamics Drive Enhanced Chromatin Motion and Mobilize Telomeres in Response to DNA Damage. Mol. Biol. Cell 2017, 28, 1701–1711. [Google Scholar] [CrossRef]
- Deng, L.; Zhou, Q.; Zhou, J.; Zhang, Q.; Jia, Z.; Zhu, G.; Cheng, S.; Cheng, L.; Yin, C.; Yang, C.; et al. 3D Organization of Regulatory Elements for Transcriptional Regulation in Arabidopsis. Genome Biol. 2023, 24, 181. [Google Scholar] [CrossRef]
- Bâcle, J.; Groizard, L.; Kumanski, S.; Moriel-Carretero, M. Nuclear Envelope-Remodeling Events as Models to Assess the Potential Role of Membranes on Genome Stability. FEBS Lett. 2023, 597, 1946–1956. [Google Scholar] [CrossRef]
- Brickner, J.H.; Walter, P. Gene Recruitment of the Activated INO1 Locus to the Nuclear Membrane. PLoS Biol. 2004, 2, e342. [Google Scholar] [CrossRef] [PubMed]
- Taddei, A.; Van Houwe, G.; Hediger, F.; Kalck, V.; Cubizolles, F.; Schober, H.; Gasser, S.M. Nuclear Pore Association Confers Optimal Expression Levels for an Inducible Yeast Gene. Nature 2006, 441, 774–778. [Google Scholar] [CrossRef]
- Padeken, J.; Heun, P. Nucleolus and Nuclear Periphery: Velcro for Heterochromatin. Curr. Opin. Cell Biol. 2014, 28, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Brickner, D.G.; Sood, V.; Tutucci, E.; Coukos, R.; Viets, K.; Singer, R.H.; Brickner, J.H. Subnuclear Positioning and Interchromosomal Clustering of the GAL1-10 Locus Are Controlled by Separable, Interdependent Mechanisms. Mol. Biol. Cell 2016, 27, 2980–2993. [Google Scholar] [CrossRef] [PubMed]
- Forer, A.; Otsuka, S. Structural Evidence for Elastic Tethers Connecting Separating Chromosomes in Crane-Fly Spermatocytes. Life Sci. Alliance 2023, 6. [Google Scholar] [CrossRef] [PubMed]
- Lawrimore, J.; Friedman, B.; Doshi, A.; Bloom, K. RotoStep: A Chromosome Dynamics Simulator Reveals Mechanisms of Loop Extrusion. Cold Spring Harb. Symp. Quant. Biol. 2017, 82, 101–109. [Google Scholar] [CrossRef]
- He, Y.; Lawrimore, J.; Cook, D.; Van Gorder, E.E.; De Larimat, S.C.; Adalsteinsson, D.; Forest, M.G.; Bloom, K. Statistical Mechanics of Chromosomes: In Vivo and In Silico Approaches Reveal High-Level Organization and Structure Arise Exclusively through Mechanical Feedback between Loop Extruders and Chromatin Substrate Properties. Nucleic Acids Res. 2020, 48, 11284–11303. [Google Scholar] [CrossRef]
- Hertwig, O. Lehrbuch Der Entwicklungsgeschichte Des Menschen Und Der Wirbeltiere (Textbook of Developmental History of Humans and Vertebrates); Smithsonian: Washington, DC, USA, 1906. [Google Scholar]
- Diaz, M.O.; Gall, J.G. Giant Readthrough Transcription Units at the Histone Loci on Lampbrush Chromosomes of the Newt Notophthalmus. Chromosoma 1985, 92, 243–253. [Google Scholar] [CrossRef]
- Cremer, T.; Cremer, C. Chromosome Territories, Nuclear Architecture and Gene Regulation in Mammalian Cells. Nat. Rev. Genet. 2001, 2, 292–301. [Google Scholar] [CrossRef]
- Pombo, A.; Branco, M.R. Functional Organisation of the Genome during Interphase. Curr. Opin. Genet. Dev. 2007, 17, 451–455. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Marti-Renom, M.A.; Mirny, L.A. Exploring the Three-Dimensional Organization of Genomes: Interpreting Chromatin Interaction Data. Nat. Rev. Genet. 2013, 14, 390–403. [Google Scholar] [CrossRef] [PubMed]
