Root Metabolism and Effects of Root Exudates on the Growth of Ralstonia solanacearum and Fusarium moniliforme Were Significantly Different between the Two Genotypes of Peanuts
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Materials and Treatments
2.2. Collection of Peanut Root Exudates
2.3. Detection and Analysis of Peanut Root Exudates
2.4. RNA Extraction, Library Construction, RNA-Sequencing and Data Analysis
2.5. qRT-PCR Analyses
2.6. Interaction between Two Pathogens and Peanut Root Exudates
3. Results
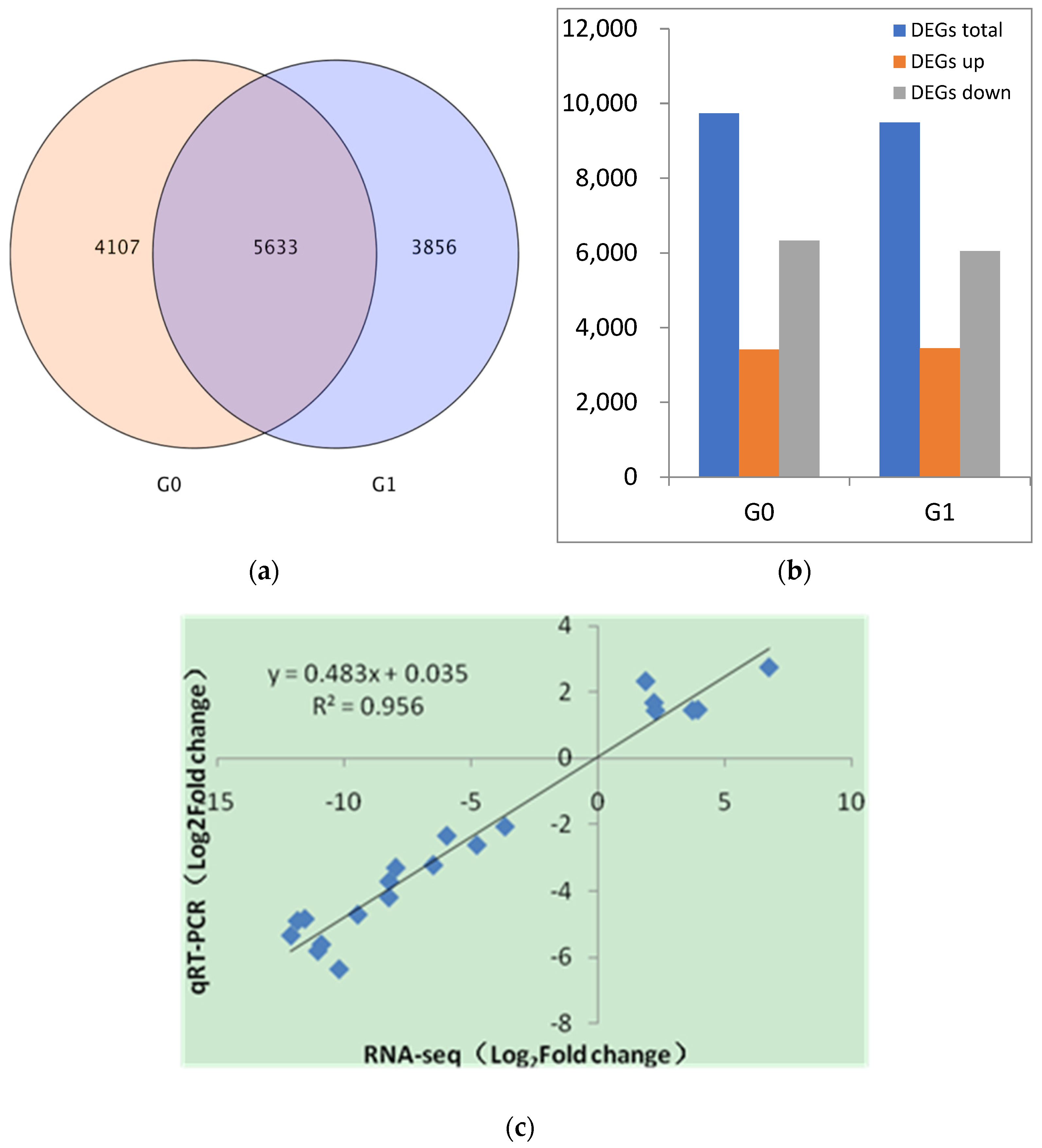
3.1. DEGs and qRT-PCR Analysis between A. correntina and GH85
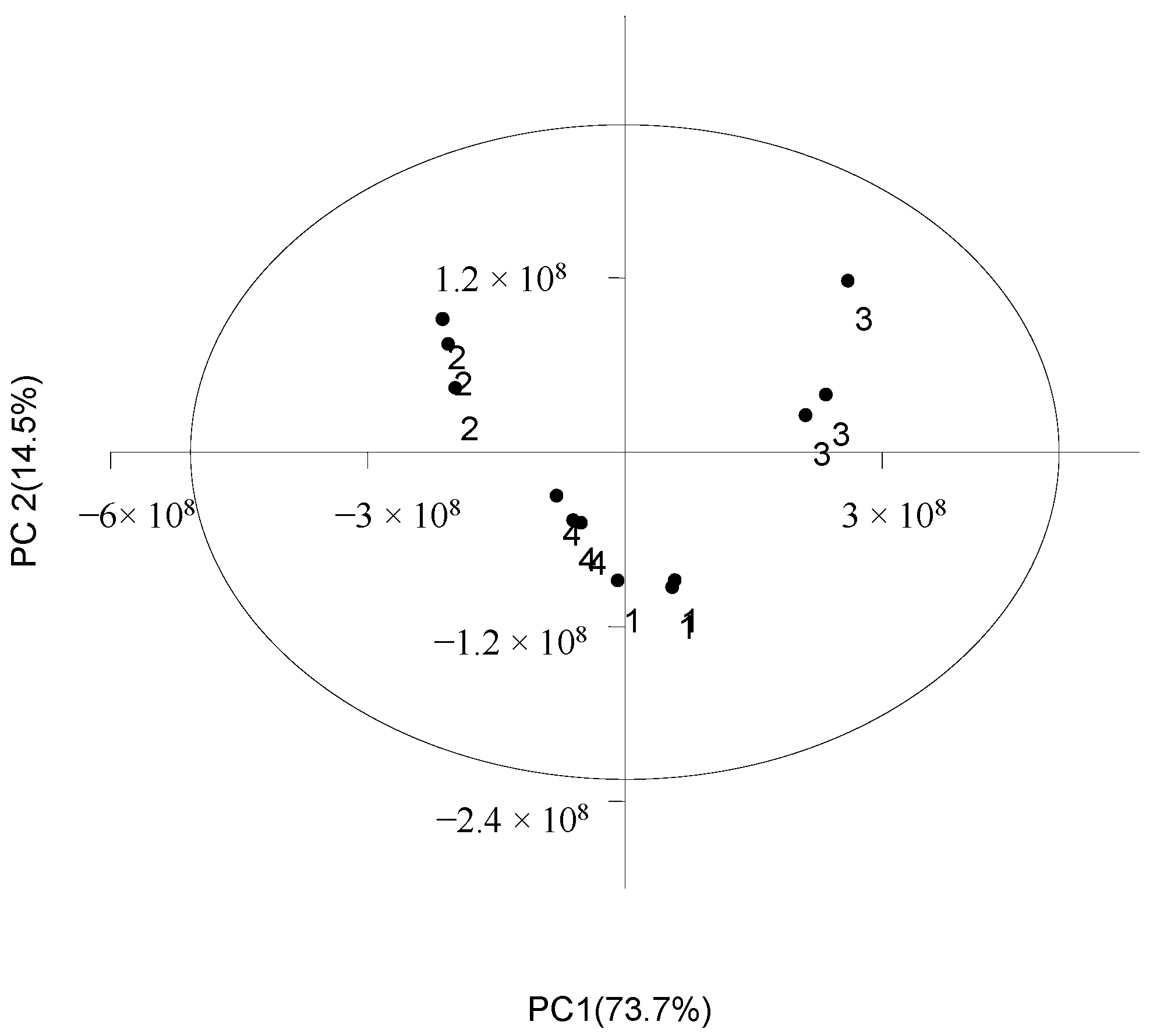
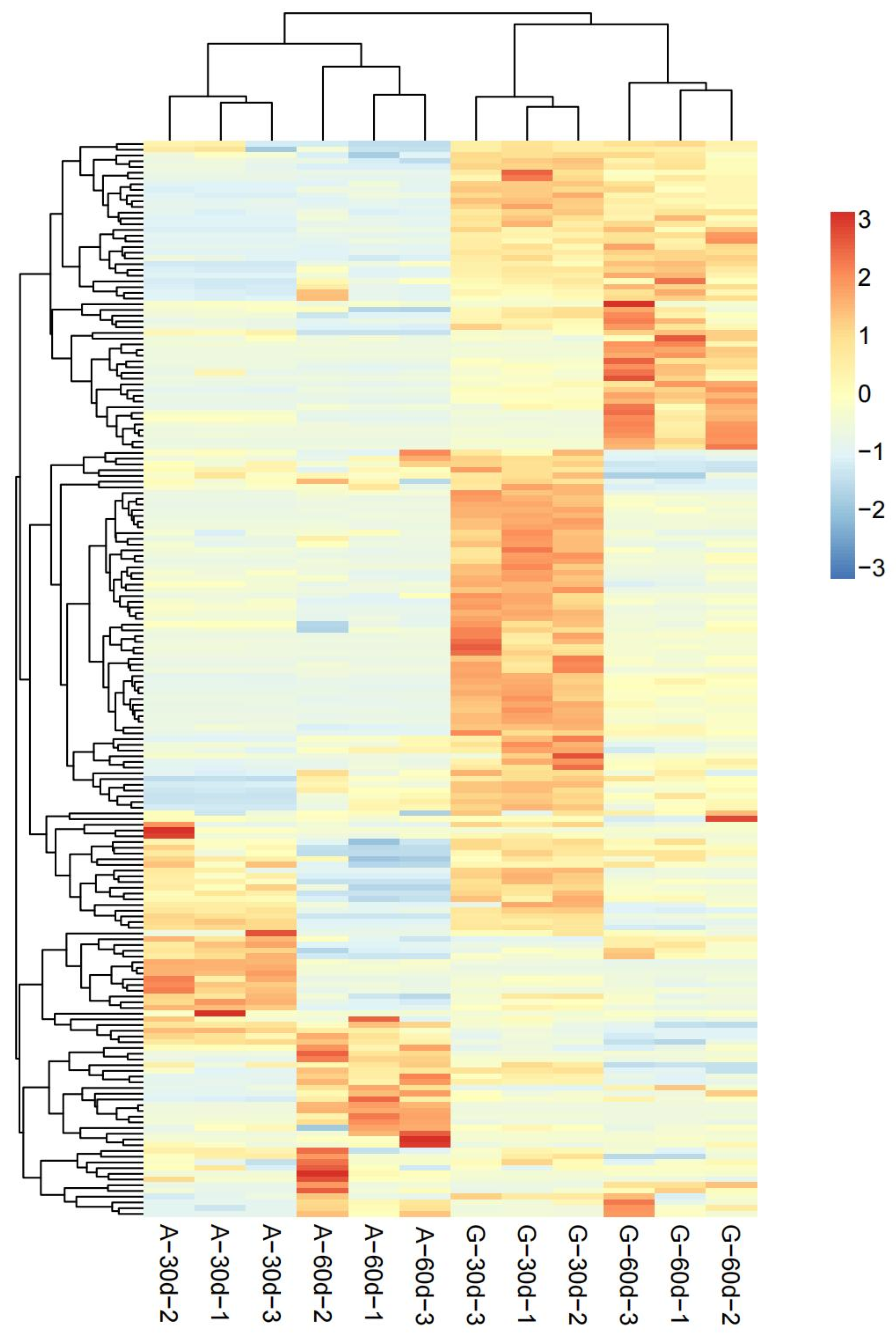
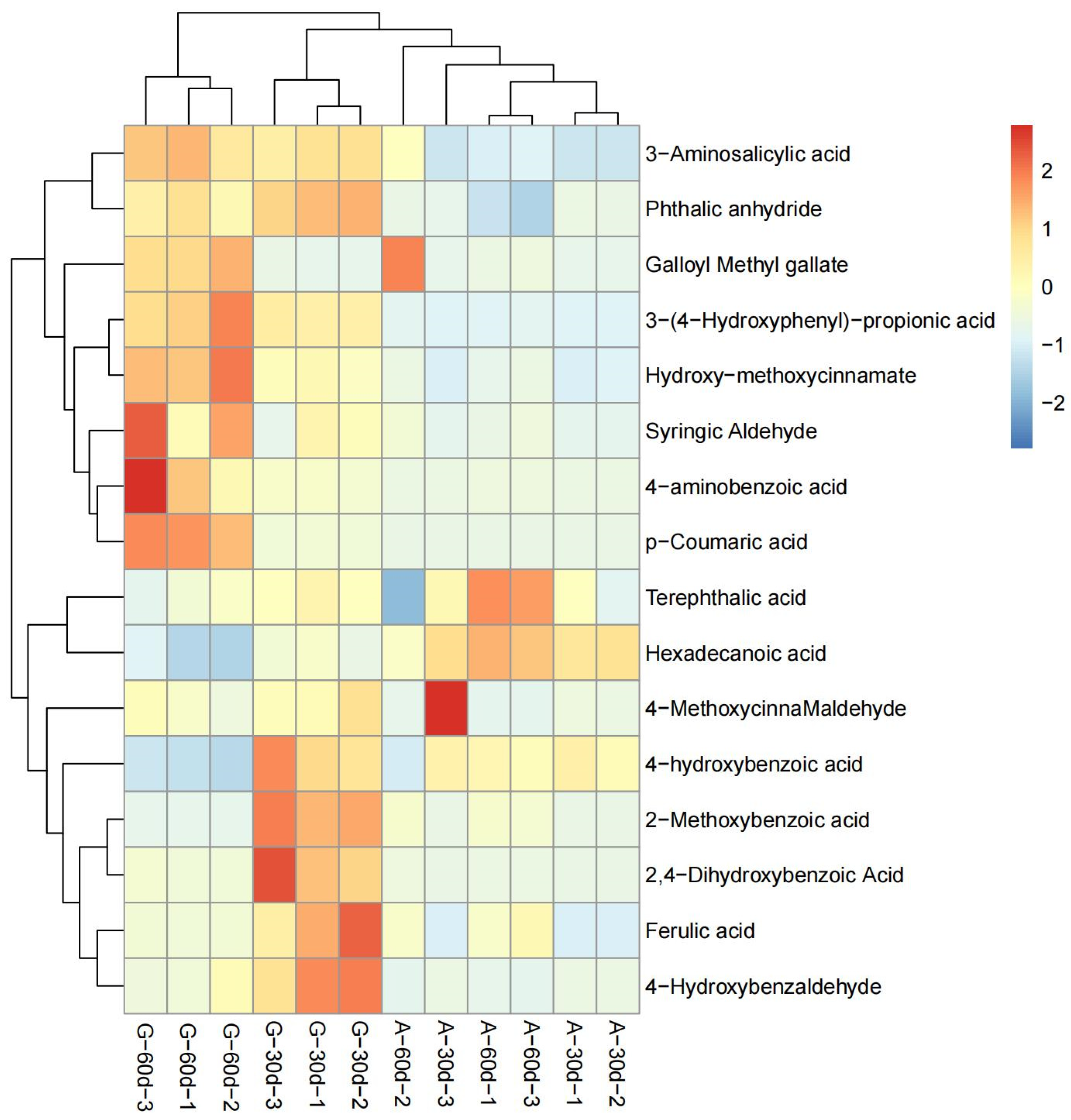
3.2. DEMs Analysis between A. correntina and GH85
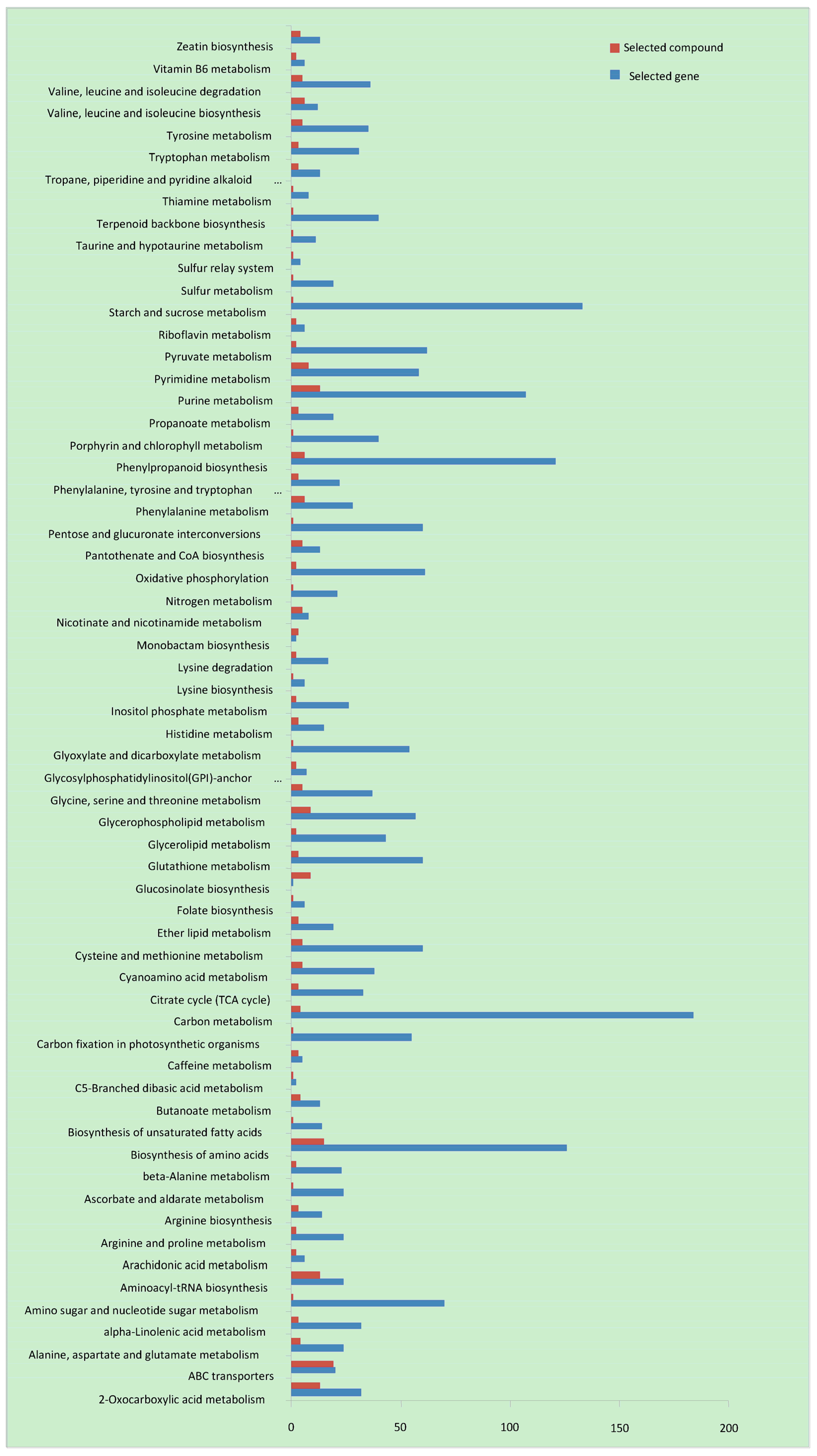
3.3. Transcriptome and Metabolomics Association Analysis of A. correntina and GH85
3.4. Interaction of Two Peanut Root Exudates and Two Pathogens
3.5. Effects of Exogenous Phenolic Acids and Amino Acids on the Growth of R. solanacearum and F. moniliforme
4. Discussion
4.1. Root Exudates of A. correntinaInhibited the Growth of R. solanacearum and F. moniliforme
4.2. Phenolic Acid and Amino Acid Might Play an Important Role in Different Effects of Root Exudates between A. correntina and GH85 on the Growth of R. solanacearum and F. moniliforme
4.3. Phenolic Acids Have a Critical Function on the Growth of Pathogens
4.4. Amino Acids Are Closely Related to Plant Disease Resistance
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Raaijmakers, J.M.; Paulitz, T.C.; Steinberg, C.; Alabouvette, C.; Moënne-Loccoz, Y. The rhizosphere: A playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil. 2008, 321, 341–361. [Google Scholar] [CrossRef] [Green Version]
- Li, P.; Wang, X.; Li, Y.; Wang, H.; Liang, F.; Dai, C. The contents of phenolic acids in continuous cropping peanut and their allelopathy. Acta Ecol. Sin. 2010, 30, 2128–2134. [Google Scholar]
- Li, X.G.; Ding, C.F.; Hua, K.; Zhang, T.L.; Zhang, Y.N.; Zhao, L.; Yang, Y.; Liu, J.G.; Wang, X.X. Soil sickness of peanuts is attributable to modifications in soil microbes induced by peanut root exudates rather than to direct allelopathy. Soil Biol. Biochem. 2014, 78, 149–159. [Google Scholar] [CrossRef]
