Abstract
The vertebrate photoperiodic neuroendocrine system uses the photoperiod as a proxy to time the annual rhythms in reproduction. The thyrotropin receptor (TSHR) is a key protein in the mammalian seasonal reproduction pathway. Its abundance and function can tune sensitivity to the photoperiod. To investigate seasonal adaptation in mammals, the hinge region and the first part of the transmembrane domain of the Tshr gene were sequenced for 278 common vole (Microtus arvalis) specimens from 15 localities in Western Europe and 28 localities in Eastern Europe. Forty-nine single nucleotide polymorphisms (SNPs; twenty-two intronic and twenty-seven exonic) were found, with a weak or lack of correlation with pairwise geographical distance, latitude, longitude, and altitude. By applying a temperature threshold to the local photoperiod–temperature ellipsoid, we obtained a predicted critical photoperiod (pCPP) as a proxy for the spring onset of local primary food production (grass). The obtained pCPP explains the distribution of the genetic variation in Tshr in Western Europe through highly significant correlations with five intronic and seven exonic SNPs. The relationship between pCPP and SNPs was lacking in Eastern Europe. Thus, Tshr, which plays a pivotal role in the sensitivity of the mammalian photoperiodic neuroendocrine system, was targeted by natural selection in Western European vole populations, resulting in the optimized timing of seasonal reproduction.
1. Introduction
Herbivores in mid to high latitudes are dependent on seasonal food availability and use the photoperiod (i.e., day length) to synchronize their reproduction with primary food production [1]. Primary production in the food web is temperature-dependent [2,3,4]. Since temperature is a notoriously noisy environmental signal, herbivores use the photoperiod as a proxy to adjust the seasonal timing of reproduction [1]. The annual relationship between the photoperiod and ambient temperature is primarily dependent on latitude [5] but also changes with altitude and longitude, depending on local climatic conditions. As a consequence, for the Northern Hemisphere, primary food production and the biological spring generally start later in the year at higher latitudes, coinciding with the longer photoperiods than at low latitudes. Therefore, selection pressure on the timing of reproduction has caused the adaptive evolution of the seasonal timing mechanisms in herbivores, such that reproduction in spring starts with the longer photoperiods in the more northern populations in the Northern Hemisphere.
Latitudinal clines in annual timing were described in many insect species and in some bird and plant species but rarely in mammals [5]. For the photoperiodic induction of diapause in insects, the critical photoperiod (CPP) increases with latitude [5,6,7,8,9,10]. Furthermore, bird species change their annual breeding frequency patterns at different latitudes, with the peak shifting to a later time point in the year at higher latitudes [1]. These findings suggest that there is a latitudinal adaptation of the photoperiodic timing mechanisms driving reproduction. A genetic basis for variation in photoperiodic responsiveness was demonstrated in the parasitoid Nasonia vitripennis, where it is associated with the clinal allelic variation of the period gene [11,12], in pitcher-plant mosquitoes Wyeomyia smithii [13,14,15], in deer mice Peromyscus leucopus [16] and Peromyscus maniculatus [17], and in Siberian hamsters Phodopus sungorus [18,19]. Although the latitude of origin influences the photoperiodic responses in deer mice [20], the underlying genetics for the adaptation of such photoperiodic mechanisms in mammals is not clear.
Laboratory experiments revealed that annual rhythms in physiology and reproduction are driven by the photoperiodic neuroendocrine system [21]. This mechanism is well-conserved among vertebrates, including the common vole, Microtus arvalis (Pallas 1778) [22,23,24,25]. The distribution of Microtus ranges from close to the equator to arctic regions (14° N–72° N) in North America, Europe, and northern Asia, which makes it an excellent genus to study the latitudinal adaptation of the photoperiodic system. Perhaps the primary logical reason why small terrestrial mammals should be ideal for using latitudinal clines as a method to study the evolution of photoperiodic adaptation is because they do not show (large-scale) migration like marine mammals, birds, and some insects do.
The photoperiodic neuroendocrine system comprises a seasonal timing mechanism that synchronizes to changes in the photoperiod using the central circadian clock (i.e., the suprachiasmatic nucleus; SCN). The photoperiod is inversely related to the duration of the nocturnal melatonin release by the pineal gland [26,27,28]. Under a short photoperiod, pineal melatonin is present in the morning hours (12h after lights off) and binds to melatonin receptors in the pars tuberalis of the pituitary, causing suppression of the thyroid-stimulating hormone β-subunit (TSHβ) [29,30]. Under a long photoperiod, melatonin is absent in the morning (12h after lights off), which allows the transcriptional coactivator Eyes absent 3 (EYA3) and, subsequently, TSHβ to increase. The TSHβ forms an active heterodimer with the glycoprotein hormone α-subunit (αGSU) [31], which then binds as TSH to its receptor, the TSHR, in the tanycytes, a key population of cells involved in the neuroendocrine control of reproduction, around the third ventricle of the brain, where it increases the production of iodothyronine deiodinase 2 (DIO2) [32,33,34,35,36]. This leads to increased active thyroid hormone levels (T3) [37], which indirectly permit gonadotropin-releasing hormone (GnRH) release into the hypophysial portal system to control reproductive behavior [33,34,35].
TSH-dependent sensitivity to the photoperiod, which is defined as a shift of the photoperiodic response curves, can be modulated by TSHR abundance and function. The TSHR is, therefore, an essential protein in the mammalian seasonal reproduction pathway. In addition, Ho and colleagues showed that Tshr mutations can change the signaling efficiency of the receptor [38]. Selection on the Tshr gene, and closely located regions, has been shown in the domestic chicken (Gallus gallus domesticus) [39] and sheep (Ovis aries), respectively [40,41], which suggests that the transmembrane domain is especially important in modulating the photoperiodic control of reproduction [39,42]. Therefore, to assess the seasonal adaptation of the photoperiodic mechanisms, we focus on Tshr.
The protein encoded by the Tshr gene belongs to the glycoprotein hormone receptor family [43]. The TSHR is a seven-transmembrane domain (TMD) G protein-coupled receptor (GPCR) with a large extracellular N-terminal part, containing leucine-rich repeats (LRRs). LRRs form a hormone-binding pocket and are responsible for TSH recognition and binding [44]. TSH binding causes a conformational change in the TSHR, which activates G-protein-dependent signaling transduction. The extracellular domain containing LRRs is connected to a large transmembrane helix, the hinge region.
The Tshr gene is very large, owing to its large introns. This offers many possibilities for cis-regulatory elements that may modulate transcription [45]. Mutations in intronic regions may disrupt transcription factor binding, which may lead to altered Tshr expression. Human TSHR mRNA splice variants encoding a TSHR without TMD have been reported [46], which may hint at alternative splice sites within intronic regions prior to the exon encoding for the TMD. Since the hinge region and the first TMD are important for ligand binding and for signaling transduction [47], and mutations in the hinge region of the TSHR are known to change the signaling efficiency of the receptor [38], we consider this region as a potential target for natural selection and functional adaptive variation.
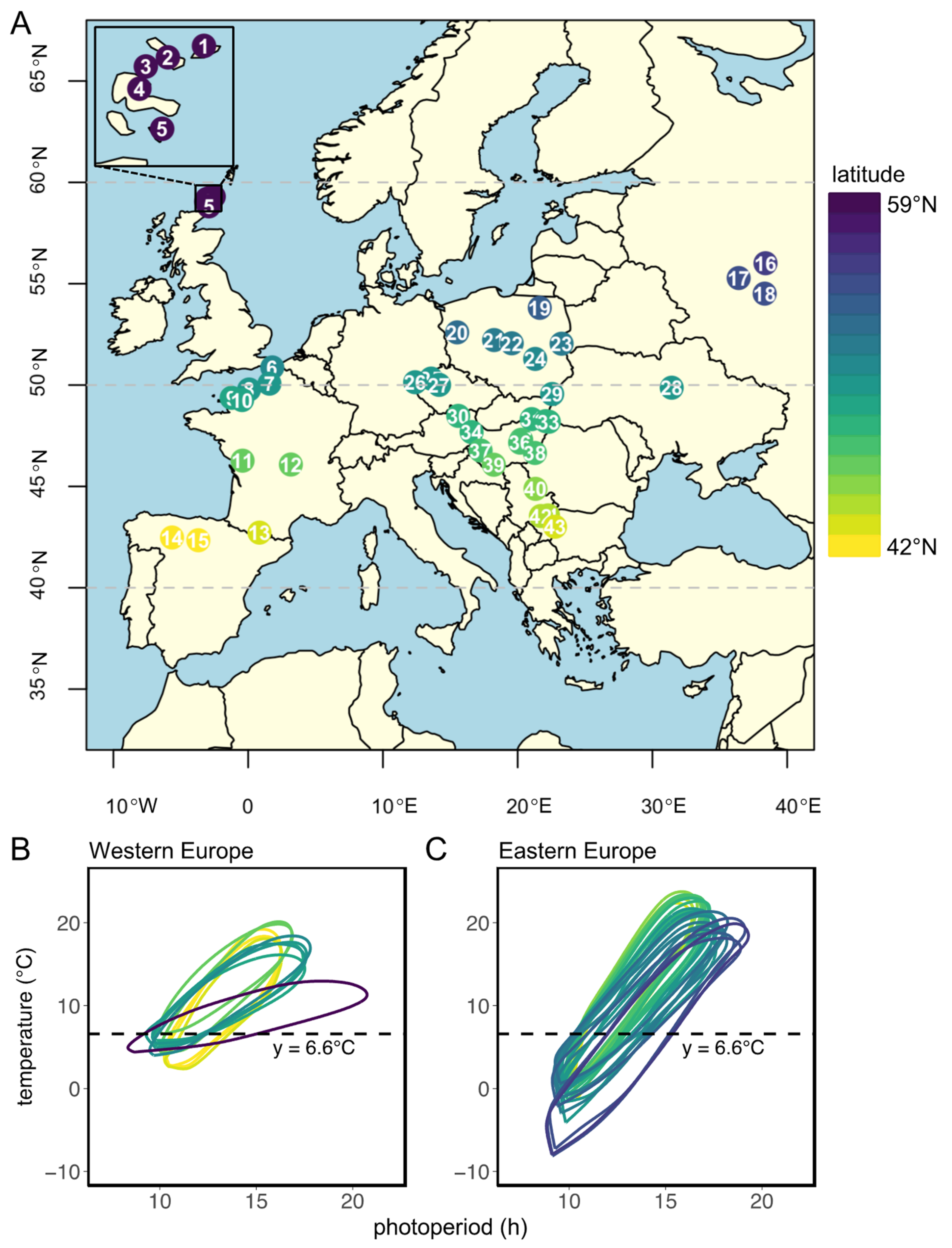
In this study, we evaluate the genetic adaptation to local climate conditions in the common vole Tshr gene by comparing genetic variance over the large European geographical range of this herbivorous rodent. For this purpose, we focus on the end of intron 8 and the beginning of exon 9, encoding for the hinge region and the first part of the transmembrane domain of the TSHR. Since the reproductive response at high latitudes in the Northern Hemisphere requires longer photoperiods, and, therefore, higher TSH levels, the allelic variation of the Tshr gene may be associated with reduced TSHR signaling at high latitudes. Tissue samples were collected from 43 different localities over a large European geographical distribution (Figure 1A, Table S1). The large variations in latitude (42°21′36″ N–59°17′60″ N), longitude (5°31′48″ W–38°23′24″ E), and altitude (4–2146 m above sea level) allowed us to assess whether location-specific annual photoperiod–temperature ellipsoid patterns can explain the distribution of the genetic variation in the coding and non-coding parts of Tshr.
