Identification of TRPC6 as a Novel Diagnostic Biomarker of PM-Induced Chronic Obstructive Pulmonary Disease Using Machine Learning Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microarray Data Acquisition
2.2. Identification of Differentially Expressed Genes (DEGs)
2.3. GO Enrichment and KEGG Pathway Analysis
2.4. Cell Culture
2.5. Extraction of RNAs and qRT-PCR
2.6. Statistical Analysis
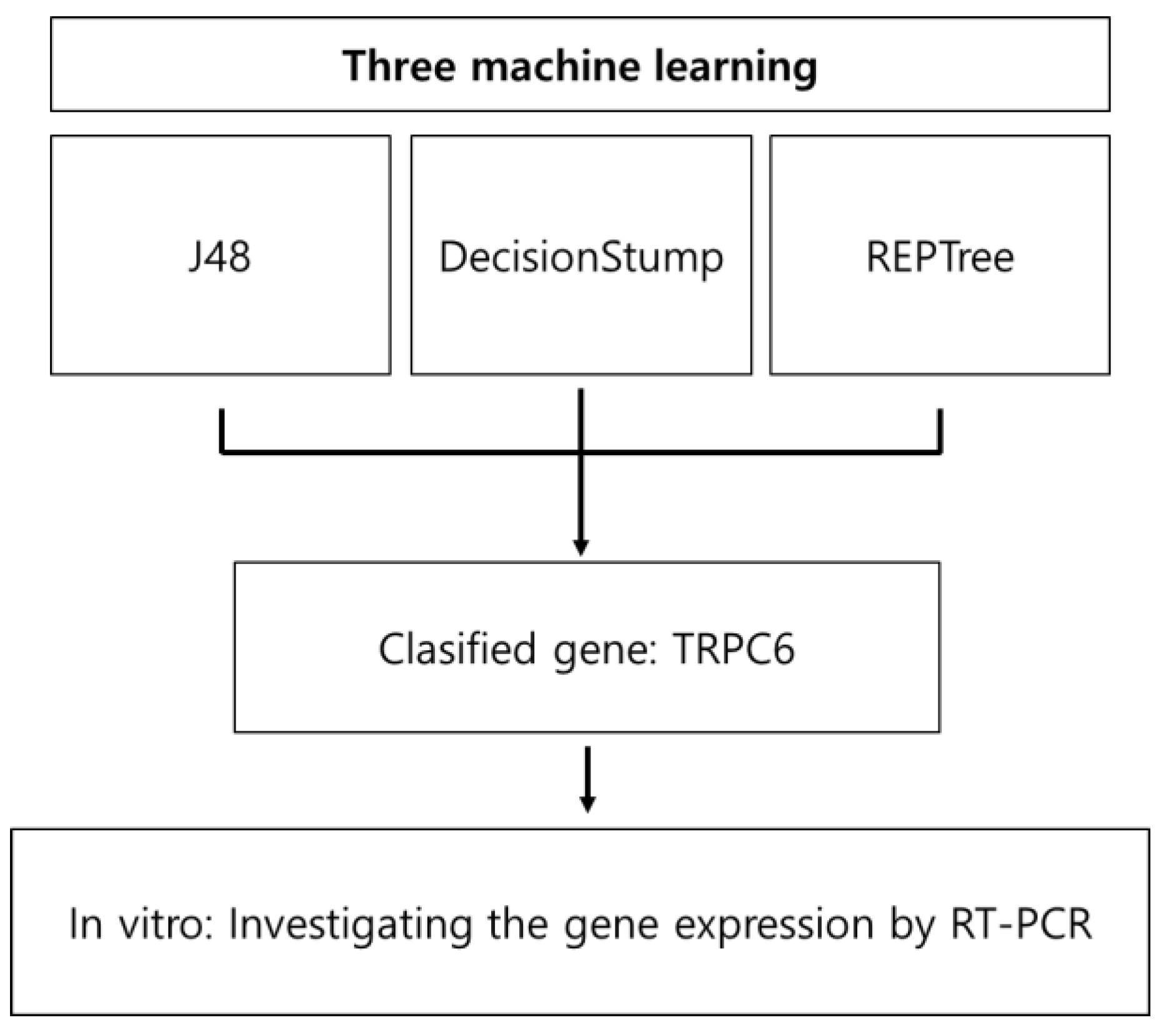
2.7. Machine Learning with Decision Tree
3. Results
3.1. Identification of TRPC6 as a Potential Biomarker for COPD Using Machine Learning Models and GEO2R
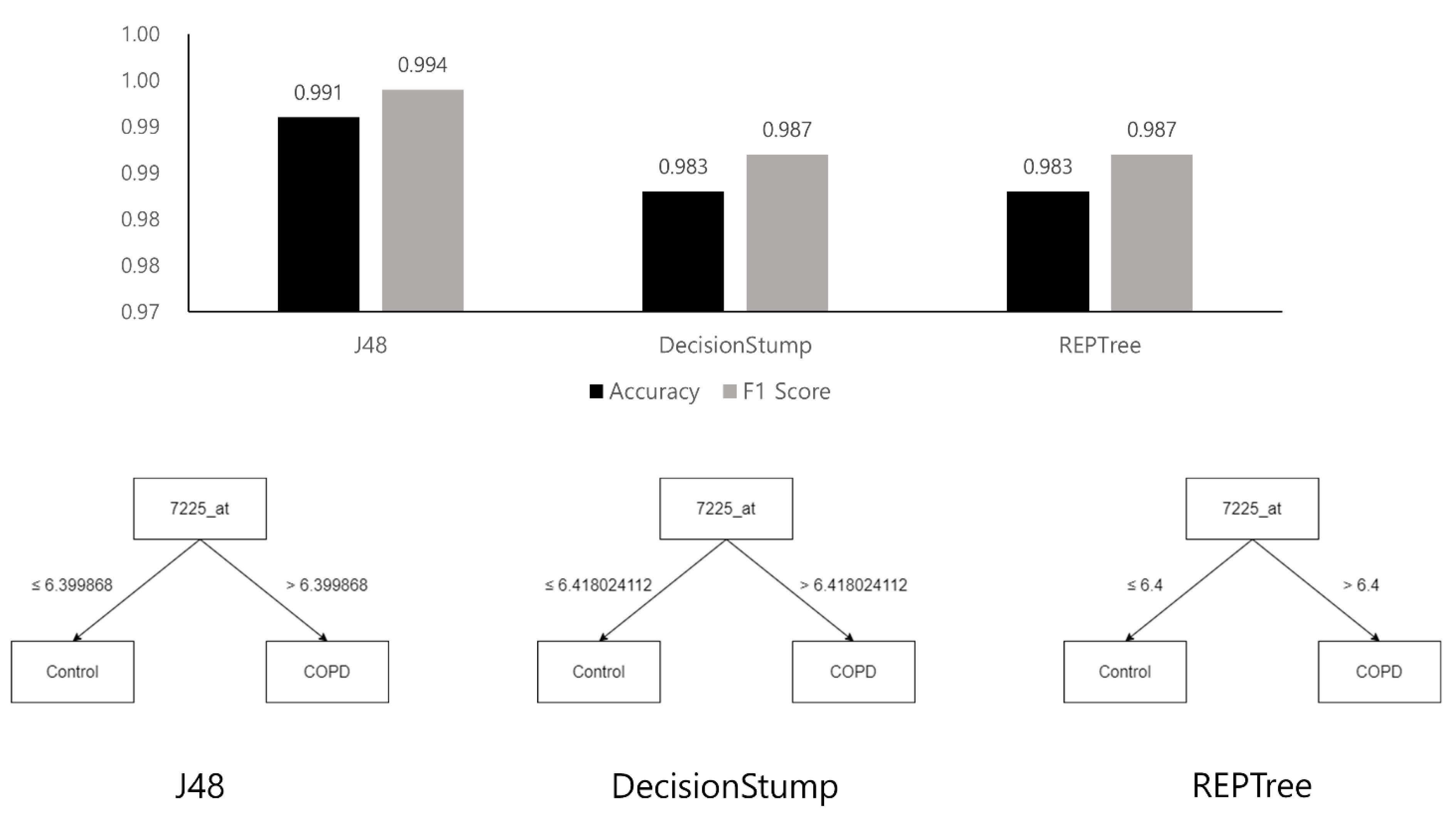
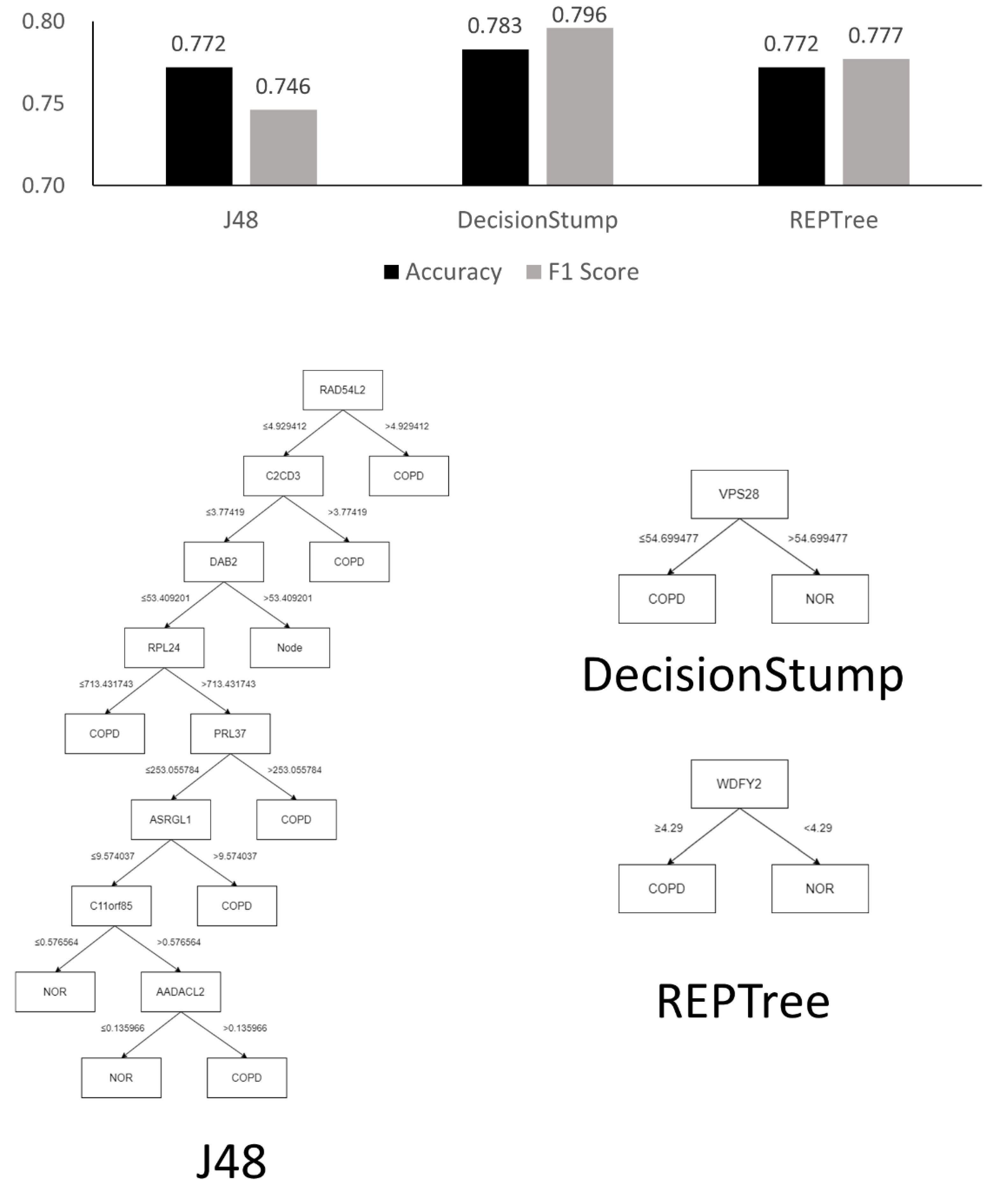
3.1.1. Analysis Using Machine Learning Models
3.1.2. GEO2R
3.2. GO Term Enrichment and KEGG Pathway Analysis
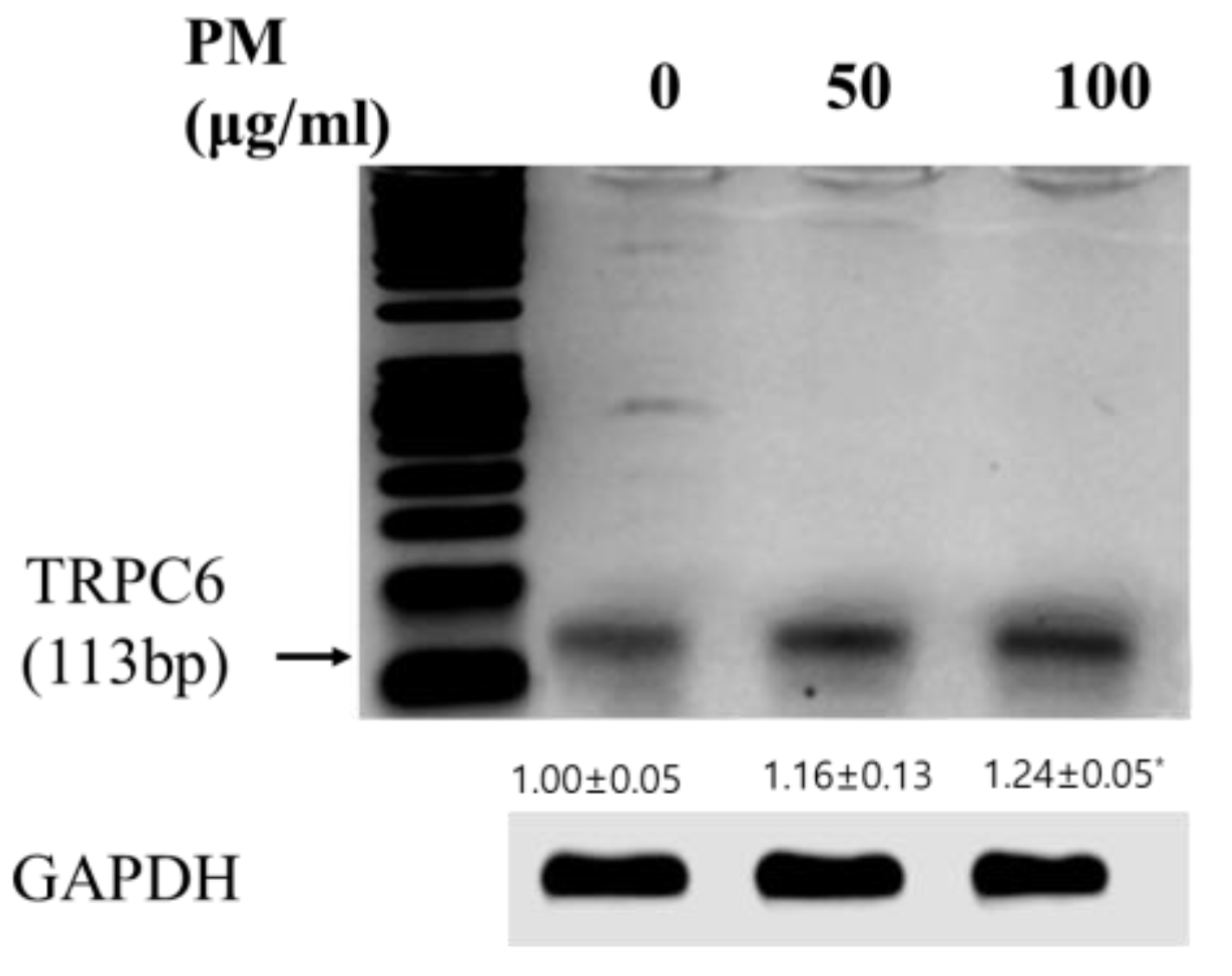
3.3. Validating the Expression and Diagnostic Value of TRPC6 in Vitro Model of COPD Using PCR Analysis
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- López-Campos, J.L.; Tan, W.; Soriano, J.B. Global burden of COPD. Respirology 2016, 21, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Polosukhin, V.V.; Richmond, B.W.; Du, R.-H.; Cates, J.M.; Wu, P.; Nian, H.; Massion, P.P.; Ware, L.B.; Lee, J.W.; Kononov, A.V. Secretory IgA deficiency in individual small airways is associated with persistent inflammation and remodeling. Am. J. Respir. Crit. Care Med. 2017, 195, 1010–1021. [Google Scholar] [CrossRef] [PubMed]
- Hogg, J.C.; Chu, F.; Utokaparch, S.; Woods, R.; Elliott, W.M.; Buzatu, L.; Cherniack, R.M.; Rogers, R.M.; Sciurba, F.C.; Coxson, H.O. The nature of small-airway obstruction in chronic obstructive pulmonary disease. New Engl. J. Med. 2004, 350, 2645–2653. [Google Scholar] [CrossRef] [PubMed]
- Barnes, P.J. Cellular and molecular mechanisms of chronic obstructive pulmonary disease. Clin. Chest Med. 2014, 35, 71–86. [Google Scholar] [CrossRef]