- Huang, L.F.; Song, L.X.; Xia, X.J.; Mao, W.H.; Shi, K.; Zhou, Y.H.; Yu, J.Q. Plant-soil feedbacks and soil sickness: From mechanisms to application in agriculture. J. Chem. Ecol. 2013, 39, 232–242. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.Q.; Wang, Q.; Zhang, W.H.; Gao, L. Reducing environmental risk of excessively fertilized soils and improving cucumber growth by Caragana microphylla-straw compost application in long-term continuous cropping systems. Sci. Total Environ. 2016, 544, 251–261. [Google Scholar] [CrossRef] [PubMed]
- Giorio, C.; Safer, A.; Sánchez-Bayo, F.; Tapparo, A.; Lentola, A.; Girolami, V.; Lexmond, M.B.; Bonmatin, J.M. An update of the Worldwide Integrated Assessment (WIA) on systemic insecticides. Part 1: New molecules, metabolism, fate, and transport. Environ. Sci. Pollut. Res. 2021, 28, 11716–11748. [Google Scholar] [CrossRef] [Green Version]
- Rojo, F.G.; Reynoso, M.M.; Ferez, M.; Chulze, S.N.; Torres, A.M. Biological control by Trichoderma species of Fusarium solani causing peanut brown root rot under field conditions. Crop Prot. 2007, 26, 549–555. [Google Scholar] [CrossRef]
- Pizzolitto, R.P.; Dambolena, J.S.; Zunino, M.P.; Larrauri, M.; Grosso, N.R.; Nepote, V.; Dalcero, A.M.; Zygadlo, J.A. Activity of natural compounds from peanut skins on Fusarium verticillioides growth and fumonisin B1 production. Ind. Crops Prod. 2013, 47, 286–290. [Google Scholar] [CrossRef]
- Mehan, V.K.; Liao, B.S.; Tan, Y.J.; Robinson-Smith, A.; Mcdonald, D.; Hayward, A.C. Bacterialwilt of Groundnut Information Bulletin No. 35; International Crops Research Institute for the Semi-Arid Tropics: Patancheru, India, 1994; pp. 1–20. [Google Scholar]
- Krapovickas, A.; Gregory, W.C. Taxonomy of the genus Arachis (Leguminosae). Bonplandia 1994, 8, 1–18. [Google Scholar]
- Li, Z.; Xiong, F.Q.; Guo, W.F.; Mo, C.M.; Wu, H.N.; Du, L. The root transcriptome analyses of peanut wild species Arachiscorrentina (Burkart) Krapov. & W.C. Gregory and cultivated variety Xiaobaisha in response to benzoic acid and p-cumaric acid stress. Genet. Resour. Crop Evol. 2020, 67, 9–20. [Google Scholar]