- Nuebler, J.; Fudenberg, G.; Imakaev, M.; Abdennur, N.; Mirny, L.A. Chromatin Organization by an Interplay of Loop Extrusion and Compartmental Segregation. Proc. Natl. Acad. Sci. USA 2018, 115, E6697–E6706. [Google Scholar] [CrossRef]
- Fudenberg, G.; Mirny, L.A. Higher-Order Chromatin Structure: Bridging Physics and Biology. Curr. Opin. Genet. Dev. 2012, 22, 115–124. [Google Scholar] [CrossRef]
- Banigan, E.J.; Mirny, L.A. Loop Extrusion: Theory Meets Single-Molecule Experiments. Curr. Opin. Cell Biol. 2020, 64, 124–138. [Google Scholar] [CrossRef] [PubMed]
- Pueschel, R.; Coraggio, F.; Meister, P. From Single Genes to Entire Genomes: The Search for a Function of Nuclear Organization. Dev. Camb. Engl. 2016, 143, 910–923. [Google Scholar] [CrossRef]
- Rowley, M.J.; Corces, V.G. Organizational Principles of 3D Genome Architecture. Nat. Rev. Genet. 2018, 19, 789–800. [Google Scholar] [CrossRef]
- van Steensel, B.; Dekker, J. Genomics Tools for Unraveling Chromosome Architecture. Nat. Biotechnol. 2010, 28, 1089–1095. [Google Scholar] [CrossRef]
- Mirny, L.A.; Imakaev, M.; Abdennur, N. Two Major Mechanisms of Chromosome Organization. Curr. Opin. Cell Biol. 2019, 58, 142–152. [Google Scholar] [CrossRef]
- Conte, M.; Esposito, A.; Vercellone, F.; Abraham, A.; Bianco, S. Unveiling the Machinery behind Chromosome Folding by Polymer Physics Modeling. Int. J. Mol. Sci. 2023, 24, 3660. [Google Scholar] [CrossRef]
- Arnould, C.; Rocher, V.; Saur, F.; Bader, A.S.; Muzzopappa, F.; Collins, S.; Lesage, E.; Le Bozec, B.; Puget, N.; Clouaire, T.; et al. Chromatin Compartmentalization Regulates the Response to DNA Damage. Nature 2023, 623, 183–192. [Google Scholar] [CrossRef] [PubMed]
- Lawrimore, J.; Bloom, K. The Regulation of Chromosome Segregation via Centromere Loops. Crit. Rev. Biochem. Mol. Biol. 2019, 54, 352–370. [Google Scholar] [CrossRef] [PubMed]
- Lawrimore, J.; Aicher, J.K.; Hahn, P.; Fulp, A.; Kompa, B.; Vicci, L.; Falvo, M.; Taylor, R.M.; Bloom, K. ChromoShake: A Chromosome Dynamics Simulator Reveals That Chromatin Loops Stiffen Centromeric Chromatin. Mol. Biol. Cell 2016, 27, 153–166. [Google Scholar] [CrossRef] [PubMed]
- Lawrimore, J.; Bloom, K. Shaping Centromeres to Resist Mitotic Spindle Forces. J. Cell Sci. 2022, 135, jcs259532. [Google Scholar] [CrossRef]
- Hult, C.; Adalsteinsson, D.; Vasquez, P.A.; Lawrimore, J.; Bennett, M.; York, A.; Cook, D.; Yeh, E.; Forest, M.G.; Bloom, K. Enrichment of Dynamic Chromosomal Crosslinks Drive Phase Separation of the Nucleolus. Nucleic Acids Res. 2017, 45, 11159–11173. [Google Scholar] [CrossRef]
- Walker, B.; Taylor, D.; Lawrimore, J.; Hult, C.; Adalsteinsson, D.; Bloom, K.; Forest, M.G. Transient Crosslinking Kinetics Optimize Gene Cluster Interactions. PLoS Comput. Biol. 2019, 15, e1007124. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.; Shendure, J. Mechanisms of Interplay between Transcription Factors and the 3D Genome. Mol. Cell 2019, 76, 306–319. [Google Scholar] [CrossRef]