- Thomsen, R.W.; Lange, P.; Hellquist, B.; Frausing, E.; Bartels, P.D.; Krog, B.R.; Hansen, A.-M.S.; Buck, D.; Bunk, A.E. Validity and underrecording of diagnosis of COPD in the Danish National Patient Registry. Respir. Med. 2011, 105, 1063–1068. [Google Scholar] [CrossRef] [Green Version]
- Nunavath, V.; Goodwin, M.; Fidje, J.T.; Moe, C.E. Deep neural networks for prediction of exacerbations of patients with chronic obstructive pulmonary disease. In Proceedings of the International Conference on Engineering Applications of Neural Networks, Bristol, UK, 3–5 September 2018; pp. 217–228. [Google Scholar]
- Tang, C.Y.; Taylor, N.F.; Blackstock, F.C. Chest physiotherapy for patients admitted to hospital with an acute exacerbation of chronic obstructive pulmonary disease (COPD): A systematic review. Physiotherapy 2010, 96, 1–13. [Google Scholar] [CrossRef]
- Almagro, P.; Calbo, E.; de Echaguïen, A.O.; Barreiro, B.; Quintana, S.; Heredia, J.L.; Garau, J. Mortality after hospitalization for COPD. Chest 2002, 121, 1441–1448. [Google Scholar] [CrossRef]
- Team, R.C. R: A language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria. Available online: http://www.R-project.org/2013 (accessed on 9 November 2022).
- Huang, D.W.; Sherman, B.T.; Tan, Q.; Kir, J.; Liu, D.; Bryant, D.; Guo, Y.; Stephens, R.; Baseler, M.W.; Lane, H.C. DAVID Bioinformatics Resources: Expanded annotation database and novel algorithms to better extract biology from large gene lists. Nucleic Acids Res. 2007, 35, W169–W175. [Google Scholar] [CrossRef] [Green Version]
- Carbon, S.; Ireland, A.; Mungall, C.J.; Shu, S.; Marshall, B.; Lewis, S.; Hub, A.; Group, W.P.W. AmiGO: Online access to ontology and annotation data. Bioinformatics 2009, 25, 288–289. [Google Scholar] [CrossRef] [Green Version]
- Faner, R.; Tal-Singer, R.; Riley, J.H.; Celli, B.; Vestbo, J.; MacNee, W.; Bakke, P.; Calverley, P.M.; Coxson, H.; Crim, C. Lessons from ECLIPSE: A review of COPD biomarkers. Thorax 2014, 69, 666–672. [Google Scholar] [CrossRef]