- Singh, B.K.; Millard, P.; Whiteley, A.S.; Murrell, J.C. Unravelling rhizosphere-microbial interactions: Opportunities and limitations. Trends Microbiol. 2004, 12, 386–393. [Google Scholar] [CrossRef] [PubMed]
- Mercado-Blanco, J.; Bakker, P.A.H.M. Interactions between plants and beneficial Pseudomonas spp.: Exploiting bacterial traits for crop protection. AntonieVan Leeuwenhoek. 2007, 92, 367–389. [Google Scholar] [CrossRef] [PubMed]
- Shaposhnikov, A.I.; Shakhnazarova, V.Y.; Vishnevskaya, N.A.; Borodina, E.V.; Strunnikova, O.K. Aromatic carboxylic acids in barley-root exudates and their influence on the growth of Fusarium culmorum and Pseudomonas fluorescens. Appl. Biochem. Microbiol. 2020, 56, 344–351. [Google Scholar] [CrossRef]
- Hein, J.W.; Wolfe, G.V.; Blee, K.A. Comparison of rhizosphere bacterial communities in Arabidopsis thaliana mutants for systemic acquired resistance. Microb. Ecol. 2008, 55, 333–343. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Park, S.W.; Weir, T.L.; Callaway, R.M.; Vivanco, J.M. How plants communicate using the underground information superhighway. Trends Plant Sci. 2004, 9, 26–32. [Google Scholar] [CrossRef]
- Bais, H.P.; Vepachedu, R.; Gilroy, S.; Callaway, R.M.; Vivanco, J.M. Allelopathy and exotic plant invasion: From molecules and genes to species interactions. Science 2003, 301, 1377–1380. [Google Scholar] [CrossRef]
- Flores, H.E.; Vivanco, J.M.; Loyola-Vargas, V.M. “Radicle” biochemistry: The biology of root-specific metabolism. Trends Plant Sci. 1999, 4, 220–226. [Google Scholar] [CrossRef]
- Li, Y.; Gu, Y.L.; Li, J.; Xu, M.; Wei, Q.; Wang, Y. Biocontrol agent Bacillus amyloliquefaciens LJ02 induces systemic resistance against cucurbits powdery mildew. Front. Microbiol. 2015, 6, 883. [Google Scholar] [CrossRef] [Green Version]
- Chen, W.; Gong, L.; Guo, Z.; Wang, W.; Zhang, H.; Liu, X.; Yu, S.; Xiong, L.; Luo, J.A. Novel Integrated Method for Large-Scale Detection, Identification, and Quantification of Widely Targeted Metabolites: Application in the Study of Rice Metabolomics. Mol. Plant. 2013, 6, 1769–1780. [Google Scholar] [CrossRef] [Green Version]