- Stadhouders, R.; Filion, G.J.; Graf, T. Transcription Factors and 3D Genome Conformation in Cell-Fate Decisions. Nature 2019, 569, 345–354. [Google Scholar] [CrossRef] [PubMed]
- Vadnala, R.N.; Hannenhalli, S.; Narlikar, L.; Siddharthan, R. Transcription Factors Organize into Functional Groups on the Linear Genome and in 3D Chromatin. Heliyon 2023, 9, e18211. [Google Scholar] [CrossRef]
- Arnould, C.; Legube, G. The Secret Life of Chromosome Loops upon DNA Double-Strand Break. J. Mol. Biol. 2020, 432, 724–736. [Google Scholar] [CrossRef]
- Sebastian, R.; Aladjem, M.I.; Oberdoerffer, P. Encounters in Three Dimensions: How Nuclear Topology Shapes Genome Integrity. Front. Genet. 2021, 12, 746380. [Google Scholar] [CrossRef] [PubMed]
- Di Stefano, M.; Cavalli, G. Integrative Studies of 3D Genome Organization and Chromatin Structure. Curr. Opin. Struct. Biol. 2022, 77, 102493. [Google Scholar] [CrossRef] [PubMed]
- Carré-Simon, À.; Fabre, E. 3D Genome Organization: Causes and Consequences for DNA Damage and Repair. Genes 2022, 13, 7. [Google Scholar] [CrossRef] [PubMed]
- Akköse, Ü.; Adebali, O. The Interplay of 3D Genome Organization with UV-Induced DNA Damage and Repair. J. Biol. Chem. 2023, 299, 104679. [Google Scholar] [CrossRef]
- Lassadi, I.; Kamgoué, A.; Goiffon, I.; Tanguy-le-Gac, N.; Bystricky, K. Differential Chromosome Conformations as Hallmarks of Cellular Identity Revealed by Mathematical Polymer Modeling. PLoS Comput. Biol. 2015, 11, e1004306. [Google Scholar] [CrossRef]
- Quail, T.; Golfier, S.; Elsner, M.; Ishihara, K.; Murugesan, V.; Renger, R.; Jülicher, F.; Brugués, J. Force Generation by Protein–DNA Co-Condensation. Nat. Phys. 2021, 17, 1007–1012. [Google Scholar] [CrossRef]
- Cui, Y.; Bustamante, C. Pulling a Single Chromatin Fiber Reveals the Forces That Maintain Its Higher-Order Structure. Proc. Natl. Acad. Sci. USA 2000, 97, 127–132. [Google Scholar] [CrossRef]
- Wedemann, G.; Langowski, J. Computer Simulation of the 30-Nanometer Chromatin Fiber. Biophys. J. 2002, 82, 2847–2859. [Google Scholar] [CrossRef]
- Bystricky, K.; Heun, P.; Gehlen, L.; Langowski, J.; Gasser, S.M. Long-Range Compaction and Flexibility of Interphase Chromatin in Budding Yeast Analyzed by High-Resolution Imaging Techniques. Proc. Natl. Acad. Sci. USA 2004, 101, 16495–16500. [Google Scholar] [CrossRef]
- Hajjoul, H.; Mathon, J.; Ranchon, H.; Goiffon, I.; Mozziconacci, J.; Albert, B.; Carrivain, P.; Victor, J.-M.; Gadal, O.; Bystricky, K.; et al. High-Throughput Chromatin Motion Tracking in Living Yeast Reveals the Flexibility of the Fiber throughout the Genome. Genome Res. 2013, 23, 1829–1838. [Google Scholar] [CrossRef]
- Arbona, J.-M.; Herbert, S.; Fabre, E.; Zimmer, C. Inferring the Physical Properties of Yeast Chromatin through Bayesian Analysis of Whole Nucleus Simulations. Genome Biol. 2017, 18, 81. [Google Scholar] [CrossRef]
