Harnessing Genomic Analysis to Explore the Role of Telomeres in the Pathogenesis and Progression of Diabetic Kidney Disease
Abstract
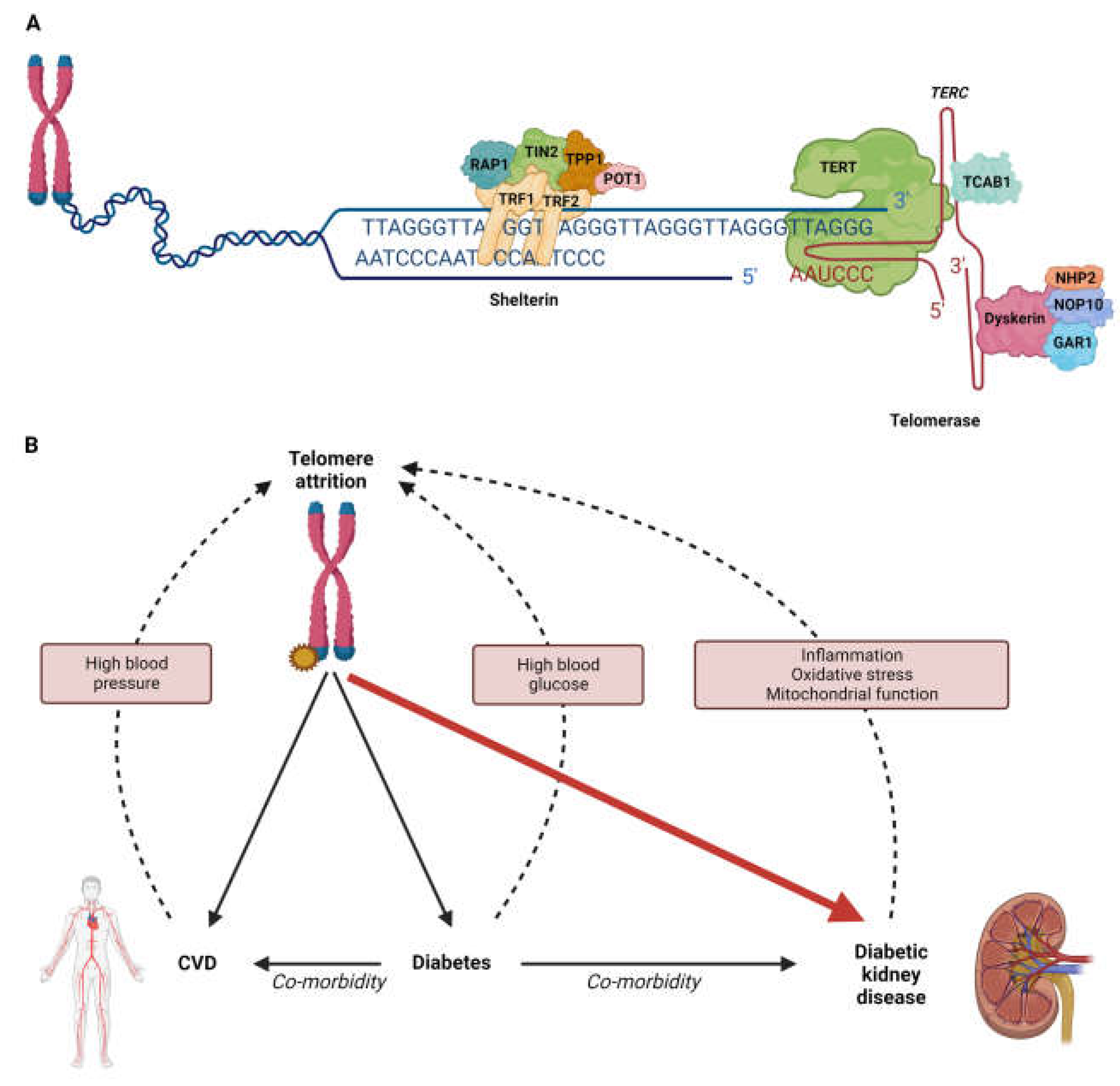
:1. Introduction
2. Genomic Analysis Provides New Insights to Improve Our Understanding of Kidney Disease
3. Diabetic Kidney Disease and Ageing
4. Associations between Genetically Determined Telomere Length and Disease
5. Genetic Variation Influencing Telomere Regulation in Diabetic Kidney Disease
6. Epigenetic Variation Influencing Telomere Regulation in Diabetic Kidney Disease
7. Therapeutic Targeting of Telomere Regulation in Diabetic Kidney Disease
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lin, X.; Xu, Y.; Pan, X.; Xu, J.; Ding, Y.; Sun, X.; Song, X.; Ren, Y.; Shan, P.F. Global, Regional, and National Burden and Trend of Diabetes in 195 Countries and Territories: An Analysis from 1990 to 2025. Sci. Rep. 2020, 10, 14790. [Google Scholar] [CrossRef] [PubMed]
- International Diabetes Federation. IDF Diabetes Atlas, 10th ed.; International Diabetes Federation: Brussels, Belgium, 2021; ISBN 9782930229874. [Google Scholar]
- Jitraknatee, J.; Ruengorn, C.; Nochaiwong, S. Prevalence and Risk Factors of Chronic Kidney Disease among Type 2 Diabetes Patients: A Cross-Sectional Study in Primary Care Practice. Sci. Rep. 2020, 10, 6205. [Google Scholar] [CrossRef] [Green Version]