- Han, J.; Wan, M.; Ma, Z.; Hu, C.; Yi, H. Prediction of targets of curculigoside A in osteoporosis and rheumatoid arthritis using network pharmacology and experimental verification. Drug Des. Dev. Ther. 2020, 14, 5235. [Google Scholar] [CrossRef] [PubMed]
- Kwon, H.-K.; Song, M.-J.; Lee, H.-J.; Park, T.-S.; Kim, M.I.; Park, H.-J. Pediococcus pentosaceus-fermented Cordyceps militaris inhibits inflammatory reactions and alleviates contact dermatitis. Int. J. Mol. Sci. 2018, 19, 3504. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhong, K.-R.; Kwon, H.-K.; Park, H.-J. Immunostimulatory Activity of Cordyceps militaris Fermented with Pediococcus pentosaceus SC11 Isolated from a Salted Small Octopus in Cyclophosphamide-Induced Immunocompromised Mice and Its Inhibitory Activity against SARS-CoV 3CL Protease. Microorganisms 2022, 10, 2321. [Google Scholar] [CrossRef] [PubMed]
- Alpaydin, E. Introduction to Machine Learning. MIT Press: Cambridge, MA, USA, 2020. [Google Scholar]
- Mahendran, N.; Durai Raj Vincent, P.; Srinivasan, K.; Chang, C.-Y. Machine learning based computational gene selection models: A survey, performance evaluation, open issues, and future research directions. Front. Genet. 2020, 11, 603808. [Google Scholar] [CrossRef] [PubMed]
- Selvaraj, S.; Natarajan, J. Microarray Data Analysis and Mining Tools. Bioinformation 2011, 6, 95–99. [Google Scholar] [CrossRef] [PubMed]
- Czajkowski, M.; Grześ, M.; Kretowski, M. Multi-test decision tree and its application to microarray data classification. Artif. Intell. Med. 2014, 61, 35–44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, M.H.; Wang, H.C.; Lee, G.W.; Lin, Y.C.; Chiu, S.H.M. A decision tree based classifier to analyze human ovarian cancer cDNA microarray datasets. J. Med. Syst. 2016, 40, 21. [Google Scholar] [CrossRef]
- Jurczuk, K.; Czajkowski, M.; Kretowski, M. Evolutionary induction of a decision tree for large-scale data: A GPU-based approach. Soft Comput. 2017, 21, 7363–7379. [Google Scholar] [CrossRef]
