Candidate Genes for Salt Tolerance in Forage Sorghum under Saline Conditions from Germination to Harvest Maturity
Abstract
:1. Introduction
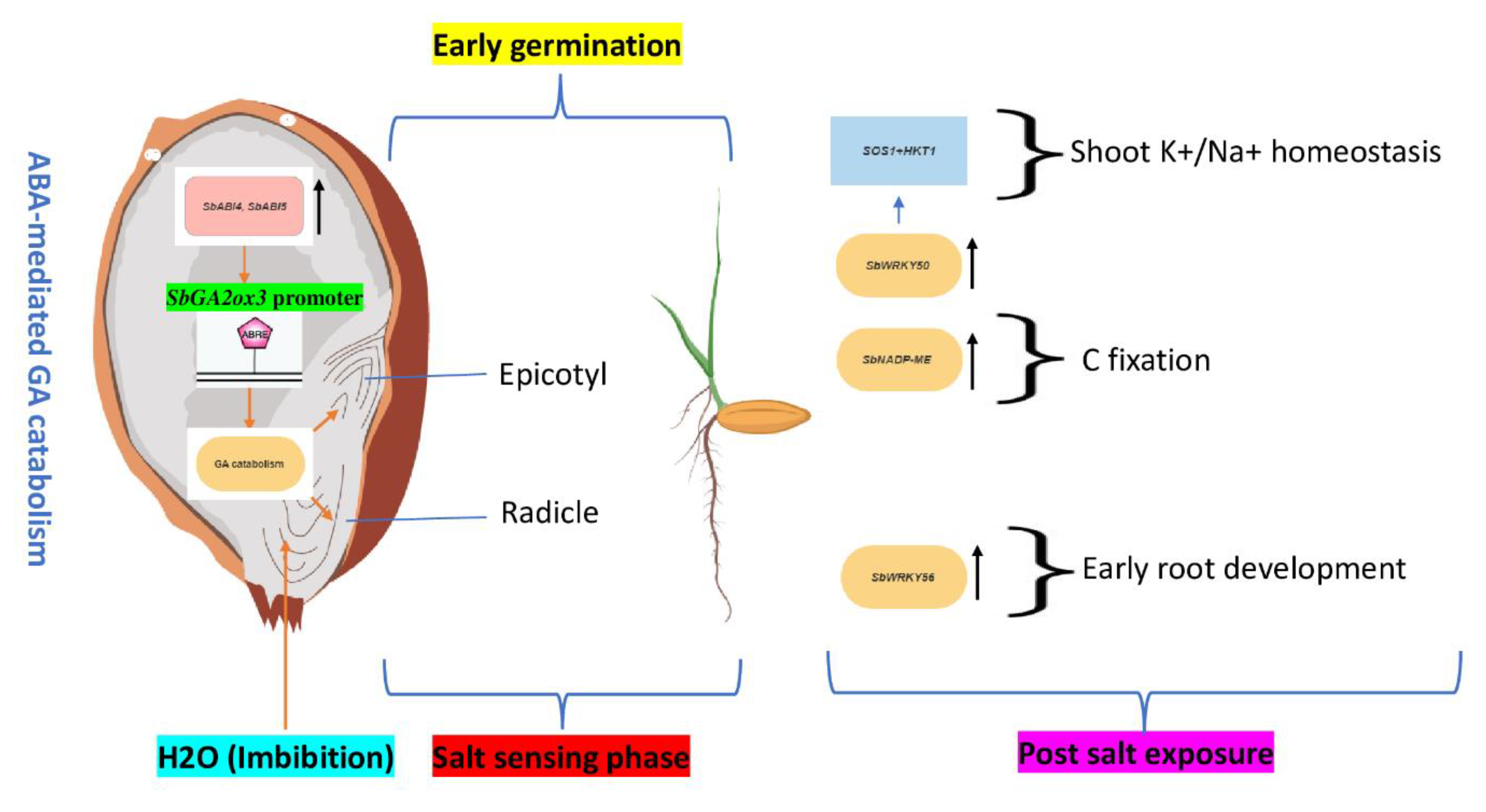
2. Seed Dormancy Release and Germination
3. Early to Late Salt Stress Signaling
3.1. Hormonal Signaling
3.2. Non-Hormonal Signaling
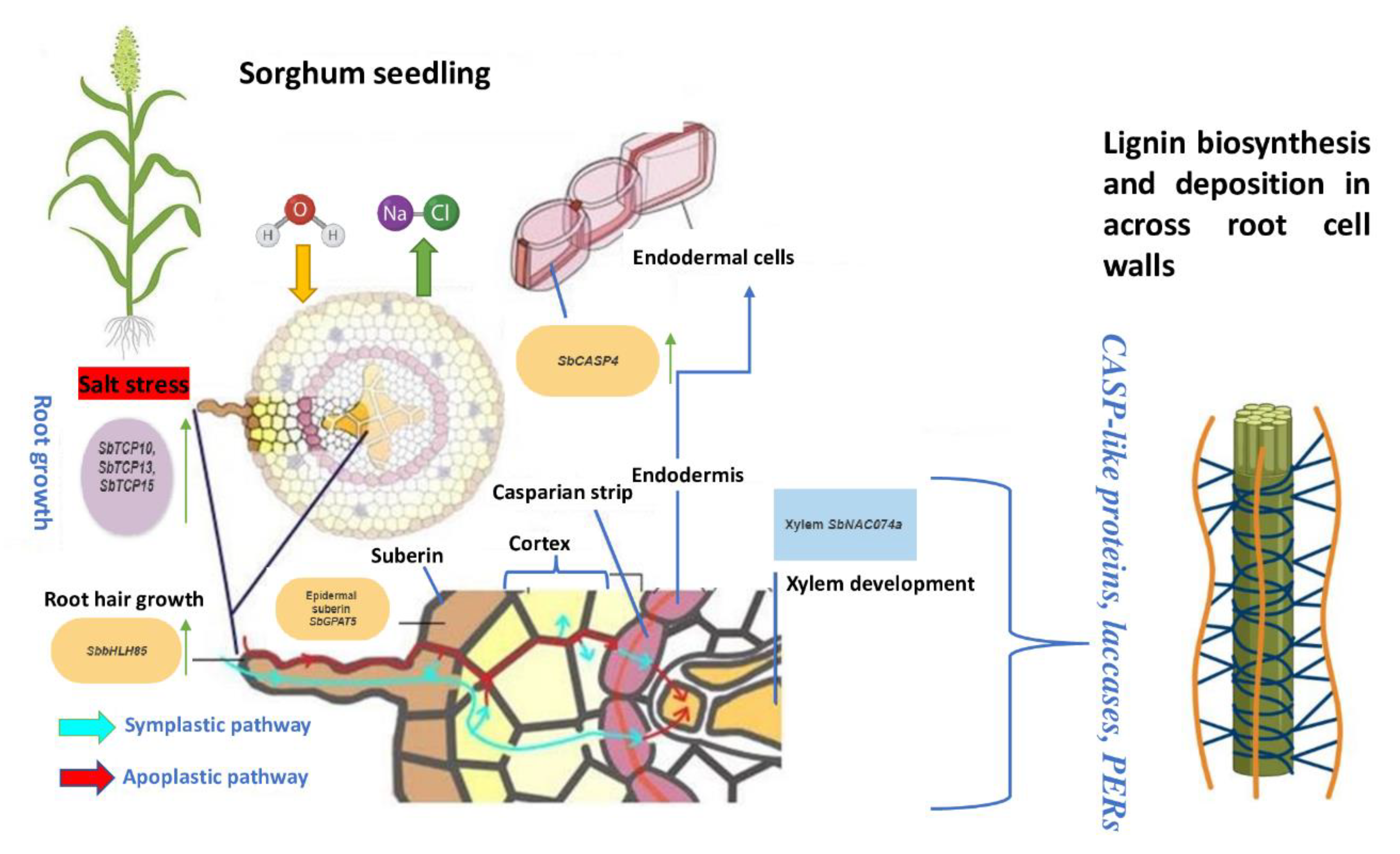
4. Root Developmental Plasticity
5. Water Absorption and Channeling
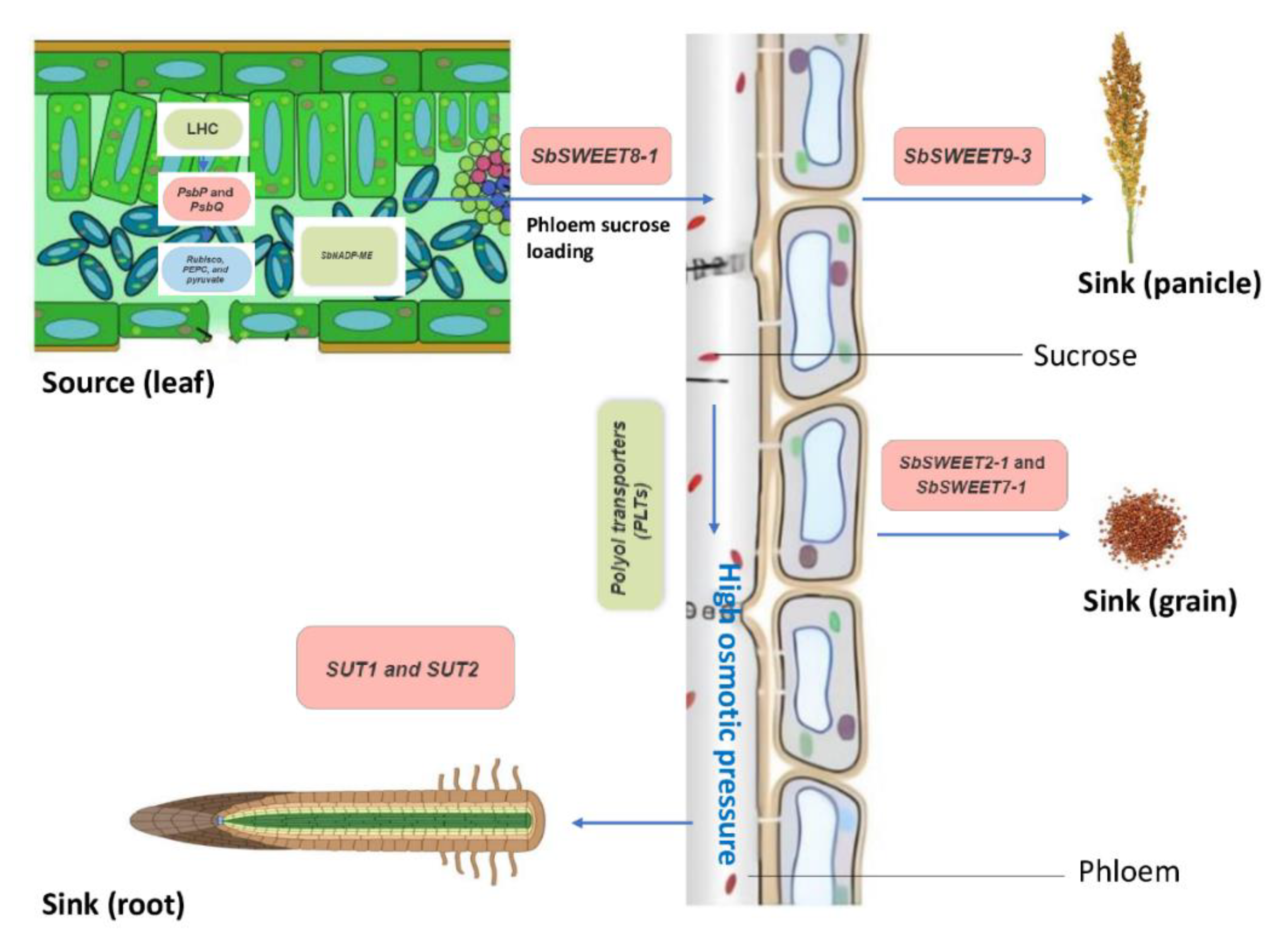
6. Photosynthesis and Carbon Partitioning
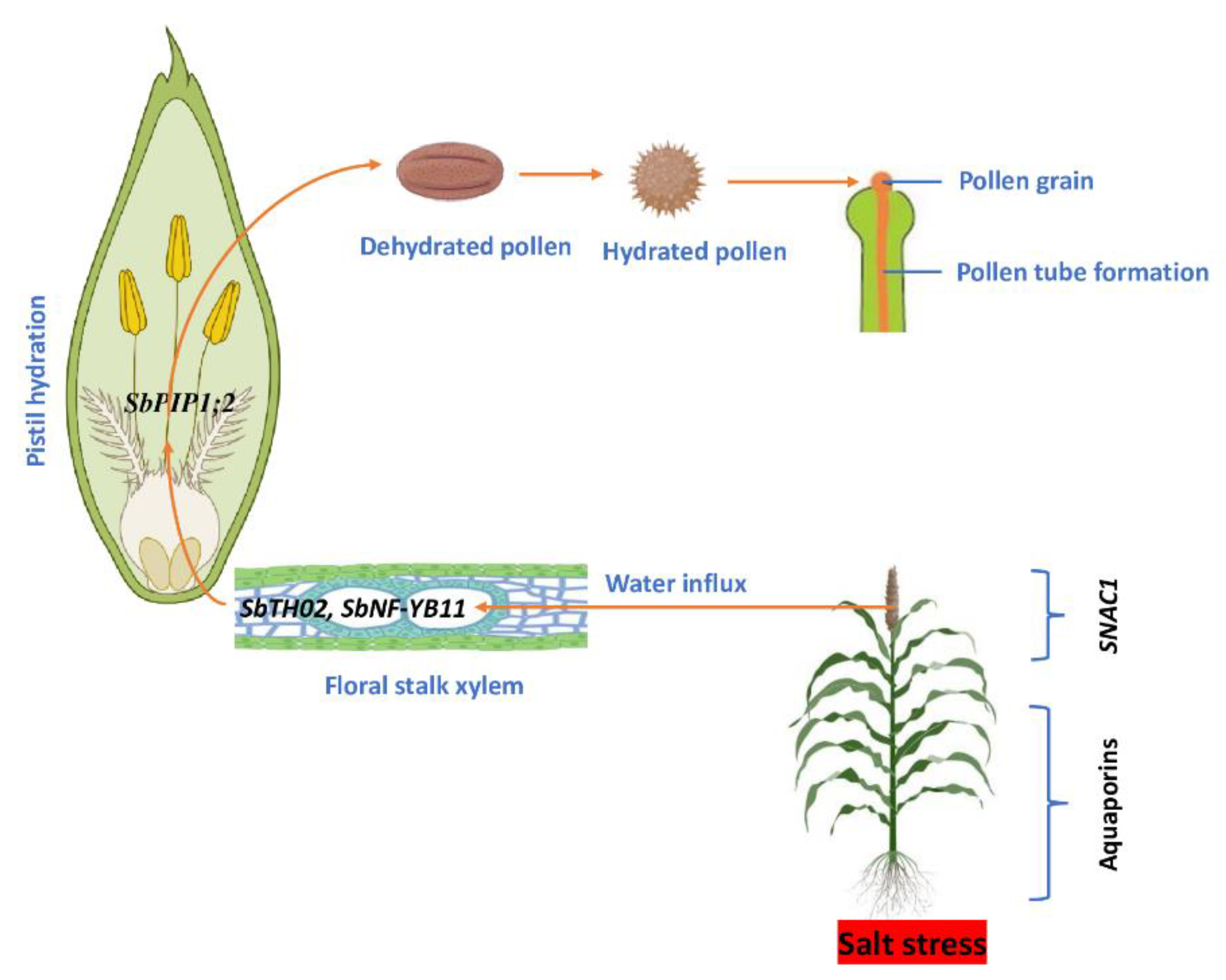
7. Flowering and Pollination
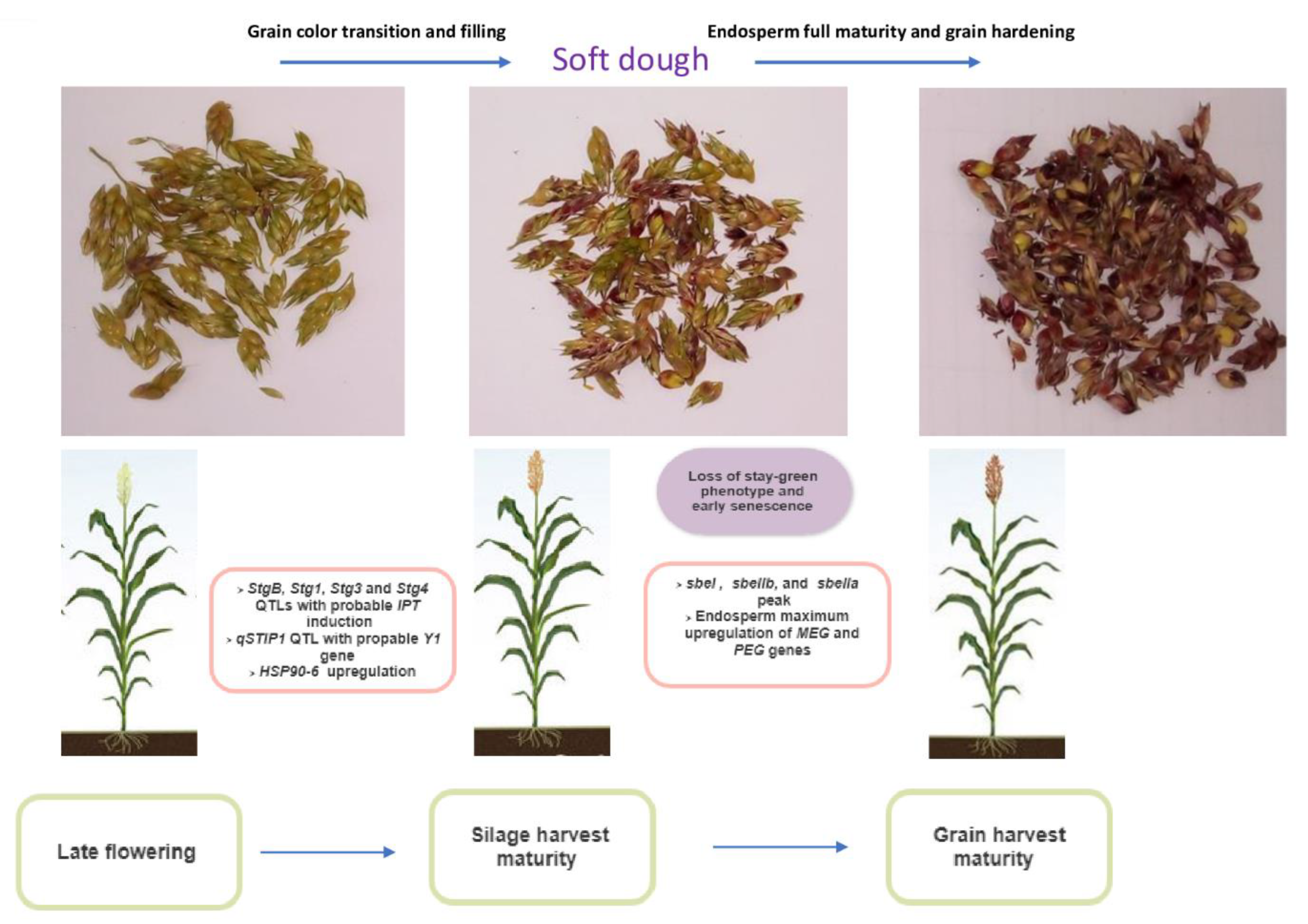
8. Silage Harvest Maturity
9. Conclusions and Future Perspective
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Szabolcs, I. Salt-Affected Soils; CRC Press: Boca Raton, FL, USA, 1989. [Google Scholar]
- Seelig, B.D. Salinity and Sodicity in North Dakota Soils; NDSU Extension Service, North Dakota State University of Agriculture and Applied Science: Fargo, ND, USA, 2000; p. 16. [Google Scholar]
- Squires, V.R.; Glenn, E.P. Salination, desertification, and soil erosion. In The Role of Food, Agriculture, Forestry and Fisheries in Human Nutrition; Squires, V.R., Ed.; UNESCO, EOLSS Publishers: Oxford, UK, 2004. [Google Scholar]
- Kovács, D.; Tóth, T.; Marth, P. Soil salinity between 1992 and 2000 in Hungary. Agrokém. Talajt. 2006, 55, 89–98. [Google Scholar] [CrossRef] [Green Version]
- Fan, X.; Pedroli, B.; Liu, G.; Liu, Q.; Liu, H.; Shu, L. Soil salinity development in the yellow river delta in relation to groundwater dynamics. Land Degrad. Dev. 2012, 23, 175–189. [Google Scholar] [CrossRef]
- Chopra, S. Techniques and tools of modern plant breeding: Field Crops. In Plant Biotech; Ricroch, A., Chopra, S., Fleischer, S., Eds.; Springer: Cham, Switzerland, 2014. [Google Scholar]
- Hameed, M.; Goher, N. Evaluation of seedling survivability and growth response as selection criteria for breeding drought tolerance in wheat. Cereal Res. Commun. 2010, 38, 193–202. [Google Scholar] [CrossRef]
- Pérez-Flores, L.; Carrari, F.; Osuna-Fernández, R.; Rodríguez, M.V.; Enciso, S. Expression analysis of a GA 20-oxidase in embryos from two sorghum lines with contrasting dormancy: Possible participation of this gene in the hormonal control of germination. J. Exp. Bot. 2003, 54, 2071–2079. [Google Scholar] [PubMed]
- Gualano, N.; Carrari, F.; Rodrıguez, M.V. Reduced embryo sensitivity to ABA in sprouting susceptible sorghum (Sorghum bicolor) variety is associated with an altered ABA signalling. Seed Sci. Res. 2007, 17, 81–90. [Google Scholar] [CrossRef]
- Cantoro, R.; Fernández, L.G.; Cervigni, G.D.L. Seed dormancy QTL identification across a Sorghum bicolor segregating population. Euphytica 2016, 211, 41–56. [Google Scholar] [CrossRef]
- Cantoro, R.; Crocco, C.D.; Benech-Arnold, R.L.; Rodríguez, M.V. In vitro binding of Sorghum bicolor transcription factors ABI4 and ABI5 to a conserved region of a GA 2-OXIDASE promoter: Possible role of this interaction in the expression of seed dormancy. J. Exp. Bot. 2013, 64, 5721–5735. [Google Scholar] [CrossRef] [Green Version]
- Rodríguez, M.V.; Mendiondo, G.M.; Cantoro, R.; Auge, G.A.; Luna, V.; Masciarelli, O.; Benech-Arnold, R.L. Expression of seed dormancy in grain Sorghum lines with contrasting pre-harvest sprouting behavior involves differential regulation of gibberellin metabolism genes. Plant Cell Physiol. 2012, 53, 64–80. [Google Scholar] [CrossRef] [Green Version]
- Collin, A.; Daszkowska-Golec, I. Updates on the role of ABSCISIC ACID INSENSITIVE 5 (ABI5) and ABSCISIC ACID-RESPONSIVE ELEMENT BINDING FACTORs (ABFs) in ABA signaling in different developmental stages in plants. Cells 2021, 10, 1996. [Google Scholar] [CrossRef]
- Rushton, P.J.; Somssich, I.E.; Ringler, P.; Shen, Q.J. WRKY transcription factors. Trends Plant Sci. 2010, 5, 247–258. [Google Scholar] [CrossRef]