- Costacou, T.; Orchard, T.J. Cumulative Kidney Complication Risk by 50 Years of Type 1 Diabetes: The Effects of Sex, Age, and Calendar Year at Onset. Diabetes Care 2018, 41, 426–433. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Perkins, B.A.; Bebu, I.; De Boer, I.H.; Molitch, M.; Tamborlane, W.; Lorenzi, G.; Herman, W.; White, N.H.; Pop-busui, R.; Paterson, A.D. Risk Factors for Kidney Disease in Type 1 Diabetes. Diabetes Care 2019, 42, 883–890. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.J.; Cardwell, C.R.; Patterson, C.C.; Maxwell, A.P.; Magee, G.M.; Young, R.J.; Matthews, B.; O’Donoghue, D.J.; Fogarty, D.G. Chronic Kidney Disease and Diabetes in the National Health Service: A Cross-Sectional Survey of the UK National Diabetes Audit. Diabet. Med. 2014, 31, 448–454. [Google Scholar] [CrossRef] [PubMed]
- Bruno, G.; Runzo, C.; Cavallo-Perin, P.; Merletti, F.; Rivetti, M.; Pinach, S.; Novelli, G.; Trovati, M.; Cerutti, F.; Pagano, G. Incidence of Type 1 and Type 2 Diabetes in Adults Aged 30-49 Years: The Population-Based Registry in the Province of Turin, Italy. Diabetes Care 2005, 28, 2613–2619. [Google Scholar] [CrossRef] [Green Version]
- Holman, N.; Young, B.; Gadsby, R. Current Prevalence of Type 1 and Type 2 Diabetes in Adults and Children in the UK. Diabet. Med. 2015, 32, 1119–1120. [Google Scholar] [CrossRef] [PubMed]
- The Renal Association. UK Renal Registry 22nd Annual Report–Data to 31/12/2018; The Renal Association: Bristol, UK, 2020; p. 3349. Available online: http://renal.org/audit-research/annual-report (accessed on 8 February 2023).