- Vlahos, R.; Bozinovski, S. Role of alveolar macrophages in chronic obstructive pulmonary disease. Front. Immunol. 2014, 5, 435. [Google Scholar] [CrossRef] [Green Version]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-validation. Encycl. Database Syst. 2009, 5, 532–538. [Google Scholar]
- Salzberg, S.L. Book Review: C4.5: Programs for Machine Learning by J. Ross Quinlan. Mach. Learn. 1993, 16, 235–240. [Google Scholar] [CrossRef]
- Hssina, B.; Merbouha, A.; Ezzikouri, H.; Erritali, M. A Comparative Study of Decision Tree ID3 and C4.5. Int. J. Adv. Comput. Sci. Appl. 2014, 4, 13–19. [Google Scholar] [CrossRef] [Green Version]
- Aljawarneh, S.; Yassein, M.B.; Aljundi, M. An Enhanced J48 Classification Algorithm for the Anomaly Intrusion Detection Systems. Clust. Comput. 2019, 22, 10549–10565. [Google Scholar] [CrossRef]
- Eibe Frank, M.A.H.; Witten, I.H. Data Mining, Fourth Edition: Practical Machine Learning Tools and Techniques, 4th ed.; Morgan kaufmann: Burlington, MA, USA, 2016. [Google Scholar]
- Eibe Frank, M.A.H.; Pal, C.J. The WEKA Workbench. Online Appendix for “Data Mining: Practical Machine Learning Tools and Techniques”, 4th ed.; Morgan Kaufmann: Burlington, MA, USA, 2016. [Google Scholar]
- Heydarian, M.; Doyle, T.E.; Samavi, R. MLCM: Multi-label confusion matrix. IEEE Access 2022, 10, 19083–19095. [Google Scholar] [CrossRef]
- Kim, W.J.; Lim, J.H.; Lee, J.S.; Lee, S.-D.; Kim, J.H.; Oh, Y.-M. Comprehensive Analysis of Transcriptome Sequencing Data in the Lung Tissues of COPD Subjects. Int. J. Genom. 2015, 2015, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Zhao, J.; Li, M.; Wang, Z.; Chen, J.; Zhao, J.; Xu, Y.; Wei, X.; Wang, J.; Xie, J. Role of PM2. 5 in the development and progression of COPD and its mechanisms. Respir. Res. 2019, 20, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Bradford, E.; Jacobson, S.; Varasteh, J.; Comellas, A.P.; Woodruff, P.; O’Neal, W.; DeMeo, D.L.; Li, X.; Kim, V.; Cho, M. The value of blood cytokines and chemokines in assessing COPD. Respir. Res. 2017, 18, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Zhou, Y.; Zhou, L.; Fu, Z.; Yang, C.; Zhao, L.; Li, S.; Chen, Y.; Wu, Y.; Ling, Z. TRPC6-dependent Ca2+ signaling mediates airway inflammation in response to oxidative stress via ERK pathway. Cell Death Dis. 2020, 11, 1–16. [Google Scholar]