- Jiang, J.; Ma, S.; Ye, N.; Jiang, M.; Cao, J.; Zhang, J. WRKY transcription factors in plant responses to stresses. J. Integr. Plant Biol. 2017, 59, 86–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baillo, E.H.; Hanif, M.S.; Guo, Y.; Zhang, Z.; Xu, P.; Algam, S.A. Genome-wide identification of WRKY transcription factor family members in sorghum (Sorghum bicolor (L.) moench). PLoS ONE 2020, 15, e0236651. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Li, J.; Sui, Y. The sweet sorghum SbWRKY50 is negatively involved in salt response by regulating ion homeostasis. Plant Mol. Biol. 2020, 102, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.T.; Ren, Z.J.; Liu, Z.Q.; Feng, X.; Guo, R.Q.; Li, B.G.; Li, L.G.; Jing, H.C. SbHKT1;4, a member of the high-affinity potassium transporter gene family from Sorghum bicolor, functions to maintain optimal Na+ /K+ balance under Na+ stress. J. Integr. Plant Biol. 2014, 56, 315–332. [Google Scholar] [CrossRef]
- Ding, Z.; Yan, J.; Li, C.; Li, G.; Yun, W.; Shao, Z. Transcription factor WRKY46 modulates the development of Arabidopsis lateral roots in osmotic/salt stress conditions via regulation of ABA signaling and auxin homeostasis. Plant J. 2015, 84, 12958. [Google Scholar] [CrossRef]
- Guo, Y.; Song, Y.; Zhang, H.; Zheng, Y.; Guo, J.; Sui, N. NADP-Malate Dehydrogenase of sweet sorghum improves salt tolerance of Arabidopsis thaliana. J. Agric. Food Chem. 2018, 66, 5992–6002. [Google Scholar] [CrossRef]
- Rivera, P.; Moya, C.; O’Brien, J.A. Low Salt Treatment Results in Plant Growth Enhancement in Tomato Seedlings. Plants 2022, 11, 807. [Google Scholar] [CrossRef]
- Javid, G.; Sorooshzadeh, M.; Moradi, A.; Modarres-Sanavy, F.; Ali Mohammad, N.; Allahdadi, I. The role of phytohormones in alleviating salt stress in crop plants. Aust. J. Crop Sci. 2011, 5, 726–734. [Google Scholar]
- Chen, K.; Li, G.J.; Bressan, R.A.; Song, C.P.; Zhu, J.K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef] [Green Version]
- Xiong, L.; Zhu, J.K. Regulation of abscisic acid biosynthesis. Plant Physiol. 2003, 133, 29–36. [Google Scholar] [CrossRef] [Green Version]
- Ma, S.L.; Meng, C.; Zhou, C.; Fu, J.; Shen, X.; Zhang, C.; Li, Y. Genome-wide analysis of abscisic acid biosynthesis, catabolism, and signaling in Sorghum Bicolor under saline-alkali stress. Biomolecules 2019, 9, 823. [Google Scholar] [CrossRef] [PubMed]
- Fujita, Y.; Yoshida, T.; Yamaguchi-Shinozaki, K. Pivotal role of the AREB/ABF-SnRK2 pathway in ABRE-mediated transcription in response to osmotic stress in plants. Physiol. Plant. 2013, 147, 15–27. [Google Scholar] [CrossRef]
- Ghate, T.; Barvkar, V.; Deshpande, S. Role of ABA signaling in regulation of stem sugar metabolism and transport under post-flowering drought stress in sweet sorghum. Plant Mol. Biol. Rep. 2019, 37, 303–313. [Google Scholar] [CrossRef]
- Kadier, Y.; Zu, Y.; Dai, Q. Genome-wide identification, classification and expression analysis of NAC family of genes in sorghum [Sorghum bicolor (L.) Moench]. Plant Growth Regul. 2017, 83, 301–312. [Google Scholar] [CrossRef]
- Zhang, X.; Long, Y.; Chen, X.; Zhang, B.; Xin, Y.; Li, L.; Cao, S.; Liu, F.; Wang, Z.; Huang, H.; et al. A NAC transcription factor OsNAC3 positively regulates ABA response and salt tolerance in rice. BMC Plant Biol. 2021, 21, 546. [Google Scholar] [CrossRef] [PubMed]
- Seok, H.Y.; Woo, D.H.; Nguyen, L.V.; Tran, H.T.; Tarte, V.; Mehdi, N. Arabidopsis AtNAP functions as a negative regulator via repression of AREB1 in salt stress response. Planta 2017, 245, 329–341. [Google Scholar] [CrossRef]
- Hu, H.; Dai, M.; Yao, J. Overexpressing a NAM, ATAF, and CUC (NAC) transcription factor enhances drought resistance and salt tolerance in rice. Proc. Natl. Acad. Sci. USA 2006, 103, 12987–12992. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.; Bai, Y.; Shen, C. Auxin-related gene families in abiotic stress response in Sorghum bicolor. Funct. Integr. Genom. 2010, 10, 533–546. [Google Scholar] [CrossRef]
- Zheng, L.; Zhou, J.; Yi, L.; Zhou, X. Identification and expression analysis of the auxin response factor (ARF) gene family in sorghum [Sorghum bicolor (L.) Moench]. Pakistan J. Bot. 2021, 53, 37. [Google Scholar] [CrossRef]
- Okushima, Y.; Overvoorde, P.J.; Arima, K.; Alonso, J.M. Functional genomic analysis of the AUXIN RESPONSE FACTOR gene family members in Arabidopsis thaliana: Unique and overlapping functions of ARF7 and ARF19. Plant Cell 2005, 17, 444–463. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.H.; Wang, S.K.; Shen, C.J.; Zhang, S.N.; Chen, Y.; Xu, Y.X.; Liu, Y.; Wu, Y.R.; Jiang, D.A. OsARF12, a transcription activator on auxin response gene, regulates root elongation and affects iron accumulation in rice (Oryza sativa). New Phytol. 2012, 193, 109–120. [Google Scholar] [CrossRef]