- Fraga, C.G.; Clowers, B.H.; Moore, R.J.; Zink, E.M. Signature-discovery approach for sample matching of a nerve-agent precursor using liquid chromatography-mass spectrometry, XCMS, and chemometrics. Anal. Chem. 2010, 82, 4165–4173. [Google Scholar] [CrossRef]
- Leng, N.; Dawson, J.A.; Thomson, J.A.; Ruotti, V.; Rissman, A.I.; Smits, B.M.G.; Haag, J.D.; Gould, M.N.; Stewart, R.M.; Kendziorski, C. EBSeq: An empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 2013, 29, 1035–1043. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, S.; Wen, Z.; Tang, T.; Liu, Y.; Dang, F.; Xie, T.; Wu, H. Study on flavonoid and bioactivity features of the pericarp of Citri Reticulatae ‘chachi’ during storage. Arab. J. Chem. 2022, 15, 103653. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, M.; Deniziak, M.A.; Barciszewski, J. Aminoacyl-tRNA synthetases database. Nucleic Acids Res. 2001, 29, 288–290. [Google Scholar] [CrossRef] [Green Version]
- Maeda, H.; Dudareva, N. Theshikimatepathwayandaromaticaminoacidbiosynthesisinplants. Annu. Rev. Plant Biol. 2012, 63, 73–105. [Google Scholar] [CrossRef] [PubMed]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [Green Version]
- Yuan, J.; Zhang, N.; Huang, Q.; Raza, W.; Li, R.; Vivanco, J.M.; Shen, Q. Organic acids from root exudates of banana help root colonization of PGPR strain Bacillus amyloliquefaciens NJN-6. Sci. Rep. 2015, 5, 13438. [Google Scholar] [CrossRef] [Green Version]
- Blum, U. Effects of Microbial Utilization of Phenolic Acids and their Phenolic Acid Breakdown Products on Allelopathic Interactions. J. Chem. Ecol. 1998, 24, 685–708. [Google Scholar] [CrossRef]
- Muscolo, A.; Panuccio, M.R.; Sidari, M. The effect of phenols on respiratory enzymes in seed germination. Plant Growth Regul. 2001, 35, 31–35. [Google Scholar] [CrossRef]
- Blum, U.; Gerig, T.M. Relationships between phenolic acid concentrations, transpiration, water utilization, leaf area expansion, and uptake of phenolic acid: Nutrient culture studies. J. Chem. Ecol. 2005, 31, 1907–1932. [Google Scholar] [CrossRef]
- Yu, J.Q.; Ye, S.F.; Zhang, M.F.; Hu, W.H. Effects of root exudates and aqueous root extracts of cucumber (Cucumissativus) and allelochemicals, on photosynthesis and antioxidant enzymes in cucumber. Biochem. Syst. Ecol. 2003, 31, 129–139. [Google Scholar] [CrossRef]