- Abramowitz, J.; Birnbaumer, L. Physiology and pathophysiology of canonical transient receptor potential channels. FASEB J. 2009, 23, 297–328. [Google Scholar] [CrossRef] [Green Version]
- Finney-Hayward, T.K.; Popa, M.O.; Bahra, P.; Li, S.; Poll, C.T.; Gosling, M.; Nicholson, A.G.; Russell, R.E.; Kon, O.M.; Jarai, G. Expression of transient receptor potential C6 channels in human lung macrophages. Am. J. Respir. Cell Mol. Biol. 2010, 43, 296–304. [Google Scholar] [CrossRef]
- Cosio, M.G.; Guerassimov, A. Chronic obstructive pulmonary disease: Inflammation of small airways and lung parenchyma. Am. J. Respir. Crit. Care Med. 1999, 160, S21–S25. [Google Scholar] [CrossRef] [PubMed]
- Keating, V.; Collins, P.; Scott, D.; Barnes, P. Differences in interleukin-8 and tumour necrosis factorinduced sputum from patients with chronic obstructive pulmonary disease or asthma. Am. J. Respir. Crit. Care Med. 1996, 153, 4. [Google Scholar]
- Ficker, J.; Dertinger, S.; Siegfried, W.; König, H.; Pentz, M.; Sailer, D.; Katalinic, A.; Hahn, E. Obstructive sleep apnoea and diabetes mellitus: The role of cardiovascular autonomic neuropathy. Eur. Respir. J. 1998, 11, 14–19. [Google Scholar] [CrossRef] [PubMed]
- Boulet, L.-P.; Milot, J.; Boutet, M.; St Georges, F.; Laviolette, M. Airway inflammation in nonasthmatic subjects with chronic cough. Am. J. Respir. Crit. Care Med. 1994, 149, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Ling, S.H.; van Eeden, S.F. Particulate matter air pollution exposure: Role in the development and exacerbation of chronic obstructive pulmonary disease. Int. J. Chronic Obstr. Pulm. Dis. 2009, 4, 233. [Google Scholar] [CrossRef] [PubMed]
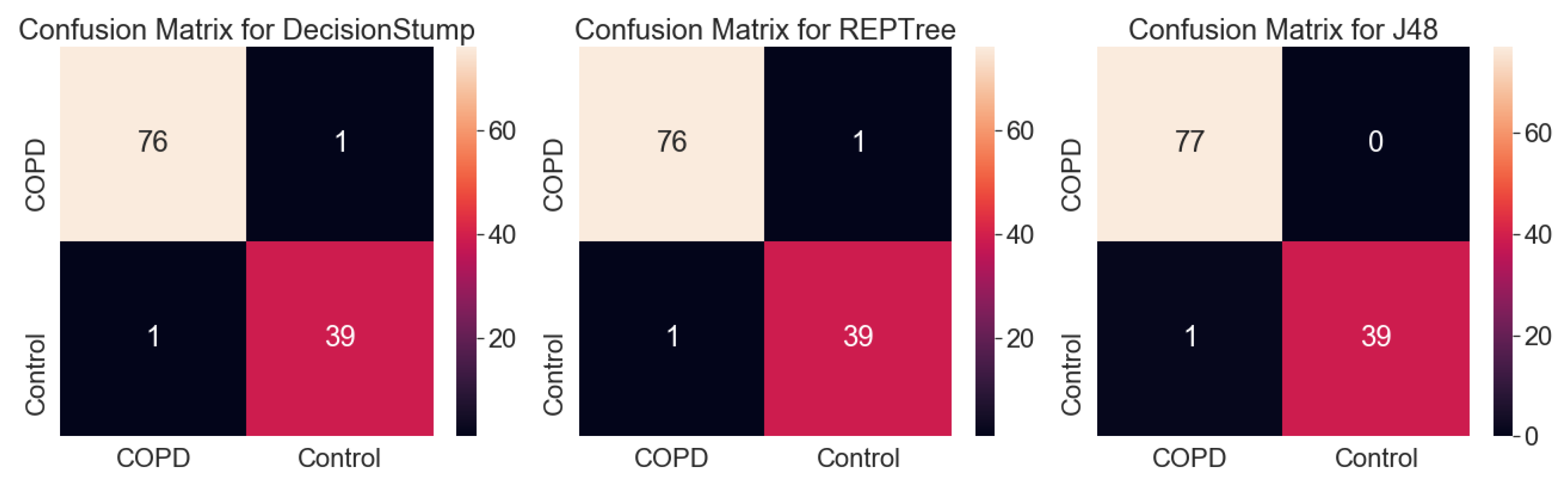
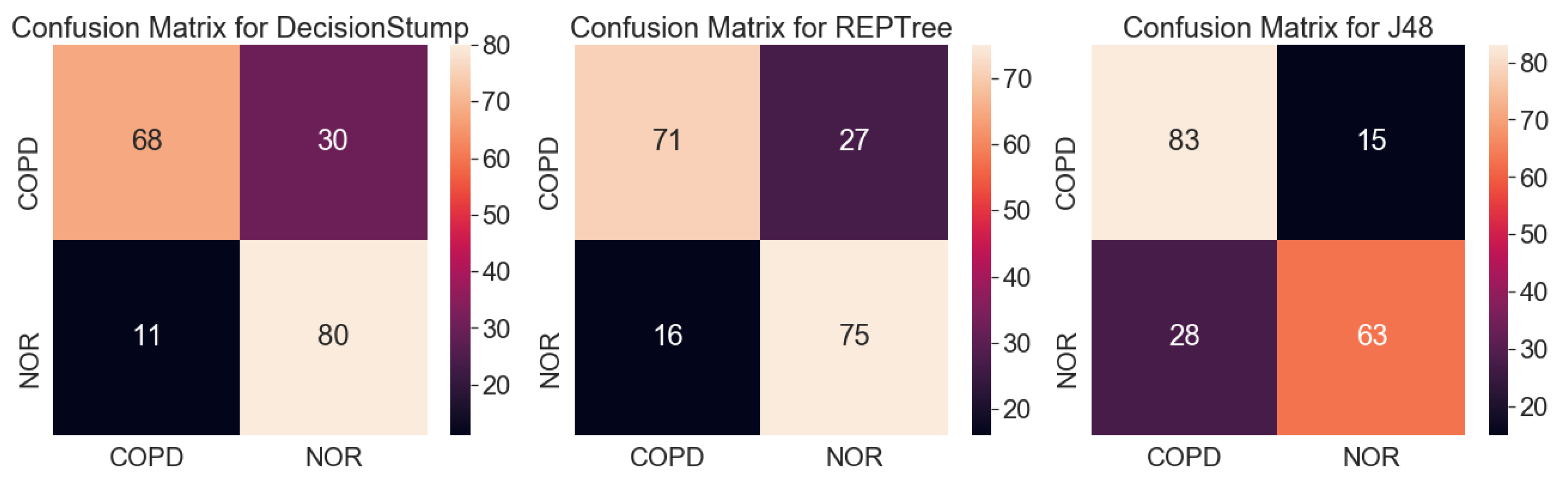