- Licausi, F.; Ohme-Takagi, M.; Perata, P. APETALA2/Ethylene Responsive Factor (AP2/ERF) transcription factors: Mediators of stress responses and developmental programs. New Phytol. 2013, 199, 639–649. [Google Scholar] [CrossRef] [PubMed]
- Mathur, S.; Priyadarshini, S.S.; Singh, V. Comprehensive phylogenomic analysis of ERF genes in sorghum provides clues to the evolution of gene functions and redundancy among gene family members. 3 Biotech 2020, 10, 139. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, R.; Caldana, C.; Mueller-Roeber, B.; Schippers, J.H. The contribution of SERF1 to root-to-shoot signaling during salinity stress in rice. Plant Signal. Behav. 2014, 9, e27540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akbudak, M.A.; Filiz, E.; Kontbay, K. DREB2 (dehydration-responsive element-binding protein 2) type transcription factor in sorghum (Sorghum bicolor): Genome-wide identification, characterization and expression profiles under cadmium and salt stresses. 3 Biotech 2018, 8, 426. [Google Scholar] [CrossRef] [PubMed]
- Herath, V. Small Family, Big Impact: In silico analysis of DREB2 transcription factor family in rice. Comput. Biol. Chem. 2012, 65, 128–139. [Google Scholar] [CrossRef] [PubMed]
- Mallikarjuna, G.; Mallikarjuna, K.; Reddy, M.K. Expression of OsDREB2A transcription factor confers enhanced dehydration and salt stress tolerance in rice (Oryza sativa L.). Biotechnol. Lett. 2011, 33, 1689–1697. [Google Scholar] [CrossRef]
- Nakano, T.K.; Suzuki, T.; Fujimura, H. Genome-wide analysis of the ERF gene family in Arabidopsis and rice. Plant Physiol. 2006, 140, 411–432. [Google Scholar] [CrossRef] [Green Version]
- Kasapoğlu, E.; Ilhan, D.; Kizilkaya, A.; Hossein’pour, K. Genome-wide analysis of BES1 transcription factor family in sorghum [Sorghum bicolor (L.) Moench] genome. Türkiye Tarımsal Araştırmalar Derg. 2020, 7, 85–95. [Google Scholar]
- Salazar-Henao, J.E.; Lehner, R.; Betegón-Putze, I.; Vilarrasa-Blasi, J.; Caño-Delgado, A.I. BES1 regulates the localization of the brassinosteroid receptor BRL3 within the provascular tissue of the Arabidopsis primary root. J. Exp. Bot. 2016, 67, 4951–4961. [Google Scholar] [CrossRef] [Green Version]
- Jia, C.; Zhao, S.; Bao, T.; Zhao, P.; Peng, K.; Guo, Q.; Gao, X.; Qin, J. Tomato BZR/BES transcription factor SlBZR1 positively regulates BR signaling and salt stress tolerance in tomato and Arabidopsis. Plant Sci. 2021, 302, 110719. [Google Scholar] [CrossRef] [PubMed]
- Friedrichsen, D.M.; Nemhauser, J.; Muramitsu, T.; Maloof, J.N.; Alonso, J.R.; Ecker, M.; Furuya, J.; Chory, J. Three redundant brassinosteroid early response genes encode putative bHLH transcription factors required for normal growth. Genetics 2002, 162, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Moreno, J.E.; Moreno-Piovano, G.; Chan, R.L. The antagonistic basic helix-loop-helix partners BEE and IBH1 contribute to control plant tolerance to abiotic stress. Plant Sci. 2018, 271, 143–150. [Google Scholar] [CrossRef] [PubMed]
- Seo, H.K.; Lee, S.H.; Lim, B.D.; Lee, J.H.; An, S.J.; Chon, G.P. The Rice Basic Helix–Loop–Helix 79 (OsbHLH079) Determines leaf angle and grain shape. Int. J. Mol. Sci. 2020, 2, 2090. [Google Scholar] [CrossRef] [Green Version]
- Colebrook, E.H.; Thomas, S.G.; Phillips, A.L.; Hedden, P. The role of gibberellin signalling in plant responses to abiotic stress. J. Exp. Biol. 2014, 217, 67–75. [Google Scholar] [CrossRef] [Green Version]
- Hedden, P. The current status of research on gibberellin biosynthesis. Plant Cell Physiol. 2020, 61, 1832–1849. [Google Scholar] [CrossRef]
- Wang, Y.L.; Ali, J.Z.; Gao, S.; Ni, L.; Li, X.N.; Wu, X.; Jiang, Y.F. Identification and expression analysis of Sorghum bicolor gibberellin oxidase genes with varied gibberellin levels involved in regulation of stem biomass. Ind. Crops Prod. 2020, 145, 11951. [Google Scholar] [CrossRef]
- Yoshida, H.; Hirano, K.; Sato, T.; Mitsuda, N.; Nomoto, M. DELLA protein functions as a transcriptional activator through the DNA binding of the indeterminate domain family proteins. Proc. Natl. Acad. Sci. USA 2014, 111, 7861–7866. [Google Scholar] [CrossRef] [Green Version]