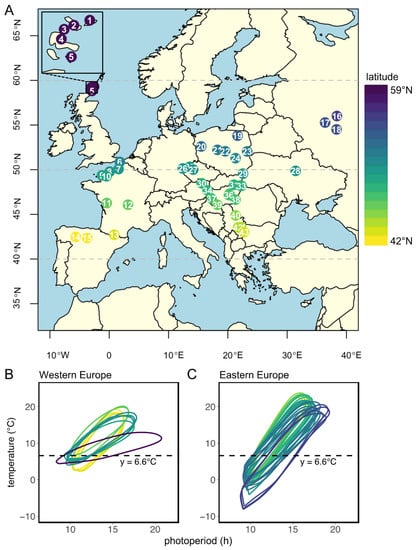
Figure 1.
Sample locations and local annual photoperiod–temperature ellipsoids. (A) Distribution map of localities of common vole samples used in this study. Location numbers with corresponding environmental proxies are listed in Table S1. (B) Annual patterns (counter-clockwise) of photoperiod and 10-year average monthly ambient temperature for each sample location in Western Europe and (C) in Eastern Europe. Temperature data were obtained from the closest weather station (always within 110 km of sample location) obtained from Wunderground (https://www.wunderground.com/ (accessed on 7 December 2022)). Photoperiod was obtained from https://www.timeanddate.com (accessed on 7 December 2022) and is based on civil twilight times at dawn and dusk, which is the timing at which log light intensities change most rapidly [5,48]. Civil twilight incorporates geographical and seasonal variation in the duration of twilight [49] and is, therefore, considered as the moment of ‘lights on’ and ‘lights off’ for biological systems [5]. Dotted lines indicate a temperature threshold at 6.6 °C, from which the corresponding predicted critical photoperiod (pCPP) in spring (ellipse crossing the 6.6 °C line for the second time) is used as a proxy for onset of grass growth and, consequently, as a proxy for onset of the favorable season. Regional pCPPs are listed in Table S1. Colors indicate latitude, ranging from 42° N (yellow) to 59° N (purple).
2. Materials and Methods
2.1. Tissue Samples
We obtained tissue samples from 278 previously collected specimens of female and male common voles from both Western Europe (France, Great Britain, and Spain) and Eastern Europe (Czech Republic, Hungary, Poland, Russia, Serbia, and Ukraine) [50,51] (Figure 1A, Table S1). Specimens were collected between 1995 and 2015 (voles from the same site were caught simultaneously) and consisted of appendages (legs, toes, ears, tail tips, and muscle tissues) preserved in 96–99% ethanol at 4–15 °C. Sex was not documented for all specimens; therefore, we could not account for sex in the model.
2.2. DNA Isolation, PCR and Sanger Sequencing
Total genomic DNA was isolated using the DNeasy Blood and Tissue Kit (Qiagen, Hilden, Germany) for Western European samples and using the Syngen Tissue DNA Mini Kit (Syngen Biotech, Wrocław, Poland) for Eastern European samples, in accordance with the protocols of the manufacturer. Primers (Table S2) were designed based on the common vole genome (NCBI:txid47230, GCA_007455615.1) using Primer-BLAST (NCBI), in two overlapping fragments, each 1100-1200 bp in length. Using those primers, the last ~829 bp of intron 8 and the first ~849 bp of exon 9 of the Tshr gene were amplified by PCR using DreamTaq (ThermoscientificTM, Waltham, MA, USA). A mastermix containing 15.7 μL ultrapure H2O, 2 μL Dreamtaq Buffer (10×), 0.4 μL dNTP mix (10 mM), 0.4 μL Forward primer (10 μM), 0.4 μL Reverse primer (10 μM), and 0.1 μL DreamTaq DNA polymerase (5 U/μL) was prepared for each reaction. Twenty μL-reactions (1 μL DNA + 19 μL mastermix) were carried out for each sample by using a thermalcycler (S1000TM, Bio-Rad, Hercules, CA, USA) (Table S3). Following PCR, an enzymatic clean-up with ExoSAP-IT reagent (Applied BiosystemsTM, Foster City, CA, USA) was performed in order to remove excess primers and nucleotides. Then, 5 μL of cleaned PCR product and 5 μL of the forward or reverse primer (5 μM) were transferred to a new 1.5 mL tube and sent out for Sanger sequencing (Eurofins Genomics, Ebersberg, Germany). The intronic fragment was sequenced in two directions, while the exonic fragment was sequenced only in the forward direction.
2.3. Data Analysis and Statistical Analysis
The distribution map of common vole samples used in this study (Figure 1A) was generated using the following R packages: ‘rworldmap’ [52], ‘rworldxtra’ [53], ‘RcolorBrewer’ [54], ‘maptools’ [55], and ‘classInt’ [56]. Ellipse-like annual relationships between temperature and photoperiod (Figure 1B,C) were built using ~10-year (between 2000 and 2019) average monthly ambient temperatures obtained from local weather stations (within 110 km of sample location) at http://www.wunderground.com (accessed on 7 December 2022). Photoperiods, based on civil twilight times at dawn and dusk at different locations, were retrieved from https://www.timeanddate.com/ (accessed on 7 December 2022). Grass growth in spring is used as a proxy for the onset of the favorable reproductive season. Grass growth is initiated at 5–10 °C air temperature [3,57,58]. To include all locations in our analysis, a temperature threshold at 6.6 °C was used to deduce, for further analysis, the corresponding predicted critical photoperiod (pCPP) that would initiate optimal timing of reproduction.
The ‘Phyre2’ web portal for protein modeling was used to predict the TSHR protein 3D structure (Figure 2D) [59]. SNPs were detected by sequence alignments using ‘CLC Sequence Viewer’ (version 8.0) (QIAGEN, Aarhus, Denmark). Chromatograms were checked for sequencing quality and heterozygosity of SNPs in the Mac OS software ‘4-peaks’ (Nucleobytes, Aalsmeer, The Netherlands). Variation in DNA sequences were classified as SNPs if >3 of the specimens contained the mutation. Putative transcription factor bindings sites were predicted using AliBaba2 [60]. To statistically test gene–environment associations, we used a population-based approach, in which an environmental variable was modeled as a linear function of population allele frequency [61]. Pearson’s correlation tests were carried out: pairwise distances of allele frequencies correlated with pairwise geographical distance, pairwise latitudinal difference, pairwise longitudinal difference, pairwise altitudinal difference, and pairwise critical photoperiod difference. p-values were adjusted according to the Benjamini–Hochberg procedure [62,63], which is one of the strongly recommended methods to use in environmental association analysis [61]. Pairwise linkage disequilibrium heatmaps (Figure S3) were generated using the R-package ‘LDheatmap’ [64]. The constructed phylogenetic tree (Figure S2) from SNP frequency data by using the neighbor-joining method [65] was generated using ‘POPTREEW’ [66]. All other analyses were performed using ‘RStudio’ (version 1.2.1335), and figures were generated using the R-package ‘ggplot2’ [67].
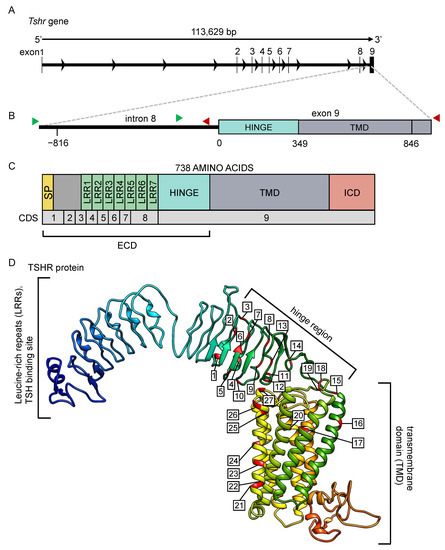
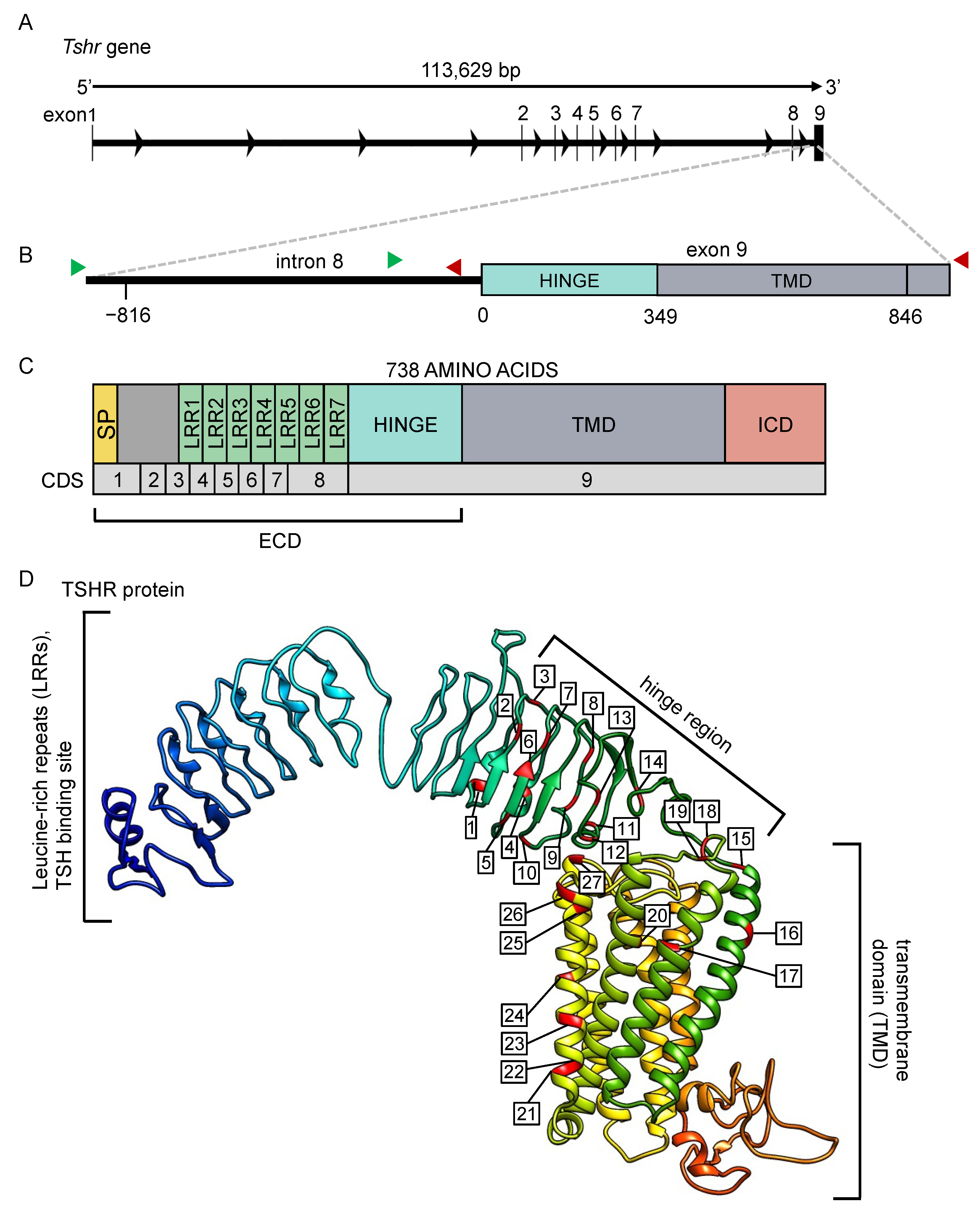
Figure 2.