| Actual | COPD | Control |
|---|---|---|
| Prediction | ||
| COPD | True Positive () | False Positive () |
| Control | False Negative () | True Negative () |
| No. | ID | Gene | Adjusted p Value | p Value | logFC |
|---|---|---|---|---|---|
| Upregulated genes (top 10) | |||||
| 1 | 55282_at | LRRC36 | 7.54 × 10−23 | 2.45 × 10−25 | 1.377 |
| 2 | 344148_at | NCKAPS | 1.87 × 10−27 | 1.71 × 10−30 | 1.251 |
| 3 | 2487_at | FRZB | 6.54 × 10−21 | 3.52 × 10−23 | 1.250 |
| 4 | 6092_at | ROBO2 | 9.24 × 10−25 | 1.64 × 10−27 | 1.159 |
| 5 | 6387_at | CXCL12 | 4.81 × 10−19 | 4.20 × 10−21 | 1.129 |
| 6 | 653_at | BMP5 | 6.71 × 10−20 | 4.46 × 10−22 | 1.128 |
| 7 | 2669_at | GEM | 3.15 × 10−19 | 2.58 × 10−21 | 1.116 |
| 8 | 7225_at | TRPC6 | 1.15 × 10−30 | 4.08 × 10−34 | 1.110 |
| 9 | 84251_at | SGIP1 | 3.26 × 10−27 | 3.31 × 10−30 | 1.062 |
| 10 | 114905_at | C1QTNF7 | 2.09 × 10−30 | 8.48 × 10−34 | 1.062 |
| Downregulated genes | |||||
| 1 | 9332_at | CD163 | 2.33 × 10−24 | 4.73 × 10−27 | −2.271 |
| 2 | 6036_at | RNASE2 | 3.00 × 10−26 | 3.65E × 10-29 | −1.567 |
| 3 | 29968_at | PSAT1 | 1.46 × 10−24 | 2.60 × 10−27 | −1.34 |
| 4 | 4830_at | NME1 | 1.05 × 10−22 | 3.71 × 10−25 | −1.318 |
| 5 | 1646_at | AKR1C2 | 8.76 × 10−20 | 6.22 × 10−22 | −1.221 |
| 6 | 1510_at | CTSE | 3.28 × 10−18 | 3.90 × 10−20 | −1.208 |
| 7 | 195814_at | SDR16C5 | 3.13 × 10−20 | 1.90 × 10−22 | −1.049 |
| 8 | 123_at | PLIN2 | 5.33 × 10−24 | 1.32 × 10−26 | −1.039 |
| 9 | 3855_at | KRT7 | 1.01 × 10−22 | 3.47 × 10−25 | −1.035 |
| 10 | 6472_at | SHMT2 | 2.58 × 10−24 | 5.37 × 10−27 | −1.019 |
| Category | Term | Count | % | p-Value | Benjamin |
|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | Transcription, DNA-templated | 14 | 15.9 | 8.40 × 10−02 | 1.00 × 100 |
| GOTERM_BP_DIRECT | Transmembrane transport | 4 | 4.5 | 9.30 × 10−02 | 1.00 × 100 |
| GOTERM_BP_DIRECT | Postembryonic development | 3 | 3.4 | 4.10 × 10−02 | 1.00 × 100 |
| GOTERM_BP_DIRECT | Ossification | 3 | 3.4 | 4.90 × 10−02 | 1.00 × 100 |