- Miné-Hattab, J.; Recamier, V.; Izeddin, I.; Rothstein, R.; Darzacq, X. Multi-Scale Tracking Reveals Scale-Dependent Chromatin Dynamics after DNA Damage. Mol. Biol. Cell 2017, 28, 3323–3332. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.-N.; Qin, L.; Wuite, G.J.L.; Dame, R.T. Unraveling the Biophysical Properties of Chromatin Proteins and DNA Using Acoustic Force Spectroscopy. In Bacterial Chromatin: Methods and Protocols; Dame, R.T., Ed.; Methods in Molecular Biology; Springer: New York, NY, USA, 2018; pp. 301–316. ISBN 978-1-4939-8675-0. [Google Scholar]
- Rivetti, C.; Walker, C.; Bustamante, C. Polymer Chain Statistics and Conformational Analysis of DNA Molecules with Bends or Sections of Different flexibility11Edited by D. Draper. J. Mol. Biol. 1998, 280, 41–59. [Google Scholar] [CrossRef] [PubMed]
- Heidinger-Pauli, J.M.; Unal, E.; Koshland, D. Distinct Targets of the Eco1 Acetyltransferase Modulate Cohesion in S Phase and in Response to DNA Damage. Mol. Cell 2009, 34, 311–321. [Google Scholar] [CrossRef]
- Gruber, S. Shaping Chromosomes by DNA Capture and Release: Gating the SMC Rings. Curr. Opin. Cell Biol. 2017, 46, 87–93. [Google Scholar] [CrossRef] [PubMed]
- Xu, X.; Yanagida, M. Suppressor Screening Reveals Common Kleisin–Hinge Interaction in Condensin and Cohesin, but Different Modes of Regulation. Proc. Natl. Acad. Sci. USA 2019, 116, 10889–10898. [Google Scholar] [CrossRef]
- Psakhye, I.; Branzei, D. SMC Complexes Are Guarded by the SUMO Protease Ulp2 against SUMO-Chain-Mediated Turnover. Cell Rep. 2021, 36, 109485. [Google Scholar] [CrossRef] [PubMed]
- Palou, R.; Dhanaraman, T.; Marrakchi, R.; Pascariu, M.; Tyers, M.; D’Amours, D. Condensin ATPase Motifs Contribute Differentially to the Maintenance of Chromosome Morphology and Genome Stability. PLoS Biol. 2018, 16, e2003980. [Google Scholar] [CrossRef]
- Elbatsh, A.M.O.; Kim, E.; Eeftens, J.M.; Raaijmakers, J.A.; van der Weide, R.H.; García-Nieto, A.; Bravo, S.; Ganji, M.; uit de Bos, J.; Teunissen, H.; et al. Distinct Roles for Condensin’s Two ATPase Sites in Chromosome Condensation. Mol. Cell 2019, 76, 724–737.e5. [Google Scholar] [CrossRef]
- Zhao, X.; Blobel, G. A SUMO Ligase Is Part of a Nuclear Multiprotein Complex That Affects DNA Repair and Chromosomal Organization. Proc. Natl. Acad. Sci. USA 2005, 102, 4777–4782. [Google Scholar] [CrossRef]
- Varejão, N.; Ibars, E.; Lascorz, J.; Colomina, N.; Torres-Rosell, J.; Reverter, D. DNA Activates the Nse2/Mms21 SUMO E3 Ligase in the Smc5/6 Complex. EMBO J. 2018, 37, e98306. [Google Scholar] [CrossRef]
- Higashi, T.L.; Pobegalov, G.; Tang, M.; Molodtsov, M.I.; Uhlmann, F. A Brownian Ratchet Model for DNA Loop Extrusion by the Cohesin Complex. eLife 2021, 10, e67530. [Google Scholar] [CrossRef]
- Ryu, J.-K.; Rah, S.-H.; Janissen, R.; Kerssemakers, J.W.J.; Bonato, A.; Michieletto, D.; Dekker, C. Condensin Extrudes DNA Loops in Steps up to Hundreds of Base Pairs That Are Generated by ATP Binding Events. Nucleic Acids Res. 2022, 50, 820–832. [Google Scholar] [CrossRef] [PubMed]
- Terakawa, T.; Bisht, S.; Eeftens, J.M.; Dekker, C.; Haering, C.H.; Greene, E.C. The Condensin Complex Is a Mechanochemical Motor That Translocates along DNA. Science 2017, 358, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Nichols, M.H.; Corces, V.G. A Tethered-Inchworm Model of SMC DNA Translocation. Nat. Struct. Mol. Biol. 2018, 25, 906–910. [Google Scholar] [CrossRef] [PubMed]