- Fan, Y.; Yan, J.; Lai, D.; Yang, H.; Xue, G.A.; He, T.; Guo, L.; Chen, X.B.; Cheng, D.B.; Xiang, J.; et al. Genome-wide identification, expression analysis, and functional study of the GRAS transcription factor family and its response to abiotic stress in sorghum [Sorghum bicolor (L.) Moench]. BMC Genom. 2020, 22, 509. [Google Scholar] [CrossRef]
- Liu, L.Y.; Ding, H.Y.; Luo, X.F. OsGRAS1 Genes Associated with Stress Resistance of Rice as Well as Promoter and Application Thereof; Shanghai Municipal Agricultural Biological Gene Center: Shanghai, China, 2010. [Google Scholar]
- Thomas, S.B.; David, M.A. Della Proteins: Master regulators of gibberellin-responsive growth and development: Gibberellins. Annu. Plant Rev. 2016, 49, 189–228. [Google Scholar]
- Gao, W.; Xu, F.C.; Guo, D.D. Calcium-dependent protein kinases in cotton: Insights into early plant responses to salt stress. BMC Plant Biol. 2018, 18, 1–15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mittal, S.; Mallikarjuna, M.G.; Atmakuri, R.; Prashant, D.; Prasanta, T. Comparative Analysis of CDPK family in maize, arabidopsis, rice, and sorghum revealed potential targets for drought tolerance improvement. Front. Chem. 2017, 5, 115. [Google Scholar] [CrossRef] [Green Version]
- Gu, L.W. Identification and Expression Analysis of MCU Protein Family Genes in Sorghum Bicolor. Botanical Res. 2020, 9, 169–179. [Google Scholar] [CrossRef]
- Teardo, E.; Carraretto, L.; Moscatiello, R. A chloroplast-localized mitochondrial calcium uniporter transduces osmotic stress in Arabidopsis. Nat. Plants 2019, 5, 581–588. [Google Scholar] [CrossRef]
- Wurzinger, B.; Mair, A.; Pfister, B.; Teige, M. Crosstalk of calcium-dependent protein kinase and MAP kinase signaling. Plant Sig. Behav. 2011, 6, 8–12. [Google Scholar] [CrossRef]
- Yu, C.H.; Dan, F.S.; Yun, H.D. Identification and analysis of the MAPK gene family in Sorghum bicolor. Master’s thesis, Harbin Normal University, Harbin, China, 2011. [Google Scholar]
- Hur, Y.; Kim, D.H. Overexpression of OsMAPK2 enhances low phosphate tolerance in rice and Arabidopsis thaliana. Am. J. Plant Sci. 2014, 5, 43254. [Google Scholar] [CrossRef] [Green Version]
- Park, H.J.; Kim, W.Y.; Yun, D.J. A New Insight of Salt Stress Signaling in Plant. Mol. Cells 2016, 39, 447–459. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Li, S.; Sui, Y. SbbHLH85, a bHLH member, modulates resilience to salt stress by regulating root hair growth in sorghum. Theor. Appl. Genet. 2022, 135, 201–216. [Google Scholar] [CrossRef]
- Francis, A.; Dhaka, N.; Bakshi, M.; Jung, K.-H.; Sharma, M.K.; Sharma, R. Comparative phylogenomic analysis provides insights into TCP gene functions in Sorghum. Sci. Rep. 2016, 6, 38488. [Google Scholar] [CrossRef] [Green Version]
- Tatematsu, K.; Nakabayashi, K.; Kamiya, Y.; Nambara, E. Transcription factor AtTCP14 regulates embryonic growth potential during seed germination in Arabidopsis thaliana. Plant J. 2008, 53, 42–52. [Google Scholar] [CrossRef]
- Jin, X.Y.; Long, Y.F.; Shi, X.; Zhen, Y.; Wei, C.; Amangul, H.; Chen, X.Y. SbNAC2 enhances abiotic stress tolerance by upregulating ROS scavenging activities and inducing stress-response genes in sorghum. Environ. Expt. Bot. 2021, 192, 104664. [Google Scholar] [CrossRef]
- Pignocchi, C.; Foyer, C. Apoplastic ascorbate metabolism and its role in the regulation of cell signaling. Curr. Opin. Plant Biol. 2003, 6, 379–389. [Google Scholar] [CrossRef] [PubMed]
- Krishnamurthy, P.; Ranathunge, K.; Franke, R.; Prakash, H.S.; Schreiber, L.; Mathew, M.K. The role of root apoplastic transport barriers in salt tolerance of rice (Oryza sativa L.). Planta 2009, 230, 119–134. [Google Scholar] [CrossRef]
- Yang, Z.; Zheng, Z.; Wei, H. Transcriptome analysis of sweet Sorghum inbred lines differing in salt tolerance provides novel insights into salt exclusion by roots. Plant Soil 2018, 430, 423–439. [Google Scholar] [CrossRef]
- Wei, X.; Yang, Z.; Han, G.; Zhao, X.; Yin, S.; Yuan, F.; Wang, B. The developmental dynamics of the sweet sorghum root transcriptome elucidate the differentiation of apoplastic barriers. Plant Signal. Behav. 2020, 15, 1724465. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Jiang, J.; Du, Q.; Luo, L.; Li, X.; Xie, X. Cytochrome P450 superfamily: Evolutionary and functional divergence in sorghum (Sorghum bicolor) stress resistance. J. Agric. Food Chem. 2021, 69, 10952–10961. [Google Scholar] [CrossRef]