The Tshr gene and predicted protein for the common vole. (A) The Tshr gene for the common vole, consisting of 8 introns and 9 exons. (B) The magnified region, including the last part of intron 8 and the first part of exon 9, were sequenced in this study. Green arrows indicate location of forward primers, and red arrows indicate location of reverse primers for sequencing. (C) The predicted TSHR protein and (D) its 3D structure. All mutations found in the current study are labeled and listed in Tables S4–S7. SP = signal peptide, LRR = leucine-rich repeat, TMD = transmembrane domain, ICD = intracellular domain, ECD = extracellular domain, CDS = coding sequence.
3. Results
The Tshr gene of the common vole is 113,629 bp long and consists of eight introns and nine exons (Figure 2A). In this study, we sequenced a ~1700 bp region around the beginning of exon 9, comprising 829 bp of intronic and 849 bp of exonic sequences (Figure 2B). The predicted TSHR protein structure, based on the M. arvalis genome (NCBI: txid47230, GCA_007455615.1), comprises seven leucine-rich repeats, a hinge region, and seven transmembrane domains (Figure 2C,D).
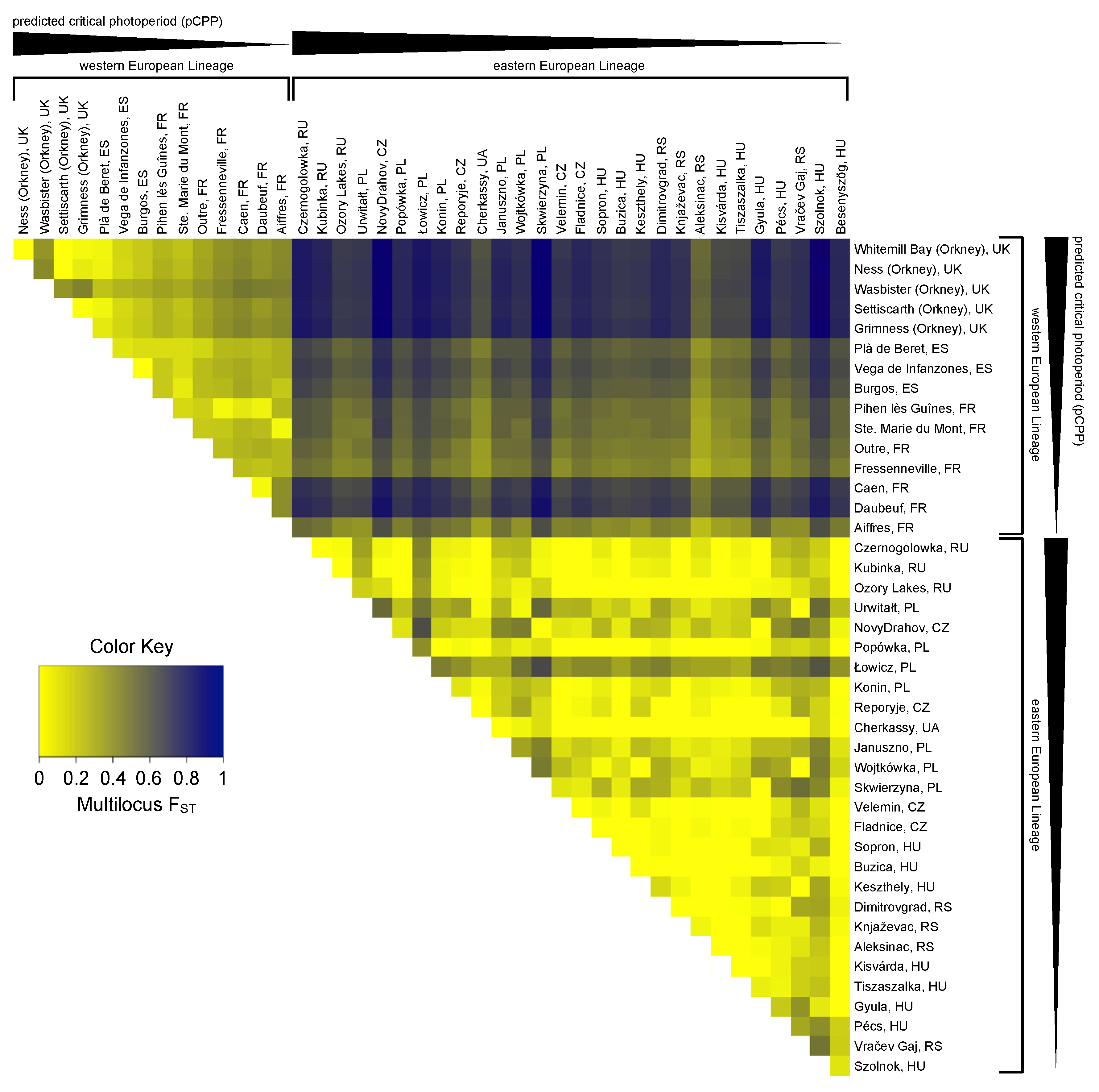
Forty-nine single nucleotide polymorphisms (SNPs) were detected (Tables S4 and S5). Twenty-two intronic SNPs and twenty-seven exonic SNPs were found, from which twenty-three were synonymous. These SNPs were used to calculate the genetic differentiation of the sampled populations. The pairwise multilocus fixation index (FST) estimates ranged from FST = 0.000 to 0.978 (mean FST = 0.366; with 0 denoting no difference and 1 referring to completely different populations) and revealed a high genetic differentiation between the sampled populations (Figure 3). The high FST values in this species are in agreement with previous studies [68,69]. The structure of the observed population differentiation suggests that not only population subdivision but also natural selection may be an explanatory factor. Therefore, it was tested whether the observed SNPs are associated with geographical location. The genetic differentiation between the Eastern and Western European populations was larger (FST = 0.247 to 0.978; mean: 0.640) than the differentiation among Western European populations (FST = 0.016 to 0.467; mean: 0.234) and among Eastern European populations (FST = 0.000 to 0.648; mean: 0.132), and strongly depended on geographical distance and longitude (Figure 3 and Figure S2, Table S5). The constructed distance tree based on Tshr haplotypes (Figure S2), together with pairwise multilocus FST analysis (Figure 3), confirmed that the Western and Eastern European populations belong to different genetic lineages. Given the genetic separation of these lineages, it is appropriate to analyze Western and Eastern European samples separately. To reveal possible patterns of association between SNPs, heatmaps of the pairwise linkage disequilibrium (LD) measurements were generated (Figure S3).
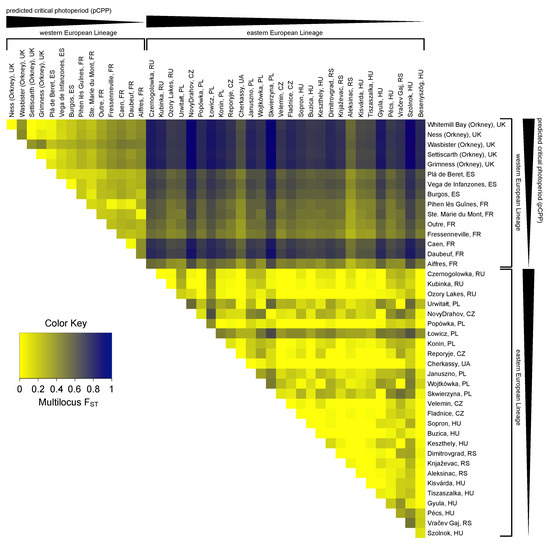
Figure 3.
Pairwise multilocus FST heatmap for Tshr haplotypes. FST values were calculated using the original method for estimation of genetic distance with correction for sample size bias [70]. Colors indicate pairwise multilocus FST values ranging from 0 (yellow) to 1 (dark blue). Western and Eastern European populations are ordered from long to short predicted critical photoperiods (pCPPs).