- United States Renal Data System. USRDS Annual Data Report: Epidemiology of Kidney Disease in the United States; National Institutes of Health, National Institute of Diabetes and Digestive and Kidney Diseases: Bethesda, MD, USA, 2020. [Google Scholar]
- Deng, Y.; Li, N.; Wu, Y.; Wang, M.; Yang, S.; Zheng, Y.; Deng, X.; Xiang, D.; Zhu, Y.; Xu, P.; et al. Global, Regional, and National Burden of Diabetes-Related Chronic Kidney Disease From 1990 to 2019. Front. Endocrinol. 2021, 12, 672350. [Google Scholar] [CrossRef]
- Smyth, L.J.; Duffy, S.; Maxwell, A.P.; McKnight, A.J. Genetic and Epigenetic Factors Influencing Chronic Kidney Disease. Am. J. Physiol. Ren. Physiol. 2014, 307, F757–F776. [Google Scholar] [CrossRef] [Green Version]
- McKnight, A.J.; Duffy, S.; Maxwell, A.P. Genetics of Diabetic Nephropathy: A Long Road of Discovery. Curr. Diab. Rep. 2015, 15, 41. [Google Scholar] [CrossRef]
- Harjutsalo, V.; Groop, P.H. Epidemiology and Risk Factors for Diabetic Kidney Disease. Adv. Chronic Kidney Dis. 2014, 21, 260–266. [Google Scholar] [CrossRef] [PubMed]
- Cañadas-Garre, M.; Anderson, K.; McGoldrick, J.; Maxwell, A.P.; McKnight, A.J. Genomic Approaches in the Search for Molecular Biomarkers in Chronic Kidney Disease. J. Transl. Med. 2018, 16, 292. [Google Scholar] [CrossRef] [Green Version]
- Cañadas-Garre, M.; Anderson, K.; Cappa, R.; Skelly, R.; Smyth, L.J.; McKnight, A.J.; Maxwell, A.P. Genetic Susceptibility to Chronic Kidney Disease—Some More Pieces for the Heritability Puzzle. Front. Genet. 2019, 10, 453. [Google Scholar] [CrossRef]
- Stanzick, K.J.; Li, Y.; Schlosser, P.; Gorski, M.; Wuttke, M.; Thomas, L.F.; Rasheed, H.; Rowan, B.X.; Graham, S.E.; Vanderweff, B.R.; et al. Discovery and Prioritization of Variants and Genes for Kidney Function in >1.2 Million Individuals. Nat. Commun. 2021, 12, 4350. [Google Scholar] [CrossRef]
- Gorski, M.; Jung, B.; Li, Y.; Matias-Garcia, P.R.; Wuttke, M.; Coassin, S.; Thio, C.H.L.; Kleber, M.E.; Winkler, T.W.; Wanner, V.; et al. Meta-Analysis Uncovers Genome-Wide Significant Variants for Rapid Kidney Function Decline. Kidney Int. 2021, 99, 926–939. [Google Scholar] [CrossRef] [PubMed]
- Hishida, A.; Nakatochi, M.; Akiyama, M.; Kamatani, Y.; Nishiyama, T.; Ito, H.; Oze, I.; Nishida, Y.; Hara, M.; Takashima, N.; et al. Genome-Wide Association Study of Renal Function Traits: Results from the Japan Multi-Institutional Collaborative Cohort Study. Am. J. Nephrol. 2018, 47, 304–316. [Google Scholar] [CrossRef]
- Hernandez-Fuentes, M.P.; Franklin, C.; Rebollo-Mesa, I.; Mollon, J.; Delaney, F.; Perucha, E.; Stapleton, C.; Borrows, R.; Byrne, C.; Cavalleri, G.; et al. Long- and Short-Term Outcomes in Renal Allografts with Deceased Donors: A Large Recipient and Donor Genome-Wide Association Study. Am. J. Transplant. 2018, 18, 1370–1379. [Google Scholar] [CrossRef] [Green Version]