- Murata, N.; Tasaka, Y. Glycerol-3-phosphate acyltransferase in plants. Biochim. Biophys. Acta 1997, 1348, 10–16. [Google Scholar] [CrossRef] [PubMed]
- Molina, Y.; Li-Beisson, F.; Beisson, J.B.; Ohlrogge, M. Pollard. Identification of an Arabidopsis feruloyl-coenzyme A transferase required for suberin synthesis. Plant Physiol. 2009, 151, 1317–1328. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- de Silva, N.D.; Murmu, J.; Chabot, D. Root Suberin Plays Important Roles in Reducing Water Loss and Sodium Uptake in Arabidopsis thaliana. Metabolites 2021, 11, 735. [Google Scholar] [CrossRef]
- Surender, R.P.; Jogeswar, G.; Rasineni, G.K.; Maheswari, M.; Reddy, A.R.; Varshney, R.K.; Kishor, P.B. Proline over-accumulation alleviates salt stress and protects photosynthetic and antioxidant enzyme activities in transgenic sorghum [Sorghum bicolor (L.) Moench]. Plant Physiol. Biochem. 2015, 94, 104–113. [Google Scholar] [CrossRef] [Green Version]
- Wood, A.J.; Saneoka, H.; Rhodes, D.; Joly, R.J.; Goldsbrough, P.B. Betaine aldehyde dehydrogenase in sorghum. Plant Physiol. 1996, 110, 1301–1308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maheswari, M.; Varalaxmi, Y.; Vijayalakshmi, A. Metabolic engineering using mtlD gene enhances tolerance to water deficit and salinity in sorghum. Biol. Plant. 2012, 54, 647–652. [Google Scholar] [CrossRef]
- Sakurai, J.; Ishikawa, F.; Yamaguchi, T.; Uemura, M.; Maeshima, M. Identification of 33 rice aquaporin genes and analysis of their expression and function. Plant Cell Physiol. 2005, 46, 1568–1577. [Google Scholar] [CrossRef] [PubMed]
- Palakolanu, R.R.; Rao, T.S.; Vincent, K.V. Genome-wide identification and characterization of the aquaporin gene family in Sorghum bicolor (L.). Plant Gene 2015, 1, 18–28. [Google Scholar]
- Martins, C.P.; Neves, D.M.; Cidade, L.C.; Mendes, A.F.; Silva, D.C. Expression of the citrus CsTIP2;1 gene improves tobacco plant growth, antioxidant capacity and physiological adaptation under stress conditions. Planta 2017, 24, 5951–5963. [Google Scholar] [CrossRef]
- Patankar, H.V.; Al-Harrasi, I.; Al-Yahyai, R.; Yaish, M.W. Functional characterization of date palm aquaporin gene PdPIP1;2 confers drought and salinity tolerance to yeast and Arabidopsis. Genes 2019, 10, 390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.; Yin, L.; Deng, X.; Wang, S.; Tanaka, K.; Zhang, S. Aquaporin-mediated increase in root hydraulic conductance is involved in silicon-induced improved root water uptake under osmotic stress in Sorghum bicolor L. J. Exp. Bot. 2014, 65, 4747–4756. [Google Scholar] [CrossRef] [Green Version]
- Jia, J.; Liang, Y.; Gou, T.; Hu, Y.; Zhu, Y.; Huo, H.; Guo, J.; Gong, H. The expression response of plasma membrane aquaporins to salt stress in tomato plants. Environ. Exp. Bot. 2020, 178, 104190. [Google Scholar] [CrossRef]
- Suorsa, M.; Sirpiö, S.; Allahverdiyeva, Y.; Paakkarinen, V.; Mamedov, F.; Styring, S. PsbR, a missing link in the assembly of the oxygen-evolving complex of plant photosystem II. J. Biol. Chem. 2006, 281, 145–150. [Google Scholar] [CrossRef] [Green Version]
- Sui, N.; Yang, Z.; Liu, M.; Wang, B. Identification and transcriptomic profiling of genes involved in increasing sugar content during salt stress in sweet sorghum leaves. BMC Genom. 2015, 16534. [Google Scholar] [CrossRef] [Green Version]
- Chang, G.G.; Tong, L. Structure and function of malic enzymes, a new class of oxidative decarboxylases. Biochem 2003, 42, 12721–12733. [Google Scholar] [CrossRef] [PubMed]
- Qazi, H.A.; Paranjpe, S.; Bhargava, S. Stem sugar accumulation in sweet sorghum-activity and expression of sucrose metabolizing enzymes and sucrose transporters. J. Plant Physiol. 2012, 169, 605–613. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann-Thoma, G.; Hinkel, K.; Nicolay, P.; Willenbrink, J. Sucrose accumulation in sweet sorghum stem internodes in relation to growth. Physiol. Plant. 2010, 97, 277–284. [Google Scholar] [CrossRef]
- Yang, Z.; Li, J.L.; Liu, L.N.; Xie, Q.; Sui, N. Photosynthetic regulation under salt stress and salt-Tolerance mechanism of sweet sorghum. Front. Plant Sci. 2020, 10, 1722. [Google Scholar] [CrossRef]
- Anjali, U.; Fatima, M.S.; Manu, S.; Ramasamy, M. Structure and regulation of SWEET transporters in plants: An update. Plant Physiol. Biochem. 2020, 156, 1–6. [Google Scholar] [CrossRef]