3.1. Western European Lineage
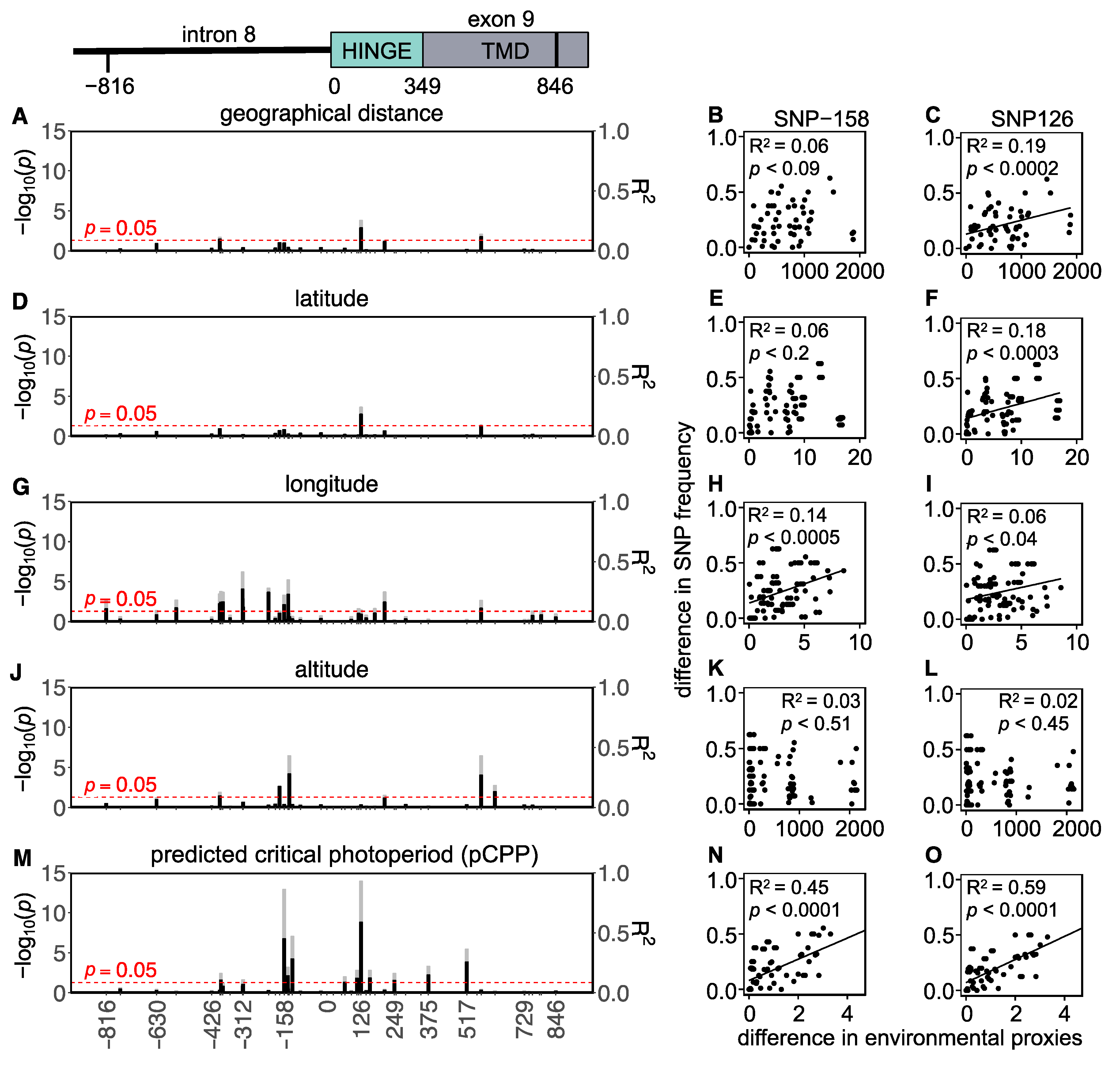
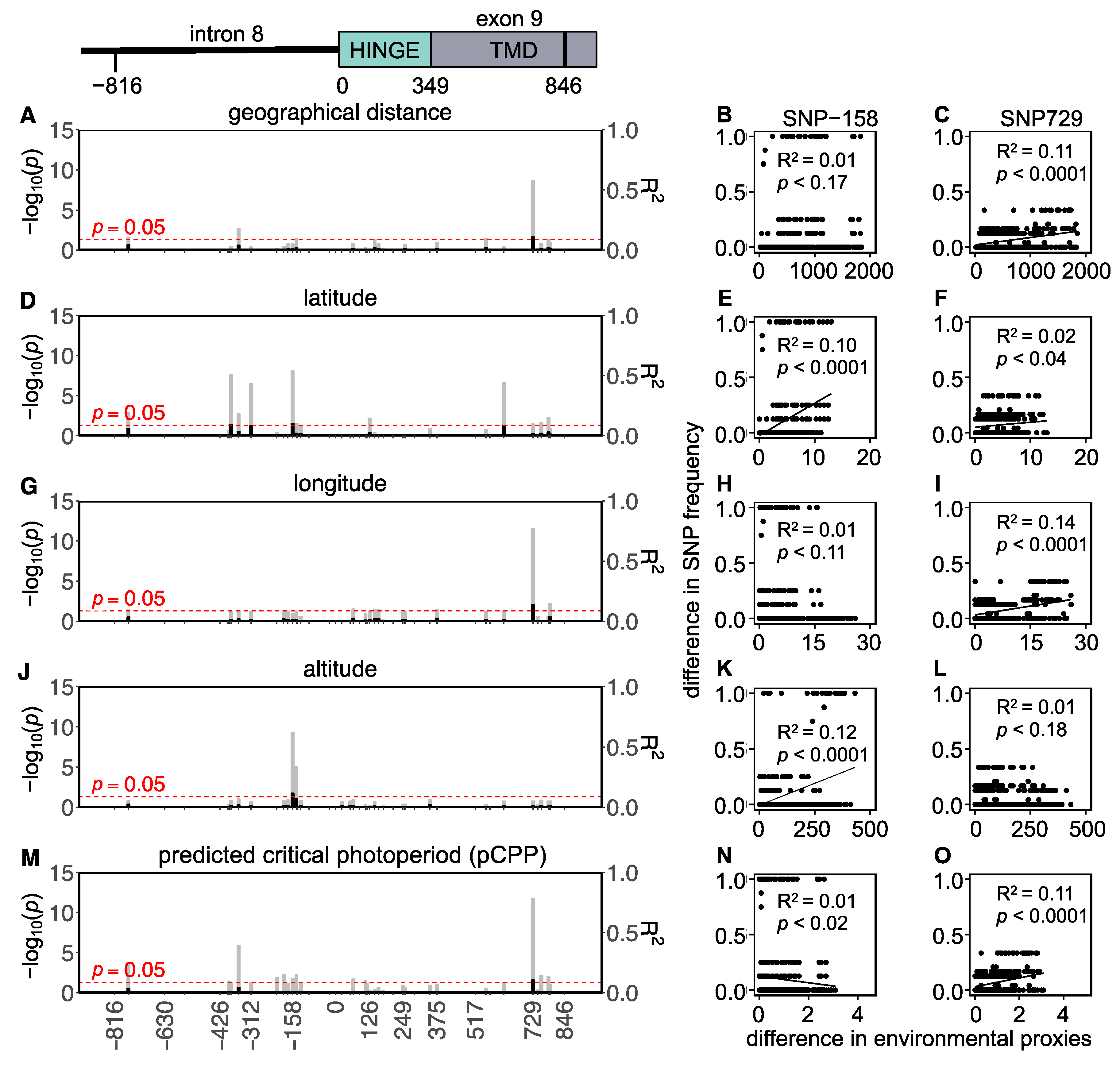
In total, 43 SNPs (22 intronic and 21 exonic) were found in the Western European populations, of which 5 were non-synonymous and 16 were synonymous (Figure 4 and Table S6). To assess whether the variation in the Tshr gene can be explained by local seasons, SNP frequencies correlated with different environmental proxies. The non-synonymous SNPs were not associated with the tested environmental proxies (Table S6). In the Western European lineage, 3/43 SNPs (exonic, synonymous) significantly correlated with pairwise geographical distance (Figure 4A,C, Table S6), indicating that there was a lack of regional equilibrium [71] and that an alternative approach may be used to detect selection, by classifying SNPs that show clinal variation [72]. SNP frequency weakly correlated with pairwise latitudinal, longitudinal, and altitudinal difference for the majority of the observed SNPs (latitude: 2 exonic SNPs; longitude: 11 intronic and 5 exonic SNPs; altitude: 3 intronic and 3 exonic SNP) (Figure 4D–L, Table S6). These findings show that geographical distance, latitude, and altitude by themselves are bad predictors for genetic variation in the Tshr gene, despite the fact that the annual photoperiod–food abundance patterns depend on all these parameters. Therefore, for each sample location, the pCPP at which grass growth is initiated in spring (at 5–10 °C ambient temperature [3,57,58]) was deduced from the local annual photoperiod–ambient temperature ellipsoids. The pCPP at 6.6 °C included ellipsoids from all sample locations. Therefore, the temperature threshold for grass-growth initiation in spring was set at 6.6 °C, and was used to deduce the corresponding pCPP, which was calculated to vary between 10.19 and 15.40 h of light/24 h (Figure 1 and Table S1). In the Western European samples, five intronic and seven exonic SNPs strongly correlated with the pairwise difference in pCPP (Figure 4M, Table S6). The FST values for these specific SNPs were high (ranging from FST = 0.032 to 0.310; mean: 0.166). All these significant mutations were, however, synonymous SNPs. The strongest associations with pCPP were found for intronic SNP-158 (G > C), −128 (T > C) and exonic SNP126 (A > G) (Figure 4M–O, Table S6). It is expected that between Orkney Island and the mainland, some of the variation reflects isolation and genetic drift. Therefore, the same analysis was performed excluding the Orkney Island populations, which revealed similar results. The pairwise multilocus FST-values were high for populations that highly differed in pCPP, while the FST-values were low for populations with a similar pCPP (Figure 3).
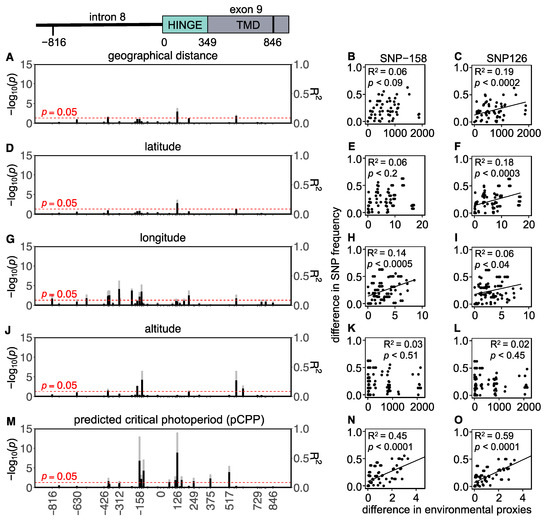
Figure 4.