| GOTERM_BP_DIRECT | Covalent chromatin modification | 3 | 3.4 | 8.90 × 10−02 | 1.00 × 100 |
| GOTERM_CC_DIRECT | Plasma membrane | 29 | 33 | 3.50 × 10−03 | 2.60 × 10−01 |
| GOTERM_CC_DIRECT | Intracellular | 11 | 12.5 | 4.90 × 10−02 | 1.00 × 100 |
| GOTERM_CC_DIRECT | Nuclear chromatin | 6 | 6.8 | 1.30 × 10−03 | 1.90 × 10−01 |
| GOTERM_CC_DIRECT | Sarcolemma | 3 | 3.4 | 4.90 × 10−02 | 1.00 × 100 |
| GOTERM_CC_DIRECT | Receptor complex | 3 | 3.4 | 9.80 × 10−02 | 1.00 × 100 |
| GOTERM_MF_DIRECT | DNA binding | 14 | 15.9 | 3.20 × 10−02 | 1.00 × 100 |
| GOTERM_MF_DIRECT | Transcription factor activity, sequence-specific DNA binding | 9 | 10.2 | 6.30 × 10−02 | 1.00 × 100 |
| GOTERM_MF_DIRECT | Calcium ion binding | 8 | 9.1 | 4.00 × 10−02 | 1.00 × 100 |
| GOTERM_MF_DIRECT | Chromatin binding | 7 | 8 | 7.80 × 10−03 | 9.90 × 10−01 |
| GOTERM_MF_DIRECT | Integrin binding | 4 | 4.5 | 1.10 × 10−02 | 9.90 × 10−01 |
| KEGG_PATHWAY | Axon guidance | 5 | 5.7 | 2.10 × 10−03 | 2.30 × 10−01 |
| KEGG_PATHWAY | Serotonergic synapse | 3 | 3.4 | 8.40 × 10−02 | 1.00 × 100 |
| KEGG_PATHWAY | Pathways in cancer | 5 | 5.7 | 8.90 × 10−02 | 1.00 × 100 |
| Category | Term | Count | % | p-Value | Benjamin |
|---|---|---|---|---|---|
| GOTERM_BP_DIRECT | Oxidation–reduction process | 15 | 10.3 | 3.20 × 10−04 | 6.70 × 10−02 |
| GOTERM_BP_DIRECT | tRNA aminoacylation for protein translation | 8 | 5.5 | 2.90 × 10−08 | 2.30 × 10−05 |
| GOTERM_BP_DIRECT | Cell–cell adhesion | 8 | 5.5 | 6.60 × 10−03 | 7.50 × 10−01 |
| GOTERM_BP_DIRECT | IRE1-mediated unfolded protein response | 7 | 4.8 | 8.00 × 10−06 | 3.20 × 10−03 |
| GOTERM_BP_DIRECT | Response to nutrient | 6 | 4.1 | 3.30 × 10−04 | 6.70 × 10−02 |
| GOTERM_CC_DIRECT | Extracellular exosome | 51 | 34.9 | 2.00 × 10−09 | 4.40 × 10−07 |
| GOTERM_CC_DIRECT | Cytoplasm | 53 | 36.3 | 1.60 × 10−02 | 2.40 × 10−01 |
| GOTERM_CC_DIRECT | Cytosol | 50 | 34.2 | 1.20 × 10−06 | 1.30 × 10−04 |
| GOTERM_CC_DIRECT | Membrane | 32 | 21.9 | 4.80 × 10−04 | 1.70 × 10−02 |