- Chou, C.H.; Waller, G.R. Phytochemical Ecology: Allelochemicals, Mycotoxins, and Insect Pheromones and Allomones; Institute of Botany, Academia Sinica: Taipei, Taiwan, 1989; p. 504. [Google Scholar]
- Chou, C.H.; LEU, L.L. Allelopathy substances and activities of Delonixregia Raf. J. Chem. Ecol. 1992, 18, 353–367. [Google Scholar] [CrossRef]
- Liu, Y.X.; Li, X.; Cai, K.; Cai, L.T.; Lu, N.; Shi, J.X. Identification of benzoic acid and 3-phenylpropanoic acid in tobacco root exudates and their role in the growth of rhizosphere microorganisms. Appl. Soil Ecol. 2015, 93, 78–87. [Google Scholar] [CrossRef]
- Hao, W.Y.; Ren, L.X.; Ran, W.; Shen, Q.R. Allelopathic effects of root exudates from watermelon and rice plants on Fusarium oxysporum f. sp. niveum. Plant Soil. 2010, 336, 485–497. [Google Scholar] [CrossRef]
- Mizutani, M.; Ohta, D.; Sato, R. Isolation of a cDNA and a genomic clone encoding cinnamate 4-hydroxylase from Arabidopsis and its expression manner in planta. Plant Physiol. 1997, 113, 755–763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weisshaa, R.B.; Jenkins, G.I. Phenylpropanoid biosynthesis and its regulation. Curr. Opin. Plant Biol. 1999, 1, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Zubieta, C.; Kota, P.; Ferrer, J.L.; Dixon, R.A.; Noel, J.P.S. Structural basis for the modulation of lignin monomer methylation by caffeic acid/5-hydroxyferulic acid 3/5-O-methyltransferase. Plant Cell. 2002, 14, 1265–1277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weng, J.K.; Li, Y.; Mo, H.; Chapple, C. Title Assembly of an evolutionarily new pathway for alpha-pyrone biosynthesis in Arabidopsis. Science 2012, 337, 960–964. [Google Scholar] [CrossRef] [PubMed]
- Kallscheuer, N.; Vogt, M.; Marienhagen, J. A novel synthetic pathway enables microbial production of polyphenols independent from the endogenous aromatic amino acid metabolism. ACS Synth. Biol. 2016, 6, 410–415. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Potential biochemical indicators of salinity tolerance in plants. Plant Sci. 2004, 166, 3–16. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Nelson, D.E.; Jensen, R.G. Adaptations to Environmental Stresses. Plant Cell. 1995, 32, 1099–1111. [Google Scholar]