Tshr mutations in Western European common vole populations. Manhattan-type plots (−log10(p)) for the sequenced Tshr region for (A) geographical distance, (D) latitude, (G) longitude, (J) altitude, and (M) predicted critical photoperiod (pCPP). Gray bars indicate Benjamini–Hochberg adjusted p-values, and black bars indicate R2-values. SNPs that meet the threshold for significant correlations (p < 0.05) cross the red dashed line. Pairwise differences in SNP frequency for two representative mutations (SNP-158 and SNP126) related to (B,C) pairwise geographical distance, (E,F) pairwise latitudinal difference, (H,I) pairwise longitudinal difference, (K,L) pairwise altitudinal difference, and (N,O) pairwise difference in pCPP. Significant correlations are indicated by linear regression lines. All statistic results of linear models for SNP frequency related to environmental proxies are found in Table S6.
3.2. Eastern European Lineage
In total, 34 SNPs (14 intronic and 20 exonic) were found in the Eastern European populations, of which 3 were non-synonymous and 17 were synonymous (Figure 5 and Table S7). Although highly significant p-values for some correlations between the Tshr SNP frequencies and environmental proxies were found in the Eastern European lineage, the R2-values were extremely low (0.1 ± 0.02; mean ± SD). This indicates that in Eastern European voles, the Tshr SNP frequencies weakly correlate with geographical distance, latitude, longitude, altitude, and pCPP (Figure 5, Table S7). This observation indicates that genetic Tshr variation in Eastern Europe is unlikely to be explained by natural selection due to seasonal variance.
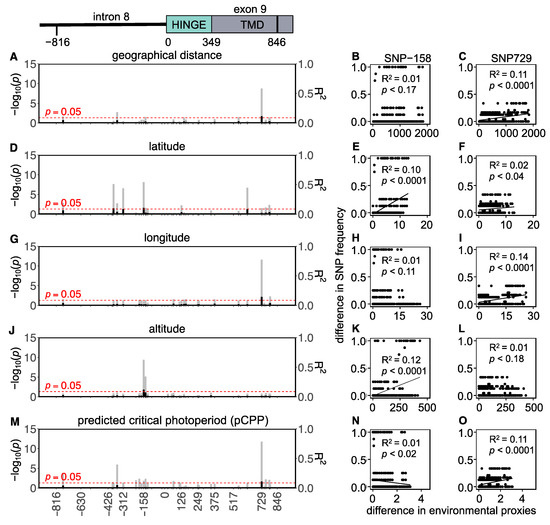
Figure 5.
Tshr mutations in Eastern European common vole populations. Manhattan-type plots (−log10(p)) for the sequenced Tshr region for (A) geographical distance, (D) latitude, (G) longitude, (J) altitude, and (M) predicted critical photoperiod (pCPP). Gray bars indicate Benjamini–Hochberg adjusted p-values, and black bars indicate R2-values. SNPs that meet the threshold for significant correlations (p < 0.05) cross the red dashed line. Pairwise differences in SNP frequency for two representative mutations (SNP-158 and SNP729) related to (B,C) pairwise geographical distance, (E,F) pairwise latitudinal difference, (H,I) pairwise longitudinal difference, (K,L) pairwise altitudinal difference, and (N,O) pairwise difference in pCPP. Significant correlations are indicated by linear regression lines. All statistic results of linear models for SNP frequency related to environmental proxies are found in Table S7.
4. Discussion
Common vole populations are characterized by large-scale genetic differentiation of Tshr, reflecting local adaptation to annual temperature–photoperiod patterns, rather than latitude per se. Variation in Tshr sequence indicates that the M. arvalis population can be subdivided into Eastern and Western European clusters, indicating that they may belong to distinct genetic lineages (Figure 3 and Figure S3). This phylogeographical structure is consistent with those found for mitochondrial cytochrome b gene sequences and microsatellite loci (representing nuclear DNA) [50,51,68,73,74,75,76], which justifies analyzing the Western and Eastern European populations separately. The Western versus Eastern European divide could well be due to the reinvasion of Northern Europe from separate glacial refugia, and, therefore, separate evolutionary events [77,78].
For insights into the geographical variation found in Tshr, the association of the SNP frequencies with the local climatic conditions was examined. Here, we showed that the genetic variation in the vole Tshr is better explained by the local photoperiod–temperature patterns than by only latitude. This may be caused by the temperature dependence of vegetation growth. In house mice, genes (but not the Tshr) that show signals of selection are also associated with the local average annual ambient temperature and are linked with clinal variation in the phenotypic aspects, such as body mass and metabolism [79,80]. Interestingly, the SNPs found in the thyroid hormone receptors, which are involved in the regulation of seasonal reproduction in the hypothalamus [35], significantly correlated with variation in the average annual temperature [80]. This suggests that the genomic evolution of seasonal adaptation in house mice and voles involves unique responses to genetic selection. Annual temperature patterns not only depend on latitude but also on longitude, altitude, and other regional climatic variables such as the Gulf Stream warming European Atlantic coastal regions. Critical photoperiods in pitcher-plant mosquitoes strongly correlated with altitude-corrected latitude (r = 0.96); however, this measurement did not integrate local temperature patterns [8,9,81]. Deriving the regional photoperiod–temperature ellipsoids may better account for such regional climatic differences than only latitude- or altitude-corrected latitude. We post hoc tested photoperiods at other temperature thresholds; however, this did not improve the results. Moreover, 6.6 °C is not an unreasonable temperature, since grass growth is initiated at 5–10 °C air temperature [3,57,58].
In addition, several SNPs correlated well with longitude and altitude (Figure 4G,J). Altitudinal gradients in the seasonal timing of breeding were observed in deer mice (P. maniculatus), with shorter breeding seasons occurring at high elevations [82]. The pCPP at which a temperature threshold for grass-growth initiation is reached can be deduced from local photoperiod–temperature patterns and is confirmed, here, to be a strong determinant for the distributional variation in the Tshr SNP frequency in Western European common vole populations (Figure 4M). Pairwise multilocus FST analysis revealed that populations that differ in pCPP also show a greater genetic distance in their Tshr haplotypes (Figure 3). These findings indicate that seasonality is likely to be a selective force for Tshr evolution in common voles and imply that Tshr is an important gene for the genetic adaptation of the photoperiodic response systems. Although the aim of the study was to establish the regional adaptation and selection, we cannot exclude the possibility that gender-specific selection is part of the mechanism driving the latitudinal selection for the alleles under study.
The observed genetic Tshr variation is unlikely to be caused by only isolation, with the possible exception of the Orkney Island populations, which are geographically isolated from each other and from mainland populations by the sea. Therefore, isolation and the related genetic drift may be more important evolutionary forces than natural selection in the Orkney Island populations. Interestingly, the same SNPs appear to be related to pCPP when the Orkney Island populations are excluded from the analysis. This indicates that the results in Western Europe are not dominated by the Orkney Island population’s data and that the observed distribution of the Tshr variation may be a sign of adaptive evolution, which is likely operating in response to the photoperiod.
In the Eastern European populations, none of the tested environmental proxies are good predictors for Tshr SNP frequencies (Figure 5). These results indicate that the Tshr in the Eastern European lineage is not linked to seasonal adaptation, as observed in the Western European lineage. Oceanic climates (Western Europe) are known for their small annual temperature amplitudes, while continental climates (Eastern Europe) are known for their large annual temperature amplitudes. These climatic differences may have led to the divergent evolutionary adaptation of the TSHR function, which may provide an explanation for the observed longitudinal separation in the genetic Tshr differentiation. Furthermore, we only sequenced a small part of one gene. Therefore, we cannot claim that this is the most important aspect of the regional adaptation of the photoperiodic neuroendocrine system. Another hypothesis is that genes other than Tshr are under selection for seasonal adaptation in vole populations.
The SNPs associated with local pCPP were all synonymous or intronic mutations. This suggests that these sites may be involved in regulatory rather than structural variation. Five intronic SNPs were strongly associated with pCPP in Western Europe (Figure 4), of which two (i.e., SNP-144 and -158) were strongly associated with altitude in Eastern Europe (Figure 5). Putative regulatory protein binding sites were predicted for the intronic region and revealed that intronic SNP-128, which strongly correlates with pCPP (Figure 4O), is located in a potential SP1 (specificity protein 1) binding site [83,84]. Interestingly, SNPs closely located to this enhancer region, such as SNP-158, are related to different environmental proxies in Eastern and Western Europe (Figure 4 and Figure 5 and Figure S2). It is tempting to speculate that variation in and around this SP1 binding site sequence may influence Tshr transcription. Furthermore, there is strong evidence that synonymous SNPs are not necessarily neutral, as they can alter mRNA expression, splicing, and structure, thus having downstream effects on protein expression [85,86]. Synonymous polymorphisms require different transfer RNAs (tRNA) to recruit the same amino acids and may cause codon bias. Synonymous tRNA vary strongly in frequency between species and tissues (i.e., codon bias) [87,88]. It is, therefore, possible that the observed synonymous mutations in the TSHR may alter translation efficiency within a species and tissue by changing the elongation rate [89]. A reduced elongation rate may, therefore, result in a lower protein abundance. Hence, synonymous SNPs in the Tshr gene could result in an altered receptor abundance, a changed sensitivity to TSH, and a modified photoperiodic response. It is, therefore, conceivable that synonymous SNPs in the Tshr gene are subject to natural selection and reflect local geographical adaptation. TSHR plays a pivotal role in photoperiodic response not only in the pars tuberalis [90] but also in the thyroid hormone metabolism in the thyroid gland [91]. The duration of the melatonin signal encodes daylength and acts on the melatonin receptors in the pars tuberalis, causing the suppression of TSHβ release [90]. Interestingly, the TSH produced by the pars tuberalis (acting on the hypothalamus to regulate seasonality) and the TSH produced by the pars distalis (stimulating the thyroid gland to produce thyroid hormones) prevent functional crosstalk by tissue-specific glycosylation [91]. The tissue-specific functions of TSHR may benefit from the genetic adaptation in photoperiodism through synonymous SNPs, since tissue-specific tRNA expression, which has been demonstrated in human and mouse tissues [88,92], may perhaps lead to altered TSHR function in the pars tuberalis but not in the thyroid gland.