- Mizuno, H.K.; Shigemitsu, K. The sorghum SWEET gene family: Stem sucrose accumulation as revealed through transcriptome profiling. Biotech 2016, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Eom, J.S.; Chen, L.; Sosso, D.; Julius, B.T.; Lin, I.; Qu, X.; Braun, D.M.; Frommer, W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015, 25, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Qu, X.; Hou, B.; Sosso, S.; Osorio, A.R.; Fernie, W.B. Sucrose efflux mediated by SWEET proteins as a key step for phloem transport. Science 2012, 335, 207–211. [Google Scholar] [CrossRef]
- Guan, Y.F.; Huang, X.Y.; Zhu, J.; Gao, J.F.; Zhang, H.X.; Yang, Z.N. RUPTURED POLLEN GRAIN1, a member of the MtN3/saliva gene family, is crucial for exine pattern formation and cell integrity of microspore in Arabidopsis. Plant Physiol. 2008, 147, 852–863. [Google Scholar] [CrossRef] [Green Version]
- Ma, L.; Zhang, D.Q.; Miao, J.; Yang, Y.; Xuan, Y.; Hu, Y. Essential role of sugar transporter OsSWEET11 during the early stage of rice grain filling. Plant Cell Physiol. 2017, 58, 863–873. [Google Scholar] [CrossRef] [Green Version]
- Zhao, H.; Wu, D.F.; Kong, K.; Lin, H.; Zhang, G. The Arabidopsis thaliana Nuclear Factor Y Transcription Factors. Front. Plant. Sci. 2017, 10, 2045. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Malviya, N.; Jaiswal, P.; Yadav, D. Genome- wide characterization of Nuclear Factor Y (NF-Y) gene family of sorghum [Sorghum bicolor (L.) Moench]: A bioinformatics approach. Physiol. Mol. Biol. Plants 2016, 22, 33–49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Siriwardana, C.L.; Gnesutta, N.; Kumimoto, R.W. NUCLEAR FACTOR Y, Subunit A (NF-YA) Proteins Positively Regulate Flowering and Act Through FLOWERING LOCUS T. PLoS Genet. 2016, 12, e1006496. [Google Scholar] [CrossRef]
- Wei, X.; Xu, J.; Guo, H.; Jiang, L.; Chen, S.; Yu, C.; Zhou, Z.; Hu, P.; Zhai, H.; Wan, J. DTH8 suppresses flowering in rice, influencing plant height and yield potential simultaneously. Plant Physiol. 2010, 153, 1747–1758. [Google Scholar] [CrossRef]
- Myers, Z.A.; Kumimoto, R.W.; Siriwardana, C.L.; Gayler, K.K.; Risinger, J.R.; Pezzetta, D.; Holt, B.F. NUCLEAR FACTOR Y, Subunit C (NF-YC) transcription factors are positive regulators of photomorphogenesis in Arabidopsis thaliana. PLoS Genet. 2016, 12, e1006333. [Google Scholar] [CrossRef] [Green Version]
- Frerichs, R.; Thoma, A.T.; Abdallah, P.; Frommolt, W.; Werr, J.W. The founder-cell transcriptome in the Arabidopsis apetala1 cauliflower inflorescence meristem. BMC Genom. 2016, 17, 855. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, J.; Qiu, Y.; Du, L.; Poovaiah, B.W. Plant-specific trihelix transcription factor AtGT2L interacts with calcium/calmodulin and responds to cold and salt stresses. Plant Sci. 2012, 274–280. [Google Scholar] [CrossRef]
- Yoo, C.Y.; Hasegawa, P.M.; Mickelbart, M.V. Regulation of stomatal density by the GTL1 transcription factor for improving water use efficiency. Plant Signal. Behav. 2011, 6, 1069–1071. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Chang, Y.; Ma, S.; Shen, J.; Hu, H.L. Genome-wide identification of SNAC1-targeted genes involved in drought response in rice. Front Plant Sci. 2019, 10, 982. [Google Scholar] [CrossRef] [Green Version]
- Sanjari, S.; Shirzadian-Khorramabad, R.; Shobbar, Z.S.; Shahbazi, M. Systematic analysis of NAC transcription factors’ gene family and identification of post-flowering drought stress responsive members in sorghum. Plant Cell Rep. 2019, 3, 361–376. [Google Scholar] [CrossRef]
- Windari, E.A.; Ando, M.; Mizoguchi, Y.; Shimada, H.; Ohira, K.; Kagaya, Y.; Higashiyama, T.; Takayama, S.M.; Watanabe, K. Two aquaporins, SIP1;1 and PIP1;2, mediate water transport for pollen hydration in the Arabidopsis pistil. Plant Biotechnol. 2021, 38, 77–87. [Google Scholar] [CrossRef]
- Borrell, K.A.; Mullet, J.E.; George-Jaeggli, B.; van Oosterom, E.J.; Hammer, G.L.; Klein, P.E.; Jordan, D.R. Drought adaptation of stay-green sorghum is associated with canopy development, leaf anatomy, root growth, and water uptake. J. Expt. Bot. 2014, 65, 6251–6263. [Google Scholar] [CrossRef] [PubMed]