- Ganji, M.; Shaltiel, I.A.; Bisht, S.; Kim, E.; Kalichava, A.; Haering, C.H.; Dekker, C. Real-Time Imaging of DNA Loop Extrusion by Condensin. Science 2018, 360, 102–105. [Google Scholar] [CrossRef]
- Kinoshita, K.; Tsubota, Y.; Tane, S.; Aizawa, Y.; Sakata, R.; Takeuchi, K.; Shintomi, K.; Nishiyama, T.; Hirano, T. A Loop Extrusion–Independent Mechanism Contributes to Condensin I–Mediated Chromosome Shaping. J. Cell Biol. 2022, 221, e202109016. [Google Scholar] [CrossRef]
- Ryu, J.-K.; Bouchoux, C.; Liu, H.W.; Kim, E.; Minamino, M.; de Groot, R.; Katan, A.J.; Bonato, A.; Marenduzzo, D.; Michieletto, D.; et al. Bridging-Induced Phase Separation Induced by Cohesin SMC Protein Complexes. Sci. Adv. 2021, 7, eabe5905. [Google Scholar] [CrossRef]
- Duan, Z.; Andronescu, M.; Schutz, K.; McIlwain, S.; Kim, Y.J.; Lee, C.; Shendure, J.; Fields, S.; Blau, C.A.; Noble, W.S. A Three-Dimensional Model of the Yeast Genome. Nature 2010, 465, 363–367. [Google Scholar] [CrossRef]
- Pouokam, M.; Cruz, B.; Burgess, S.; Segal, M.R.; Vazquez, M.; Arsuaga, J. The Rabl Configuration Limits Topological Entanglement of Chromosomes in Budding Yeast. Sci. Rep. 2019, 9, 6795. [Google Scholar] [CrossRef]
- Agmon, N.; Liefshitz, B.; Zimmer, C.; Fabre, E.; Kupiec, M. Effect of Nuclear Architecture on the Efficiency of Double-Strand Break Repair. Nat. Cell Biol. 2013, 15, 694–699. [Google Scholar] [CrossRef] [PubMed]
- Chubb, J.R.; Boyle, S.; Perry, P.; Bickmore, W.A. Chromatin Motion Is Constrained by Association with Nuclear Compartments in Human Cells. Curr. Biol. 2002, 12, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Strecker, J.; Gupta, G.D.; Zhang, W.; Bashkurov, M.; Landry, M.-C.; Pelletier, L.; Durocher, D. DNA Damage Signalling Targets the Kinetochore to Promote Chromatin Mobility. Nat. Cell Biol. 2016, 18, 281–290. [Google Scholar] [CrossRef] [PubMed]
- Davidson, I.F.; Barth, R.; Zaczek, M.; van der Torre, J.; Tang, W.; Nagasaka, K.; Janissen, R.; Kerssemakers, J.; Wutz, G.; Dekker, C.; et al. CTCF Is a DNA-Tension-Dependent Barrier to Cohesin-Mediated DNA Loop Extrusion. Nature 2023, 616, 822–827. [Google Scholar] [CrossRef] [PubMed]
- Rubinstein, M.; Colby, R.H. Polymer Physics; Oxford University Press: Oxford, UK; New York, NY, USA, 2003; ISBN 978-0-19-852059-7. [Google Scholar]
- Fisher, J.K.; Ballenger, M.; O’Brien, E.T.; Haase, J.; Superfine, R.; Bloom, K. DNA Relaxation Dynamics as a Probe for the Intracellular Environment. Proc. Natl. Acad. Sci. USA 2009, 106, 9250–9255. [Google Scholar] [CrossRef] [PubMed]
- Gerguri, T.; Fu, X.; Kakui, Y.; Khatri, B.S.; Barrington, C.; Bates, P.A.; Uhlmann, F. Comparison of Loop Extrusion and Diffusion Capture as Mitotic Chromosome Formation Pathways in Fission Yeast. Nucleic Acids Res. 2021, 49, 1294–1312. [Google Scholar] [CrossRef]
- Lawrimore, J.; de Larminat, S.C.; Cook, D.; Friedman, B.; Doshi, A.; Yeh, E.; Bloom, K. Polymer Models Reveal How Chromatin Modification Can Modulate Force at the Kinetochore. Mol. Biol. Cell 2022, 33, ar97. [Google Scholar] [CrossRef]