The photoperiodic regulation of the reproductive system in deer mice was shown to vary with latitude, with weaker photoperiodic responses in animals originating from lower latitudes [20]. Moreover, photoperiodic sensitivity in pitcher-plant mosquitoes correlated with global warming, indicating the importance of the season-length driving evolution (genetic change) of photoperiodism during the recent, rapid climate change [93,94,95,96]. Our findings confirm that the Tshr gene is under selection, which was previously reported in chicken domestication in relation to photoperiodic responsiveness [39,42]. Future studies should determine whether the SNPs identified as a seasonal-timing-dependent genetic variation in the vole Tshr can indeed alter genetically based photoperiodic responses. Such an approach should confirm whether habitat-specific photoperiodic responses are indeed regulated by means of functional TSHR adaptation. In vole populations with later onsets of reproduction and shorter breeding seasons [97], our results predict lower concentrations in the tanycytes of Tshr or the lower TSH-binding affinities of Tshr haplotypes.
Optimal timing of reproduction, enhancing energetically demanding pregnancy, and parental care are necessary to maximize fitness in temperate and northern seasonal environments. Tshr is an essential gene in the pathway programming seasonal reproduction in mammals. Herein, we show how the onset of the favorable season, over a wide geographical range of the common vole, M. arvalis, explains much of the genetic variation in the TSH binding site, hinge region, and transmembrane domain of Tshr in Western Europe but not Eastern Europe. Yet vole populations thrive in both regions. We, therefore, conclude that different genetic mechanisms have been important in enabling vole populations to exploit geographically distinct regions. Such distinctions, regarding how the genetic underpinnings of seasonal timing have evolved over climatic gradients in nature, are important in predicting how animals will adapt to new seasonal environments during the ongoing, rapid climate change [98].
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14020292/s1. Figure S1: Tshr mutations in Western and Eastern European common vole populations; Figure S2: A distance-based phylogenetic tree for the sequenced Tshr region, inferred with the neighbor-joining method; Figure S3: Pairwise linkage disequilibrium heatmaps; Table S1: List of locations where M. arvalis specimens were obtained; Table S2: Primer sequences; Table S3: Thermal cycling conditions for PCR; Table S4: Nucleotide and predicted amino acid sequences; Table S5: Statistical output for linear models relating pairwise distances of allele frequencies to pairwise differences in environmental proxies (geographical distance, latitude, longitude, altitude, and pCPP) in Eastern and Western Europe; Table S6: Statistical output for linear models relating pairwise distances of allele frequencies to pairwise differences in environmental proxies (geographical distance, latitude, longitude, altitude, and pCPP) in Western Europe; Table S7: Statistical output for linear models relating pairwise distances of allele frequencies to pairwise differences in environmental proxies (geographical distance, latitude, longitude, altitude, and pCPP) in Eastern Europe.
Author Contributions
L.v.R., L.v.d.Z. and R.A.H. conceived and designed the experiments. J.S. and J.M.W. provided the Eastern European samples. J.S.H. and J.B.S. provided the Western European samples. L.v.R., R.S., W.H. and A.S.P.-P. conducted the experiments. L.v.R. analyzed and visualized the data. L.v.R. wrote the manuscript. L.v.d.Z., J.B.S. and R.A.H. revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Ecology Fund of the Royal Netherlands Academy of Arts and Sciences (KNAW Fonds Ecologie). This research was supported by an Adaptive Life Programme grant awarded to R.A.H. and L.v.d.Z. made possible by the Board of the University of Groningen, the Faculty of Science and Engineering and the Groningen Institute for Evolutionary Life Sciences (GELIFES). The sample collection was financed by the National Science Centre in Poland (UMO-2013/09/N/NZ8/03205 to J.S. and N N304 058340 to J.M.W.).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The raw sequence reads and metadata will be deposited in a public repository (FigShare, https://doi.org/10.6084/m9.figshare.14695356.v1 (accessed on 7 December 2022)) after acceptance.
Acknowledgments
We would like to thank B. Kryštufek, J. Purger, N. Bulatova, and J. Uhlíková for their help in collecting samples. We thank W.E. Bradshaw, C.M. Holzapfel, D.G. Hazlerigg, and the several anonymous reviewers for their critical and valuable comments on the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Baker, J. The Evolution of Breeding Seasons. In Evolution, Essays on Aspects of Ev. Biology; Oxford University Press: New York, NY, USA, 1938; pp. 161–177. [Google Scholar]
- Robson, M.J. A Comparison of British and North African Varieties of Tall Fescue (Festuca arundinacea). I. Leaf Growth during Winter and the Effects on it of Temperature and Daylength. J. Appl. Ecol. 1967, 4, 475–484. [Google Scholar] [CrossRef]
- Peacock, J.M. Temperature and Leaf Growth in Four Grass Species. J. Appl. Ecol. 1976, 13, 225–232. [Google Scholar] [CrossRef]
- Malyshev, A.V.; Henry, H.A.L.; Kreyling, J. Relative Effects of Temperature vs Photoperiod on Growth and Cold Acclimation of Northern and Southern Ecotypes of the Grass Arrhenatherum elatius. Environ. Exp. Bot. 2014, 106, 189–196. [Google Scholar] [CrossRef]
- Hut, R.A.; Paolucci, S.; Dor, R.; Kyriacou, C.P.; Daan, S. Latitudinal Clines: An Evolutionary View on Biological Rhythms. Proc. R. Soc. B Biol. Sci. 2013, 280, 20130433. [Google Scholar] [CrossRef] [PubMed]
- Paolucci, S.; van de Zande, L.; Beukeboom, L.W. Adaptive Latitudinal Cline of Photoperiodic Diapause Induction in the Parasitoid Nasonia vitripennis in Europe. J. Evol. Biol. 2013, 26, 705–718. [Google Scholar] [CrossRef]
- Saunders, D.S. Thermoperiodic Control of Diapause in an Insect: Theory of Internal Coincidence. Science 1973, 181, 358–360. [Google Scholar] [CrossRef]
- Bradshaw, W.E.; Holzapfel, C.M.; Mathias, D. Circadian Rhythmicity and Photoperiodism in the Pitcher-Plant Mosquito: Can the Seasonal Timer Evolve Independently of the Circadian Clock? Am. Nat. 2006, 167, 601–605. [Google Scholar] [CrossRef]
- Bradshaw, W.E.; Lounibos, L.P. Evolution of Dormancy and Its Photoperiodic Control in Pitcher-Plant Mosquitoes. Evolution 1977, 31, 546–567. [Google Scholar]
- Bradshaw, W.E.; Holzapfel, C.M. What Season Is It Anyway? Circadian Tracking vs. Photoperiodic Anticipation in Insects. J. Biol. Rhythm. 2010, 25, 155–165. [Google Scholar] [CrossRef]
- Paolucci, S.; Salis, L.; Vermeulen, C.J.; Beukeboom, L.W.; van de Zande, L. QTL Analysis of the Photoperiodic Response and Clinal Distribution of Period Alleles in Nasonia vitripennis. Mol. Ecol. 2016, 25, 4805–4817. [Google Scholar] [CrossRef]
- Benetta, E.D.; Beukeboom, L.W.; van de Zande, L. Adaptive Differences in Circadian Clock Gene Expression Patterns and Photoperiodic Diapause Induction in Nasonia vitripennis. Am. Nat. 2019, 193, 881–896. [Google Scholar] [CrossRef] [PubMed]
- Mathias, D.; Jacky, L.; Bradshaw, W.E.; Holzapfel, C.M. Quantitative Trait Loci Associated with Photoperiodic Response and Stage of Diapause in the Pitcher-Plant Mosquito, Wyeomyia smithii. Genetics 2007, 176, 391–402. [Google Scholar] [CrossRef] [PubMed]
- Mathias, D.; Jacky, L.; Bradshaw, W.E.; Holzapfel, C.M. Geographic and Developmental Variation in Expression of the Circadian Rhythm Gene, Timeless, in the Pitcher-Plant Mosquito, Wyeomyia smithii. J. Insect Physiol. 2005, 51, 661–667. [Google Scholar] [CrossRef]
- Bradshaw, W.E.; Holzapfel, C.M. Phenotypic Evolution and Genetic Architecture Underlying Photoperiodic Time Measurement. J. Insect Physiol. 2001, 47, 809–820. [Google Scholar] [CrossRef]
- Heideman, P.; Bronson, F. Characteristics of a Genetic Polymorphism for Reproductive Photoresponsiveness in the White-Footed Mouse (Peromyscus leucopus). Biol. Reprod. 1991, 44, 1189–1196. [Google Scholar] [CrossRef]
- Desjardines, C.; Bronson, F.H.; Blank, J.L. Genetic Selection for Reproductive Photoresponsiveness in Deer Mice. Nature 1986, 322, 172–173. [Google Scholar] [CrossRef]
- Kliman, R.M.; Lynch, R.G. Evidence for Genetic Variation in the Occurrence of the Photoresponse of the Djungarian Hamster, Phodopus sungorus. J. Biol. Rhythm. 1992, 7, 161–173. [Google Scholar] [CrossRef]