| GOTERM_CC_DIRECT | Mitochondrion | 27 | 18.5 | 7.70 × 10−06 | 5.50 × 10−04 |
| GOTERM_MF_DIRECT | NADP binding | 6 | 4.1 | 7.60 × 10−06 | 2.50 × 10−03 |
| GOTERM_MF_DIRECT | Poly(A) RNA binding | 21 | 14.4 | 5.30 × 10−04 | 7.60 × 10−02 |
| GOTERM_MF_DIRECT | ATP binding | 21 | 14.4 | 1.30 × 10−02 | 5.50 × 10−01 |
| GOTERM_MF_DIRECT | Cadherin binding involved in cell–cell adhesion | 8 | 5.5 | 8.10 × 10−03 | 3.90 × 10−01 |
| GOTERM_MF_DIRECT | Protein kinase binding | 7 | 4.8 | 7.70 × 10−02 | 1.00 × 100 |
| KEGG_PATHWAY | Biosynthesis of antibiotics | 16 | 11 | 1.20 × 10−08 | 1.50 × 10−06 |
| KEGG_PATHWAY | Metabolic pathways | 33 | 22.6 | 1.50 × 10−06 | 6.10 × 10−05 |
| KEGG_PATHWAY | Biosynthesis of amino acids | 9 | 6.2 | 1.40 × 10−06 | 6.10 × 10−05 |
| KEGG_PATHWAY | Carbon metabolism | 9 | 6.2 | 4.10 × 10−05 | 1.20 × 10−03 |
| KEGG_PATHWAY | Aminoacyl–tRNA biosynthesis | 7 | 4.8 | 9.90 × 10−05 | 2.40 × 10−03 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Dhong, K.-R.; Lee, J.-H.; Yoon, Y.-R.; Park, H.-J. Identification of TRPC6 as a Novel Diagnostic Biomarker of PM-Induced Chronic Obstructive Pulmonary Disease Using Machine Learning Models. Genes 2023, 14, 284. https://doi.org/10.3390/genes14020284
Dhong K-R, Lee J-H, Yoon Y-R, Park H-J. Identification of TRPC6 as a Novel Diagnostic Biomarker of PM-Induced Chronic Obstructive Pulmonary Disease Using Machine Learning Models. Genes. 2023; 14(2):284. https://doi.org/10.3390/genes14020284
Chicago/Turabian StyleDhong, Kyu-Ree, Jae-Hyeong Lee, You-Rim Yoon, and Hye-Jin Park. 2023. "Identification of TRPC6 as a Novel Diagnostic Biomarker of PM-Induced Chronic Obstructive Pulmonary Disease Using Machine Learning Models" Genes 14, no. 2: 284. https://doi.org/10.3390/genes14020284
APA StyleDhong, K.-R., Lee, J.-H., Yoon, Y.-R., & Park, H.-J. (2023). Identification of TRPC6 as a Novel Diagnostic Biomarker of PM-Induced Chronic Obstructive Pulmonary Disease Using Machine Learning Models. Genes, 14(2), 284. https://doi.org/10.3390/genes14020284