- Zhang, P.; Fu, J.; Hu, L. Effects of alkali stress on growth, free amino acids and carbohydrates metabolism in Kentucky bluegrass (Poapratensis). Ecotoxicology 2012, 21, 1911–1918. [Google Scholar] [CrossRef]
- Park, S.G.; Schimmel, P.; Kim, S. Aminoacyl tRNA synthetases and their connections to disease. Proc. Natl. Acad. Sci. USA 2008, 105, 11043–11049. [Google Scholar] [CrossRef] [Green Version]
- Pavlou, G.C.; Vakalounakis, D.J. Biological control of root and stem rot of greenhouse cucumber, caused by Fusarium oxysporum f. sp. radicis-cucumerinum, by lettuce soil amendment. Crop Prot. 2005, 24, 135–140. [Google Scholar] [CrossRef]

| F. moniliforme | R. solanacearum | ||||||
|---|---|---|---|---|---|---|---|
| Treatment | Concentration (%) | 3d (cm) | Inhibition (%) | 4d (cm) | Inhibition (%) | 1d (106 cfu/mL) | Inhibition (%) |
| Control | 1 | 4.17 ± 0.06 | - | 5.20 ± 0.10 | - | 8.79 ± 0.20 | - |
| 5 | 4.03 ± 0.06 | - | 5.03 ± 0.06 | - | 8.58 ± 0.32 | - | |
| 30 | 4.00 ± 0.10 | - | 4.93 ± 0.15 | - | 7.87 ± 0.39 | - | |
| GH85 30d | 1 | 4.37 ± 0.06 | −4.8 | 5.40 ± 0.10 | −3.85 | 9.31 ± 0.36 | −5.99 |
| 5 | 4.50 ± 0.10 | −11.66 | 5.33 ± 0.06 | −5.96 | 9.08 ± 0.24 | −5.75 | |
| 30 | 3.70 ± 0.10 | 7.5 | 4.50 ± 0.20 | 8.72 | 6.50 ± 0.30 | 17.4 | |
| GH85 30–60d | 1 | 4.50 ± 0.10 | −7.91 | 5.50 ± 0.10 | −5.77 | 9.36 ± 0.30 | −6.49 |
| 5 | 4.40 ± 0.10 | −9.18 | 5.30 ± 0.10 | −5.37 | 8.68 ± 0.23 | −1.09 | |
| 30 | 3.60 ± 0.10 | 10.00 | 4.57 ± 0.15 | 7.3 | 6.10 ± 0.26 * | 22.49 | |
| A. correntina 30d | 1 | 4.07 ± 0.06 | 2.4 | 5.07 ± 0.06 | 2.5 | 8.51 ± 0.39 | 3.11 |
| 5 | 3.77 ± 0.12 | 6.45 | 4.73 ± 0.06 | 5.96 | 8.01 ± 0.05 | 6.72 | |
| 30 | 3.37 ± 0.12 | 15.75 | 4.00 ± 0.10 * | 18.86 | 7.51 ± 0.44 | 4.57 | |
| A. correntina 30–60d | 1 | 4.17 ± 0.06 | 0.00 | 5.00 ± 0.10 | 3.85 | 8.48 ± 0.24 | 3.53 |
| 5 | 3.8 ± 0.10 | 5.70 | 4.83 ± 0.15 | 3.98 | 8.07 ± 0.21 | 5.98 | |
| 30 | 3.23 ± 0.06 * | 19.25 | 4.13 ± 0.06 | 16.23 | 7.39 ± 0.12 | 6.14 | |
| F. moniliforme | R. solanacearum | ||||||
|---|---|---|---|---|---|---|---|
| Treatment | Concentration (nM) | 3d (cm) | Inhibition Rate (%) | 4d (cm) | Inhibition Rate (%) | 1d (106 cfu/mL) | Inhibition Rate (%) |