- Lynch, G.R.; Lynch, C.B.; Kliman, R.M. Genetic Analyses of Photoresponsiveness in the Djungarian Hamster, Phodopus sungorus. J. Comp. Physiol. A 1989, 164, 475–481. [Google Scholar] [CrossRef]
- Dark, J.; Johnston, P.G.; Healy, M.; Zucker, I. Latitude of Origin Influences Photoperiodic Control of Reproduction of Deer Mice (Peromyscus maniculatus). Biol. Reprod. 1983, 28, 213–220. [Google Scholar] [CrossRef]
- Nakane, Y.; Yoshimura, T. Photoperiodic Regulation of Reproduction in Vertebrates. Annu. Rev. Anim. Biosci. 2019, 7, 173–194. [Google Scholar] [CrossRef]
- Król, E.; Douglas, A.; Dardente, H.; Birnie, M.J.; van der Vinne, V.; Eijer, W.G.; Gerkema, M.P.; Hazlerigg, D.G.; Hut, R.A. Strong Pituitary and Hypothalamic Responses to Photoperiod but Not to 6-Methoxy-2-Benzoxazolinone in Female Common Voles (Microtus arvalis). Gen. Comp. Endocrinol. 2012, 179, 289–295. [Google Scholar] [CrossRef] [PubMed]
- Van Rosmalen, L.; van Dalum, J.; Hazlerigg, D.G.; Hut, R.A. Gonads or Body? Differences in Gonadal and Somatic Photoperiodic Growth Response in Two Vole Species. J. Exp. Biol. 2020, 223, jeb.230987. [Google Scholar] [CrossRef] [PubMed]
- Van Rosmalen, L.; van Dalum, J.; Appenroth, D.; Roodenrijs, R.; de Witt, L.; Hazlerigg, D.; Hut, R. Mechanisms of Temperature Modulation in Mammalian Seasonal Timing. FASEB J. 2021, 35, e21605. [Google Scholar] [CrossRef] [PubMed]
- Van Rosmalen, L.; Riedstra, B.; Beemster, N.; Dijkstra, C.; Hut, R.A. Differential Temperature Effects on Photoperiodism in Female Voles: A Possible Explanation for Declines in Vole Populations. Mol. Ecol. 2022, 31, 3360–3373. [Google Scholar] [CrossRef]
- Hoffman, R.A.; Reiter, R.J. Pineal Gland: Influence on Gonads of Male Hamsters. Science 1965, 148, 1609–1611. [Google Scholar] [CrossRef]
- Carter, D.S.; Goldman, B.D. Antigonadal Effects of Timed Melatonin Infusion in Pinealectomized Male Djungarian Hamsters (Phodopus sungorus): Duration Is the Critical Parameter. Endocrinology 1983, 113, 1261–1267. [Google Scholar] [CrossRef]
- Bittman, E.L.; Dempsey, R.J.; Karsch, F.J. Pineal Melatonin Secretion Drives the Reproductive Response to Daylength in the Ewe. Endocrinology 1983, 113, 2276–2283. [Google Scholar] [CrossRef]
- Masumoto, K.H.; Ukai-Tadenuma, M.; Kasukawa, T.; Nagano, M.; Uno, K.D.; Tsujino, K.; Horikawa, K.; Shigeyoshi, Y.; Ueda, H.R. Acute Induction of Eya3 by Late-Night Light Stimulation Triggers TSHβ Expression in Photoperiodism. Curr. Biol. 2010, 20, 2199–2206. [Google Scholar] [CrossRef]
- Dardente, H.; Wyse, C.A.; Birnie, M.J.; Dupré, S.M.; Loudon, A.S.I.; Lincoln, G.A.; Hazlerigg, D.G. A Molecular Switch for Photoperiod Responsiveness in Mammals. Curr. Biol. 2010, 20, 2193–2198. [Google Scholar] [CrossRef]
- Magner, J.A. Thyroid-Stimulating Hormone: Biosynthesis, Cell Biology, and Bioactivity. Endocr. Rev. 1990, 11, 354–385. [Google Scholar] [CrossRef]
- Guerra, M.; Blázquez, J.L.; Peruzzo, B.; Peláez, B.; Rodríguez, S.; Toranzo, D.; Pastor, F.; Rodríguez, E.M. Cell Organization of the Rat Pars Tuberalis. Evidence for Open Communication between Pars Tuberalis Cells, Cerebrospinal Fluid and Tanycytes. Cell Tissue Res. 2010, 339, 359–381. [Google Scholar] [CrossRef] [PubMed]
- Hanon, E.A.; Lincoln, G.A.; Fustin, J.-M.; Dardente, H.; Masson-Pévet, M.; Morgan, P.J.; Hazlerigg, D.G. Ancestral TSH Mechanism Signals Summer in a Photoperiodic Mammal. Curr. Biol. 2008, 18, 1147–1152. [Google Scholar] [CrossRef] [PubMed]
- Nakao, N.; Ono, H.; Yamamura, T.; Anraku, T.; Takagi, T.; Higashi, K.; Yasuo, S.; Katou, Y.; Kageyama, S.; Uno, Y.; et al. Thyrotrophin in the Pars Tuberalis Triggers Photoperiodic Response. Nature 2008, 452, 317–322. [Google Scholar] [CrossRef]
- Yoshimura, T.; Yasuo, S.; Watanabe, M.; Iigo, M.; Yamamura, T.; Hirunagi, K.; Ebihara, S. Light-Induced Hormone Conversion of T4 to T3 Regulates Photoperiodic Response of Gonads in Birds. Nature 2003, 426, 178–181. [Google Scholar] [CrossRef] [PubMed]
- Ono, H.; Hoshino, Y.; Yasuo, S.; Watanabe, M.; Nakane, Y.; Murai, A.; Ebihara, S.; Korf, H.-W.; Yoshimura, T. Involvement of Thyrotropin in Photoperiodic Signal Transduction in Mice. Proc. Natl. Acad. Sci. USA 2008, 105, 18238–18242. [Google Scholar] [CrossRef] [PubMed]
- Lechan, R.M.; Fekete, C. Role of Thyroid Hormone Deiodination in the Hypothalamus. Thyroid 2005, 15, 883–897. [Google Scholar] [CrossRef]
- Ho, S.C.; Sande, J.V.A.N.; Lefort, A.; Vassart, G.; Costagliola, S. Effects of Mutations Iinvolving the Highly Conserved S281HCC Motif in the Extracellular Domain of the Thyrotropin (TSH) Receptor on TSH Binding and Constitutive Activity. Endocrinology 2001, 142, 2760–2767. [Google Scholar] [CrossRef]
- Rubin, C.J.; Zody, M.C.; Eriksson, J.; Meadows, J.R.S.; Sherwood, E.; Webster, M.T.; Jiang, L.; Ingman, M.; Sharpe, T.; Ka, S.; et al. Whole-Genome Resequencing Reveals Loci under Selection during Chicken Domestication. Nature 2010, 464, 587–591. [Google Scholar] [CrossRef]
- Kijas, J.W.; Lenstra, J.A.; Hayes, B.; Boitard, S.; Neto, L.R.; Cristobal, M.S.; Servin, B.; McCulloch, R.; Whan, V.; Gietzen, K.; et al. Genome-Wide Analysis of the World’s Sheep Breeds Reveals High Levels of Historic Mixture and Strong Recent Selection. PLoS Biol. 2012, 10, e1001258. [Google Scholar] [CrossRef]
- Fariello, M.I.; Servin, B.; Tosser-Klopp, G.; Rupp, R.; Moreno, C.; Cristobal, M.S.; Boitard, S.; Arranz, J.J.; Banos, G.; Barendse, W.; et al. Selection Signatures in Worldwide Sheep Populations. PLoS ONE 2014, 9, e103813. [Google Scholar] [CrossRef]
- Karlsson, A.C.; Fallahshahroudi, A.; Johnsen, H.; Hagenblad, J.; Wright, D.; Andersson, L.; Jensen, P. A Domestication Related Mutation in the Thyroid Stimulating Hormone Receptor Gene (TSHR) Modulates Photoperiodic Response and Reproduction in Chickens. Gen. Comp. Endocrinol. 2016, 228, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Smits, G.; Campillo, M.; Govaerts, C.; Janssens, V.; Richter, C.; Vassart, G.; Pardo, L.; Costagliola, S. Glycoprotein Hormone Receptors: Determinants in Leucine-Rich Repeats Responsible for Ligand Specificity. EMBO J. 2003, 22, 2692–2703. [Google Scholar] [CrossRef] [PubMed]
- Kleinau, G.; Neumann, S.; Grüters, A.; Krude, H.; Biebermann, H. Novel Insights on Thyroid-Stimulating Hormone Receptor Signal Transduction. Endocr. Rev. 2013, 34, 691–724. [Google Scholar] [CrossRef]
- Wittkopp, P.J.; Kalay, G. Cis-Regulatory Elements: Molecular Mechanisms and Evolutionary Processes Underlying Divergence. Nat. Rev. Genet. 2012, 13, 59–69. [Google Scholar] [CrossRef] [PubMed]
- Graves, P.N.; Tomer, Y.; Davies, T.F. Cloning and Sequencing of a 1.3 Kb Variant of Human Thyrotropin Receptor mRNA Lacking the Transmembrane Domain. Biochem. Biophys. Res. Commun. 1992, 187, 1135–1143. [Google Scholar] [CrossRef]
- Mizutori, Y.; Chen, C.R.; McLachlan, S.M.; Rapoport, B. The Thyrotropin Receptor Hinge Region Is Not Simply a Scaffold for the Leucine-Rich Domain but Contributes to Ligand Binding and Signal Transduction. Mol. Endocrinol. 2008, 22, 1171–1182. [Google Scholar] [CrossRef]
- Daan, S.; Aschoff, J. Circadian Rhythms of Locomotor Activity in Captive Birds and Mammals: Their Variations with Season and Latitude. Oecologia 1975, 18, 269–316. [Google Scholar] [CrossRef]
- Nielsen, E.T. Twilight and the “crep” Unit. Nature 1961, 190, 878–879. [Google Scholar] [CrossRef]
- Stojak, J.; McDevitt, A.D.; Herman, J.S.; Searle, J.B.; Wójcik, J.M. Post-Glacial Colonization of Eastern Europe from the Carpathian Refugium: Evidence from Mitochondrial DNA of the Common Vole Microtus arvalis. Biol. J. Linn. Soc. 2015, 115, 927–939. [Google Scholar] [CrossRef]
- Haynes, S.; Jaarola, M.; Searle, J.B. Phylogeography of the Common Vole (Microtus arvalis) with Particular Emphasis on the Colonzaition of the Orkney Archipelago. Mol. Ecol. 2003, 12, 951–956. [Google Scholar] [CrossRef]
- South, A. Rworldmap: A New R Package for Mapping Global Data. R J. 2011, 3, 35–43. [Google Scholar] [CrossRef]
- South, A. Rworldxtra: Country Boundaries at High Resolution, R Package Version 1.01. 2012. Available online: https://rdrr.io/cran/rworldxtra/ (accessed on 7 December 2022).
- Neuwirth, E. RColorBrewer: ColorBrewer Palettes, R Package Version 1.1-2. 2014. Available online: https://cran.r-project.org/web/packages/RColorBrewer/index.html (accessed on 7 December 2022).
- Bivand, R.; Lewin-Koh, N. Maptools: Tools for Handling Spatial Objects, R Package Version 0.9-5. 2019. Available online: https://cran.r-project.org/web/packages/maptools/index.html (accessed on 7 December 2022).
- Bivand, R. ClassInt: Choose Univariate Class Intervals, R Package Version 0.3-3. 2019. Available online: https://r-spatial.github.io/classInt/reference/classIntervals.html (accessed on 7 December 2022).