- Gorski, M.; Van Der Most, P.J.; Teumer, A.; Chu, A.Y.; Li, M.; Mijatovic, V.; Nolte, I.M.; Cocca, M.; Taliun, D.; Gomez, F.; et al. 1000 Genomes-Based Metaanalysis Identifies 10 Novel Loci for Kidney Function. Sci. Rep. 2017, 7, 45040. [Google Scholar] [CrossRef] [Green Version]
- Pattaro, C.; Teumer, A.; Gorski, M.; Chu, A.Y.; Li, M.; Mijatovic, V.; Garnaas, M.; Tin, A.; Sorice, R.; Li, Y.; et al. Genetic Associations at 53 Loci Highlight Cell Types and Biological Pathways Relevant for Kidney Function. Nat. Commun. 2016, 7, 10023. [Google Scholar] [CrossRef] [Green Version]
- Parsa, A.; Kanetsky, P.A.; Xiao, R.; Gupta, J.; Mitra, N.; Limou, S.; Xie, D.; Xu, H.; Anderson, A.H.; Ojo, A.; et al. Genome-Wide Association of CKD Progression: The Chronic Renal Insufficiency Cohort Study. J. Am. Soc. Nephrol. 2017, 28, 923–934. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morris, A.P.; Le, T.H.; Wu, H.; Akbarov, A.; van der Most, P.J.; Hemani, G.; Smith, G.D.; Mahajan, A.; Gaulton, K.J.; Nadkarni, G.N.; et al. Trans-Ethnic Kidney Function Association Study Reveals Putative Causal Genes and Effects on Kidney-Specific Disease Aetiologies. Nat. Commun. 2019, 10, 29. [Google Scholar] [CrossRef] [Green Version]
- Sinnott-Armstrong, N.; Tanigawa, Y.; Amar, D.; Mars, N.; Benner, C.; Aguirre, M.; Venkataraman, G.R.; Wainberg, M.; Ollila, H.M.; Kiiskinen, T.; et al. Genetics of 35 Blood and Urine Biomarkers in the UK Biobank. Nat. Genet. 2021, 53, 185–194. [Google Scholar] [CrossRef]
- Wuttke, M.; Li, Y.; Li, M.; Sieber, K.B.; Feitosa, M.F.; Gorski, M.; Tin, A.; Wang, L.; Chu, A.Y.; Hoppmann, A.; et al. A Catalog of Genetic Loci Associated with Kidney Function from Analyses of a Million Individuals. Nat. Genet. 2019, 51, 957–972. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turchin, M.C.; Stephens, M. Bayesian Multivariate Reanalysis of Large Genetic Studies Identifies Many New Associations. PLoS Genet. 2019, 15, e1008431. [Google Scholar] [CrossRef] [Green Version]
- Wojcik, G.L.; Graff, M.; Nishimura, K.K.; Tao, R.; Haessler, J.; Gignoux, C.R.; Highland, H.M.; Patel, Y.M.; Sorokin, E.P.; Avery, C.L.; et al. Genetic Analyses of Diverse Populations Improves Discovery for Complex Traits. Nature 2019, 570, 514–518. [Google Scholar] [CrossRef]
- Lin, B.M.; Nadkarni, G.N.; Tao, R.; Graff, M.; Fornage, M.; Buyske, S.; Matise, T.C.; Highland, H.M.; Wilkens, L.R.; Carlson, C.S.; et al. Genetics of Chronic Kidney Disease Stages across Ancestries: The PAGE Study. Front. Genet. 2019, 10, 494. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graham, S.E.; Nielsen, J.B.; Zawistowski, M.; Zhou, W.; Fritsche, L.G.; Gabrielsen, M.E.; Skogholt, A.H.; Surakka, I.; Hornsby, W.E.; Fermin, D.; et al. Sex-Specific and Pleiotropic Effects Underlying Kidney Function Identified from GWAS Meta-Analysis. Nat. Commun. 2019, 10, 1847. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yun, S.; Han, M.; Kim, H.J.; Kim, H.; Kang, E.; Kim, S.; Ahn, C.; Oh, K.H. Genetic Risk Score Raises the Risk of Incidence of Chronic Kidney Disease in Korean General Population-Based Cohort. Clin. Exp. Nephrol. 2019, 23, 995–1003. [Google Scholar] [CrossRef]