- Emebiri, L.C. QTL dissection of the loss of green colour during postanthesis grain maturation in two-rowed barley. Theor. App. Genet. 2013, 126, 1873–1884. [Google Scholar] [CrossRef] [PubMed]
- Ougham, H.; Hörtensteiner, S.; Armstead, I.; Donnison, I.; King, I.; Thomas, H.; Mur, L. The control of chlorophyll catabolism and the status of yellowing as a biomarker of leaf senescence. Plant Biol. 2008, 10, 4–14. [Google Scholar] [CrossRef] [PubMed]
- Vadez, V.; Deshpande, S.; Kholova, K.; Ramu, P.; Hash, C.T. Molecular breeding for stay-green: Progress and challenges in sorghum. In Genomic Applications to Crop Breeding; Varshney, R., Tuberosa, R., Eds.; Weily Online Library: Hoboken, NJ, USA, 2013; Volume 2, pp. 125–141. [Google Scholar]
- Jordan, D.R.; Hunt, C.H.; Cruickshank, A.W.; Borrell, A.K.; Henzell, R.G. The relationship between the stay-green trait and grain yield in elite sorghum hybrids grown in a range of environment. Crop Sci. 2012, 52, 1153–1161. [Google Scholar] [CrossRef]
- Subudhi, P.K.; Rosenow, D.T.; Nguyen, H.T. Quantitative trait loci for the stay green trait in sorghum (Sorghum bicolor L. Moench): Consistency across genetic backgrounds and environments. Theor. Appl. Genet. 2000, 101, 733–741. [Google Scholar] [CrossRef]
- Wilkinson, S.; Kudoyarova, G.R.; Veselov, D.S.; Arkhipova, T.N.; Davies, W.J. Plant hormone interactions: Innovative targets for crop breeding and management. J. Exp. Bot. 2012, 63, 3499–3509. [Google Scholar] [CrossRef]
- Rosa, M.; Rivero, J.G.; Van Deynze, A.; Harkamal, W.; Eduardo, B. Enhanced cytokinin synthesis in tobacco plants expressing PSARK::IPT prevents the degradation of photosynthetic protein complexes during drought. Plant Cell Physiol. 2010, 51, 1929–1941. [Google Scholar]
- Boddu, J.; Jiang, C.Z.; Sangar, V.; Olson, T.; Terry, P.; Thomas, C. Comparative structural and functional characterization of sorghum and maize duplications containing orthologous Myb transcription regulators of 3-Deoxyflavonoid biosynthesis. Plant Mol. Biol. 2006, 60, 185–199. [Google Scholar] [CrossRef]
- Takanashi, H.; Shichijo, M.; Sakamoto, L. Genetic dissection of QTLs associated with spikelet-related traits and grain size in sorghum. Sci. Rep. 2021, 11, 9398. [Google Scholar] [CrossRef]
- Sapkota, S.; Boatwright, J.L.; Jordan, K.; Boyles, R.; Kresovich, S. Identification of novel genomic associations and gene candidates for grain starch content in sorghum. Genes 2020, 11, 1448. [Google Scholar] [CrossRef] [PubMed]
- Kamal, N.M.; Alnor Gorafi, Y.S.; Abdelrahman, M.; Abdellatef, E.; Tsujimoto, H. Stay-green trait: A prospective approach for yield potential, and drought and heat stress adaptation in globally important cereals. Int. J. Mol. Sci. 2019, 20, 5837. [Google Scholar] [CrossRef] [PubMed]

| Predicted | |||||||
|---|---|---|---|---|---|---|---|
| Measured | Salinity Level | Non | Moderately | Highly | Extremely | Total | |
| Non | 90 | 10 | 0 | 0 | 0 | 100 | |
| Slightly | 10 | 87 | 1 | 0 | 0 | 100 | |
| Moderately | 11 | 26 | 69 | 0 | 0 | 100 | |
| Highly | 15 | 34 | 3 | 47 | 0 | 100 | |
| Extremely | 18 | 29 | 1 | 0 | 49 | 100 | |
| Total | 144 | 186 | 74 | 47 | 49 | 500 | |
| Gene Family | Gene Name | Prediction Method | Growth Stage | Organ | Mode of Action | Reference | Phenotype |
|---|---|---|---|---|---|---|---|
| Gibberellins biosynthesis (GA) | SbGA2ox3 | Transgenic | dormancy breaking and early germination | Seed | SbABI4 and SbABI5 mediated ABA signaling | Rodrguez et al. 2009 | Promote early seed germination |
| WRKY | SbWRKY50 | Transgenic | Seedling | Root | directly bind to the upstream promoters of SOS1 and HKT1. | Song et al. 2020 | Promote K/Na homeostasis |
| SbWRKY56 | Orthology | Seedling | Root | Promotion of ABA-mediated auxin homeostasis | Ding et al. 2015 | Root growth | |
| bHLH | SbbHLH050 | Transgenic | Seedling | Root | Salt induced induction of root hairs | Friedrichsen et al. 2002 | Root hair growth |
| SbBHLH079 | Orthologs | Post flowering | Grain | early response BR signaling components | Seo, Hyoseob et al. 2020 | Shapes the grain architecture | |
| SbTCP10, SbTCP13, SbTCP15 | Orthologs | Throughout life cycle | Root | Radicle growth promotion | Tatematsu et al. 2008 | Early sorghum root development | |
| NAC | SbNAC074a | Orthologs | Seedling | Root | Differentiation of xylem tissue | Promotion of water transport | |