| Control | 3.47 ± 0.12 | 4.73 ± 0.06 | 2.90 ±0.06 | ||||
| 3-Aminosalicylic Acid | 0.001 | 3.90 ± 0.20 | −12.39 | 4.80 ± 0.20 | −1.48 | 2.86 ± 0.09 | 1.72 |
| 0.01 | 3.43 ± 0.12 | 1.15 | 4.87 ± 0.12 | −2.96 | 1.88 ± 0.16 * | 35.17 | |
| 0.1 | 3.21 ± 0.10 | 7.49 | 4.60 ± 0.10 | 2.75 | 1.34 ± 0.30 * | 53.79 | |
| 2,4-Dihydroxybenzoic Acid | 0.001 | 3.67 ± 0.06 | −5.76 | 4.90 ± 0.10 | −3.89 | 2.44 ± 0.21 | 15.86 |
| 0.01 | 3.77 ± 0.06 | −8.65 | 5.17 ± 0.06 | −9.3 | 0.38 ± 0.18 * | 87.24 | |
| 0.1 | 3.33 ± 0.06 | 4.03 | 4.37 ± 0.12 | 7.61 | 0.00 ± 0.00 | - | |
| 3-(4-Hydroxyphenyl)-propionic Acid | 0.001 | 3.67 ± 0.06 | −5.76 | 4.97 ± 0.15 | −5.07 | 3.00 ± 0.15 | −3.45 |
| 0.01 | 3.80 ± 0.10 | −9.51 | 5.13 ± 0.15 | −8.46 | 1.56 ± 19.52 * | 46.21 | |
| 0.1 | 3.57 ± 0.06 | −2.88 | 4.87 ± 0.06 | −2.96 | 0.00 ± 0.00 | - | |
| Syringic Aldehyde | 0.001 | 3.67 ± 0.06 | −5.76 | 4.90 ± 0.10 | −3.59 | 2.95 ± 0.08 | −1.72 |
| 0.01 | 3.33 ± 0.06 | 4.03 | 4.80 ± 0.10 | −1.48 | 1.60 ± 0.04 * | 44.94 | |
| 0.1 | 2.63 ± 0.06 | 24.21 | 3.33 ± 0.06 * | 29.6 | 1.18 ± 0.14 * | 59.31 | |
| 2-Methoxybenzoic acid | 0.001 | 3.63 ± 0.06 | −4.61 | 4.87 ± 0.06 | −2.96 | 1.85 ± 0.11 * | 36.21 |
| 0.01 | 3.33 ± 0.06 | 4.03 | 4.60 ± 0.10 | 2.75 | 46.67 ± 0.09 * | 83.91 | |
| 0.1 | 3.43 ± 0.15 | 1.15 | 4.70 ± 0.10 | 0.63 | 0.00 ± 0.00 | - | |
| p-Coumaric Acid | 0.001 | 3.80 ± 0.10 | −9.51 | 5.10 ± 0.10 | −7.82 | 3.13 ± 0.10 | −7.93 |
| 0.01 | 3.13 ± 0.21 | 9.8 | 3.83 ± 0.06 | 19.03 | 1.22 ± 0.14 * | 57.82 | |
| 0.1 | 0.50 ± 0.00 | - | 0.50 ± 0.00 | - | 0.000 ± 0.00 | - | |
| Ferulic Acid | 0.001 | 3.83 ± 0.06 | −10.37 | 5.13 ± 0.06 | −8.46 | 3.04 ± 0.12 | −4.71 |
| 0.01 | 3.37 ± 0.12 | 2.88 | 4.30 ± 0.17 | 9.09 | 1.77 ± 0.12 * | 39.08 | |
| 0.1 | 0.50 ± 0.00 | - | 0.50 ± 0.00 | - | 0.00 ± 0.00 | - | |
| Tryptophan | 0.001 | 3.70 ± 0.00 | −6.62 | 4.83 ± 0.06 | −2.11 | 3.00 ± 0.12 | −3.45 |
| 0.01 | 3.87 ± 0.06 | −11.53 | 5.13 ± 0.06 | −8.46 | 2.13 ± 0.18 | 26.78 | |
| 0.1 | 3.47 ± 0.06 | 0 | 4.67 ± 0.06 | 1.28 | 1.16 ± 0.13 * | 60.11 | |
| L-Proline | 0.001 | 3.47 ± 0.06 | 0 | 4.90 ± 0.02 | −3.59 | 2.97 ± 0.20 | −2.52 |
| 0.01 | 3.77 ± 0.06 | −8.65 | 4.93 ± 0.06 | −4.23 | 2.16 ± 0.21 | 25.52 | |