- Mary, H.; Rueden, C.; Ferreira, T. KymographBuilder: Release 1.2.4; Zenodo: Geneva, Switzerland, 2016. [Google Scholar] [CrossRef]
- Yang, S.S.; Yeh, E.; Salmon, E.D.; Bloom, K. Identification of a Mid-Anaphase Checkpoint in Budding Yeast. J. Cell Biol. 1997, 136, 345–354. [Google Scholar] [CrossRef]
- Lawrimore, J.; Kolbin, D.; Stanton, J.; Khan, M.; de Larminat, S.C.; Lawrimore, C.; Yeh, E.; Bloom, K. The rDNA Is Biomolecular Condensate Formed by Polymer-Polymer Phase Separation and Is Sequestered in the Nucleolus by Transcription and R-Loops. Nucleic Acids Res. 2021, 49, 4586–4598. [Google Scholar] [CrossRef]




| Persistence Length, Lp (nm) | Kuhn’s Length, b (nm) | Segments, N | (nm) | Measured Mean (nm) * | Hinge Factor (Unitless) |
|---|---|---|---|---|---|
| 5 | 10 | 100 | 40.82 | 39.97 | 0 |
| 50 | 100 | 10 | 129.10 | 130.68 | 3.25 |
| 150 | 300 | 3.333 | 223.61 | 221.80 | 16 |
| Time | Value |
|---|---|
| Time conversion factor ** | 14,100× |
| Maximum computed simulation time | 35 ms |
| Converted simulation time (14,100 × 0.035 s) | ~8 min |
| Time-lapse experiment duration (max. 600 s) | 6–10 min |
| Feature 1 | Value | Reference |
|---|---|---|
| Translocation rate (due to directional stepping/extrusion) | 60 bps/s | [17,66] |
| Throttling/stalling/tension sensing | N/A | [18,68] |
| Unbinding and rebinding rates, and reversal rate | 0.01135 events/s; 0.02 events/s | [68] |
| Diffusion capture | N/A | [18,79] |
| Condensin spring strength and flexibility | 2.0 GPa; 0.2 GPa; 0.02 GPa | [17], This study |
| Condensin spring critical length | 30 nm | [17,18] |
| Described as | Strain | Genotype |
|---|---|---|
| Tubulin-GFP (dicentric) | KBY7004 | KBY4137/J178D dicentric URA3::Hg + TUB1:GFP:URA3 pAFS125 |
| WT (dicentric) | KBY6201a | ade2-1; can1-100; his3-11,15::GFP-LacI-HIS3; leu2-3,112::lacO-LEU2; trp1-1; ura3-1::URA3GALCEN3; Spc29RFP:Hb |
| brn1-9 (dicentric) | KBY8182 | ade2-1; can1-100; his3-11,15::GFP-LacI-HIS3; leu2-3,112::lacO-LEU2; trp1-1; ura3-1::URA3GALCEN3; Spc29RFP:Hb; brn1-9:Nat |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kolbin, D.; Walker, B.L.; Hult, C.; Stanton, J.D.; Adalsteinsson, D.; Forest, M.G.; Bloom, K. Polymer Modeling Reveals Interplay between Physical Properties of Chromosomal DNA and the Size and Distribution of Condensin-Based Chromatin Loops. Genes 2023, 14, 2193. https://doi.org/10.3390/genes14122193
Kolbin D, Walker BL, Hult C, Stanton JD, Adalsteinsson D, Forest MG, Bloom K. Polymer Modeling Reveals Interplay between Physical Properties of Chromosomal DNA and the Size and Distribution of Condensin-Based Chromatin Loops. Genes. 2023; 14(12):2193. https://doi.org/10.3390/genes14122193
Chicago/Turabian StyleKolbin, Daniel, Benjamin L. Walker, Caitlin Hult, John Donoghue Stanton, David Adalsteinsson, M. Gregory Forest, and Kerry Bloom. 2023. "Polymer Modeling Reveals Interplay between Physical Properties of Chromosomal DNA and the Size and Distribution of Condensin-Based Chromatin Loops" Genes 14, no. 12: 2193. https://doi.org/10.3390/genes14122193