- Peacock, J.M. Temperature and Leaf Growth in Lolium perenne. II. The Site of Temperature Perception. J. Appl. Ecol. 1975, 12, 115–123. [Google Scholar] [CrossRef]
- Cooper, J.P. Climatic Variation in Forage Grasses. I. Leaf Development in Climatic Races of Lolium and Dactylis. J. Appl. Ecol. 1964, 1, 45–61. [Google Scholar] [CrossRef]
- Kelley, L.A.; Mezulis, S.; Yates, C.M.; Wass, M.N.; Sternberg, M.J. The Phyre2 Web Portal for Protein Modeling, Prediction and Analysis. Nat. Protoc. 2015, 10, 845–858. [Google Scholar] [CrossRef] [PubMed]
- Grabe, N. AliBaba2: Context Specific Identification of Transcription Factor Binding Sites. In Silico Biol. 2002, 2, S1–S15. [Google Scholar] [PubMed]
- Rellstab, C.; Gugerli, F.; Eckert, A.J.; Hancock, A.M.; Holderegger, R. A Practical Guide to Environmental Association Analysis in Landscape Genomics. Mol. Ecol. 2015, 24, 4348–4370. [Google Scholar] [CrossRef]
- Benjamini, Y.; Hochberg, Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J. R. Stat. Soc. Ser. B 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Yekutieli, D.; Benjamini, Y. Resampling-Based False Discovery Rate Controlling Multiple Test Procedures for Correlated Test Statistics. J. Stat. Plan. Inference 1999, 82, 171–196. [Google Scholar] [CrossRef]
- Shin, J.-H.; Blay, S.; McNeney, B.; Graham, J. LDheatmap: An R Function for Graphical Display of Pairwise Linkage Disequilibria between Single Nucleotide Polymorphisms. J. Stat. Softw. 2006, 16, 1–9. [Google Scholar] [CrossRef]
- Saitou, N.; Nei, M. The Neighbor-Joining Method: A New Method for Reconstructing Phylogenetic Trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar]
- Takezaki, N.; Nei, M.; Tamura, K. POPTREEW: Web Version of POPTREE for Constructing Population Trees from Allele Frequency Data and Computing Some Other Quantities. Mol. Biol. Evol. 2014, 31, 1622–1624. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Martínková, N.; Barnett, R.; Cucchi, T.; Struchen, R.; Pascal, M.; Pascal, M.; Fischer, M.C.; Higham, T.; Brace, S.; Ho, S.Y.W.; et al. Divergent Evolutionary Processes Associated with Colonization of Offshore Islands. Mol. Ecol. 2013, 22, 5205–5220. [Google Scholar] [CrossRef] [PubMed]
- Heckel, G.; Burri, R.; Fink, S.; Desmet, J.-F.; Excoffier, L. Genetic Structure and Colonization Processes in European Populations of the Common Vole, Microtus arvalis. Evolution 2005, 59, 2231. [Google Scholar] [CrossRef] [PubMed]
- Nei, M. Estimation of Average Heterozygosity and Genetic Distance from a Small Number of Individuals. Genetics 1978, 89, 583–590. [Google Scholar] [CrossRef] [PubMed]
- Hutchison, D.W.; Templeton, A.R. Correlation of Pairwise Genetic and Geographic Distance Measures: Inferring the Relative Influences of Gene Flow and Drift on the Distribution of Genetic Variability. Evolution 1999, 53, 1898–1914. [Google Scholar] [CrossRef] [PubMed]
- Endler, J.A. Geographic Variation, Speciation, and Clines; Princeton University Press: Princeton, NJ, USA, 1977. [Google Scholar]
- Stojak, J.; McDevitt, A.D.; Herman, J.S.; Krystufek, B.; Uhlikova, J.; Purger, J.J.; Lavrenchenko, L.A.; Searle, J.B.; Wójcik, J.M. Between the Balkans and the Baltic: Phylogeography of a Common Vole Mitochondrial DNA Lineage Limited to Central Europe. PLoS ONE 2016, 11, e0168621. [Google Scholar] [CrossRef] [PubMed]
- Stojak, J.; Borowik, T.; Górny, M.; McDevitt, A.D.; Wójcik, J.M. Climatic Influences on the Genetic Structure and Distribution of the Common Vole and Field Vole in Europe. Mammal Res. 2019, 64, 19–29. [Google Scholar] [CrossRef]
- Baca, M.; Popović, D.; Lemanik, A.; Bañuls-Cardona, S.; Conard, N.J.; Cuenca-Bescós, G.; Desclaux, E.; Fewlass, H.; Garcia, J.T.; Hadravova, T.; et al. Ancient DNA Reveals Interstadials as a Driver of Common Vole Population Dynamics during the Last Glacial Period. J. Biogeogr. 2022, 50, 183–196. [Google Scholar] [CrossRef]
- Stojak, J.; Wójcik, J.M.; Ruczyńska, I.; Searle, J.B.; McDevitt, A.D. Contrasting and Congruent Patterns of Genetic Structuring in Two Microtus Vole Species Using Museum Specimens. Mammal Res. 2016, 61, 141–152. [Google Scholar] [CrossRef]
- Hewitt, G.M. Post-Glacial Re-Colonization of European Biota. Biol. J. Linn. Soc. 1999, 68, 87–112. [Google Scholar] [CrossRef]
- Stojak, J.; Tarnowska, E. Polish Suture Zone as the Goblet of Truth in Post-Glacial History of Mammals in Europe. Mammal Res. 2019, 64, 463–475. [Google Scholar] [CrossRef]
- Phifer-Rixey, M.; Bi, K.; Ferris, K.G.; Sheehan, M.J.; Lin, D.; Mack, K.L.; Keeble, S.M.; Suzuki, T.A.; Good, J.M.; Nachman, M.W. The Genomic Basis of Environmental Adaptation in House Mice. PLoS Genet. 2018, 14, e1007672. [Google Scholar] [CrossRef] [PubMed]
- Ferris, K.G.; Chavez, A.S.; Suzuki, T.A.; Beckman, E.J.; Bi, K.; Nachman, M.W. The Genomics of Rapid Climatic Adaptation and Parallel Evolution in North American House Mice. PLoS Genet. 2021, 17, e1009495. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, W.E. Geography of Photoperiodic Response in Diapausing Mosquito. Nature 1976, 262, 384–386. [Google Scholar] [CrossRef]
- Millar, J.S.; Innes, D.G.L. Breeding by Peromyscus maniculatus over an Elevational Gradient. Can. J. Zool. 1985, 63, 124–129. [Google Scholar] [CrossRef]
- Ji, C.; Casinghino, S.; McCarthy, T.L.; Centrella, M. Multiple and Essential Sp1 Binding Sites in the Promoter for Transforming Growth Factor-β Type I Receptor. J. Biol. Chem. 1997, 272, 21260–21267. [Google Scholar] [CrossRef]
- Höller, M.; Westin, G.; Jiricny, J.; Schaffner, W. Sp1 Transcription Factor Binds DNA and Activates Transcription Even When the Binding Site Is CpG Methylated. Genes Dev. 1988, 2, 1127–1135. [Google Scholar] [CrossRef]
- Chamary, J.V.; Parmley, J.L.; Hurst, L.D. Hearing Silence: Non-Neutral Evolution at Synonymous Sites in Mammals. Nat. Rev. Genet. 2006, 7, 98–108. [Google Scholar] [CrossRef]
- Hunt, R.; Sauna, Z.E.; Ambudkar, S.V.; Gottesman, M.M.; Kimchi-Sarfaty, C. Silent (Synonymous) SNPs: Should We Care about Them? Methods Mol. Biol. 2009, 578, 23–39. [Google Scholar] [CrossRef]
- Goodenbour, J.M.; Pan, T. Diversity of tRNA Genes in Eukaryotes. Nucleic Acids Res. 2006, 34, 6137–6146. [Google Scholar] [CrossRef]
- Dittmar, K.A.; Goodenbour, J.M.; Pan, T. Tissue-Specific Differences in Human Transfer RNA Expression. PLoS Genet. 2006, 2, e221. [Google Scholar] [CrossRef] [PubMed]
- Quax, T.E.F.; Claassens, N.J.; Soll, D.; Oost, J. van der Codon Bias as a Means to Fine-Tune Gene Expression. Cell Press 2015, 59, 149–161. [Google Scholar] [CrossRef]
- Shinomiya, A.; Shimmura, T.; Nishiwaki-Ohkawa, T.; Yoshimura, T. Regulation of Seasonal Reproduction by Hypothalamic Activation of Thyroid Hormone. Front. Endocrinol. 2014, 5, 12. [Google Scholar] [CrossRef]
- Ikegami, K.; Liao, X.H.; Hoshino, Y.; Ono, H.; Ota, W.; Ito, Y.; Nishiwaki-Ohkawa, T.; Sato, C.; Kitajima, K.; Iigo, M.; et al. Tissue-Specific Posttranslational Modification Allows Functional Targeting of Thyrotropin. Cell Rep. 2014, 9, 801–809. [Google Scholar] [CrossRef] [PubMed]
- Pinkard, O.; McFarland, S.; Sweet, T.; Coller, J. Quantitative tRNA-Sequencing Uncovers Metazoan Tissue-Specific tRNA Regulation. Nat. Commun. 2020, 11, 4104. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, W.E.; Holzapfel, C.M. Genetic Shift in Photoperiodic Response Correlated with Global Warming. Proc. Natl. Acad. Sci. USA 2001, 98, 14187–14744. [Google Scholar] [CrossRef]
- Bradshaw, W.E.; Holzapfel, C.M. Evolutionary Response to Rapid Climate Change. Science 2006, 312, 1477–1478. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, W.E.; Holzapfel, C.M. Genetic Response to Rapid Climate Change: It’s Seasonal Timing That Matters. Mol. Ecol. 2008, 17, 157–166. [Google Scholar] [CrossRef]
- Bradshaw, W.E.; Zani, P.A.; Holzapfel, C.M. Adaptation to Temperate Climates. Evolution 2004, 58, 1748–1762. [Google Scholar] [CrossRef]
- Tkadlec, E. The Effects of Seasonality on Variation in the Length of Breeding Season in Arvicoline Rodents. Folia Zool. 2000, 49, 269–286. [Google Scholar]
- Stojak, J.; Jędrzejewska, B. Extinction and Replacement Events Shaped the Historical Biogeography of Arctic Mammals in Europe: New Models of Species Response. Mamm. Rev. 2022, 52, 507–518. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).