| 0.1 | 3.70 ± 0.00 | −6.63 | 5.07 ± 0.06 | −7.19 | 2.72 ± 0.14 | 6.21 | |
| L-Valine | 0.001 | 3.50 ± 0.10 | −0.86 | 4.77 ± 0.15 | −0.85 | 2.68 ± 0.14 | 7.59 |
| 0.01 | 3.93 ± 0.12 | −13.26 | 5.33 ± 0.06 | −12.68 | 3.04 ± 0.20 | −4.83 | |
| 0.1 | 3.67 ± 0.12 | −5.76 | 5.03 ± 0.06 | −6.34 | 1.39 ± 0.16 * | 52.07 | |
| L-Methionine | 0.001 | 3.60 ± 0.10 | −3.75 | 4.77 ± 0.06 | −0.85 | 2.87 ± 0.06 | 0.92 |
| 0.01 | 3.50 ± 0.10 | −0.86 | 4.50 ± 0.10 | 4.86 | 3.26 ± 0.0 8 | −12.3 | |
| 0.1 | 3.30 ± 0.00 | 4.9 | 4.03 ± 0.15 | 14.8 | 2.19 ± 0.21 | 24.6 | |
| L-Aspartic Acid | 0.001 | 3.60 ± 0.10 | −3.75 | 4.80 ± 0.10 | −1.48 | 2.79 ± 0.08 | 3.68 |
| 0.01 | 3.53 ± 0.06 | −0.73 | 4.77 ± 0.06 | −0.85 | 1.24 ± 0.13 * | 57.36 | |
| 0.1 | 3.23 ± 0.06 | 6.92 | 4.60 ± 0.10 | 2.75 | 0.57 ± 0.14 * | 80.46 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Z.; Guo, W.; Mo, C.; Tang, R.; He, L.; Du, L.; Li, M.; Wu, H.; Tang, X.; Huang, Z.; et al. Root Metabolism and Effects of Root Exudates on the Growth of Ralstonia solanacearum and Fusarium moniliforme Were Significantly Different between the Two Genotypes of Peanuts. Genes 2023, 14, 528. https://doi.org/10.3390/genes14020528
Li Z, Guo W, Mo C, Tang R, He L, Du L, Li M, Wu H, Tang X, Huang Z, et al. Root Metabolism and Effects of Root Exudates on the Growth of Ralstonia solanacearum and Fusarium moniliforme Were Significantly Different between the Two Genotypes of Peanuts. Genes. 2023; 14(2):528. https://doi.org/10.3390/genes14020528
Chicago/Turabian StyleLi, Zhong, Wenfeng Guo, Changming Mo, Ronghua Tang, Liangqiong He, Lin Du, Ming Li, Haining Wu, Xiumei Tang, Zhipeng Huang, and et al. 2023. "Root Metabolism and Effects of Root Exudates on the Growth of Ralstonia solanacearum and Fusarium moniliforme Were Significantly Different between the Two Genotypes of Peanuts" Genes 14, no. 2: 528. https://doi.org/10.3390/genes14020528
APA StyleLi, Z., Guo, W., Mo, C., Tang, R., He, L., Du, L., Li, M., Wu, H., Tang, X., Huang, Z., & Wu, X. (2023). Root Metabolism and Effects of Root Exudates on the Growth of Ralstonia solanacearum and Fusarium moniliforme Were Significantly Different between the Two Genotypes of Peanuts. Genes, 14(2), 528. https://doi.org/10.3390/genes14020528