| SbNAC56 | Transgenic | Seedling | ABA mediated hypersensitive to NaCl | Kadier et al. 2017 | Root and shoot growth | ||
| SbNAC58 | Transgenic | Seedling | ABA mediated sensitivity to osmotic stress | Seok et al. 2017 Hu et al. 2006 | Improved water intake | ||
| SbNAC005, SbNAC021 and SbNAC052 | Transgenic and orthology | Post flowering | Flower | Osmotic response | Sanjari et al. 2019, Hu et al. 2006 | Improved water intake | |
| Cytochrome | SORBI3006G148800, SORBI3006G148900 | Orthologs | Emergence | Root | Conversion of phenylalanine to cinnamic acid and tyrosine to p-cinnamic acid | Yang et al. 2017 | Casparian strip development |
| SbCASP4 | Transgenic | Germination | root | Involved in the phenylpropanoid pathway | Wei et al. 2021 | Lignin biosynthesis | |
| SbGPAT5 | Orthologs | Germination | Root | Catalyzes the transfer of an acyl group from an acyl donor to the sn-1 position of glycerol 3-phosphate | Murata et al. 1997 | Suberin biosynthesis | |
| ARF | SbARF16, SbARF7 | Orthologs | Flowering | Flower | Floral organ abscission | Qi et al. 2012 | Sorghum panicle development |
| ERF | SbERF080, SbERF094 | Orthologs | Early to late | Ethylene mediated root to shoot salt signaling | Schmidt et al. 2014 | Increased osmotic adjustment and water absorption | |
| SbDREB2A | Orthologs | Early to late | Leaves | ABA-mediated transcriptional regulation of drought responsive elements | Herath (2016) | Shoot growth | |
| BES1 | SbBES1-4 and SbBES1-9 | Orthologs | Early to late | Roots | Work synergistically with BHLH family members under salt stress | Jia et al. 2021 | Work synergistically to positively regulate BR signaling and salt stress tolerance |
| MCU | SbMCU5.2 | Orthologs | Early to late | Root | Activation of mitogen-activated protein kinases (MAPK) | Teardo et al. 2019 | Intracellular Ca2+ signal transduction and cationic homeostasis |
| MAPKs | SbMAPK13 | Orthologs | Early to late | Root | Late ABA-mediated salt response | Yu et al. 2011 | Stress-stimulus-specific Ca2+ dynamics in the chloroplast |
| Aquaporins | SbTIP2;1 | Transgenics | Early to late | Shoot | Regulating the water and oxidative status | Martins et al. 2017 | Increases in relative water content |
| SbPIP1;2 | Transgenic | Early to late | Leaves | Codes for plasma membrane intrinsic proteins | Increased root and leaf water | ||
| SbPIP2.8, | Transgenic | Early to late | Root | Improves root permeability to water | Sun et al. 2017 | Increasing the ability to retain water | |
| Trihelix | SbTH02 | Orthologs | Flowering | Flower | Stamen development | Shabalina et al. 2010, Frerichs et al. 2016 | Leaf inflorescence development |
| SbTH15 | Orthologs | Flowering | Flower | Salt-induced floral differentiation | Xi et al. 2012 | Flower development | |
| Nuclear factor Y | SbNF-YBs, SbNF-YB11 | Orthologs | Flowering | Regulates photoperiodic flowering | Wei et al. 2010 | Flower development | |
| SWEET | SbSWEET8-1 | Orthologs | Soft dough | Shoot | Bidirectional sugar transporters | Eom et al. 2015, Chen et al. 2012 | Phloem loading and sugar partitioning |
| SbSWEET9-3 | Orthologs | Flowering | Panicle | plasma membrane integrity | Guan et al. 2008 | Source-sink (panicle) sugar transportation | |
| SbSWEET2-1, SbSWEET7-1 | Orthologs | Soft dough | Grain | Sucrose release from maternal tissue to the maternal-filial interface | Ma et al. 2017 | Source-sink (seed) sugar transportation |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fan, S.; Chen, J.; Yang, R. Candidate Genes for Salt Tolerance in Forage Sorghum under Saline Conditions from Germination to Harvest Maturity. Genes 2023, 14, 293. https://doi.org/10.3390/genes14020293
Fan S, Chen J, Yang R. Candidate Genes for Salt Tolerance in Forage Sorghum under Saline Conditions from Germination to Harvest Maturity. Genes. 2023; 14(2):293. https://doi.org/10.3390/genes14020293
Chicago/Turabian StyleFan, Shugao, Jianmin Chen, and Rongzhen Yang. 2023. "Candidate Genes for Salt Tolerance in Forage Sorghum under Saline Conditions from Germination to Harvest Maturity" Genes 14, no. 2: 293. https://doi.org/10.3390/genes14020293
APA StyleFan, S., Chen, J., & Yang, R. (2023). Candidate Genes for Salt Tolerance in Forage Sorghum under Saline Conditions from Germination to Harvest Maturity. Genes, 14(2), 293. https://doi.org/10.3390/genes14020293