- Yamada, Y.; Kato, K.; Oguri, M.; Horibe, H.; Fujimaki, T.; Yasukochi, Y.; Takeuchi, I.; Sakuma, J. Identification of 13 Novel Susceptibility Loci for Early-Onset Myocardial Infarction, Hypertension, or Chronic Kidney Disease. Int. J. Mol. Med. 2018, 42, 2415–2436. [Google Scholar] [CrossRef] [Green Version]
- Langefeld, C.D.; Comeau, M.E.; Ng, M.C.Y.; Guan, M.; Dimitrov, L.; Mudgal, P.; Spainhour, M.H.; Julian, B.A.; Edberg, J.C.; Croker, J.A.; et al. Genome-Wide Association Studies Suggest That APOL1-Environment Interactions More Likely Trigger Kidney Disease in African Americans with Nondiabetic Nephropathy than Strong APOL1–Second Gene Interactions. Kidney Int. 2018, 94, 599–607. [Google Scholar] [CrossRef] [Green Version]
- Taira, M.; Imamura, M.; Takahashi, A.; Kamatani, Y.; Yamauchi, T.; Araki, S.I.; Tanaka, N.; Van Zuydam, N.R.; Ahlqvist, E.; Toyoda, M.; et al. A Variant within the FTO Confers Susceptibility to Diabetic Nephropathy in Japanese Patients with Type 2 Diabetes. PLoS ONE 2018, 13, e0208654. [Google Scholar] [CrossRef] [PubMed]
- Sandholm, N.; Van Zuydam, N.; Ahlqvist, E.; Juliusdottir, T.; Deshmukh, H.A.; Rayner, N.W.; Di Camillo, B.; Forsblom, C.; Fadista, J.; Ziemek, D.; et al. The Genetic Landscape of Renal Complications in Type 1 Diabetes. J. Am. Soc. Nephrol. 2017, 28, 557–574. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guan, M.; Keaton, J.M.; Dimitrov, L.; Hicks, P.J.; Xu, J.; Palmer, N.D.; Ma, L.; Das, S.K.; Chen, Y.D.I.; Coresh, J.; et al. Genome-Wide Association Study Identifies Novel Loci for Type 2 Diabetes-Attributed End-Stage Kidney Disease in African Americans. Hum. Genomics 2019, 13, 21. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salem, R.M.; Todd, J.N.; Sandholm, N.; Cole, J.B.; Chen, W.M.; Andrews, D.; Pezzolesi, M.G.; Mc, P.M.K.; Hiraki, L.T.; Qiu, C.; et al. Genome-Wide Association Study of Diabetic Kidney Disease Highlights Biology Involved in Glomerular Basement Membrane Collagen. J. Am. Soc. Nephrol. 2019, 30, 2000–2016. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vujkovic, M.; Keaton, J.M.; Lynch, J.A.; Miller, D.R.; Zhou, J.; Tcheandjieu, C.; Huffman, J.E.; Assimes, T.L.; Lorenz, K.; Zhu, X.; et al. Discovery of 318 New Risk Loci for Type 2 Diabetes and Related Vascular Outcomes among 1.4 Million Participants in a Multi-Ancestry Meta-Analysis. Nat. Genet. 2020, 52, 680–691. [Google Scholar] [CrossRef]
- Ustinova, M.; Peculis, R.; Rescenko, R.; Rovite, V.; Zaharenko, L.; Elbere, I.; Silamikele, L.; Konrade, I.; Sokolovska, J.; Pirags, V.; et al. Novel Susceptibility Loci Identified in a Genome-Wide Association Study of Type 2 Diabetes Complications in Population of Latvia. BMC Med. Genomics 2021, 14, 18. [Google Scholar] [CrossRef]
- van Zuydam, N.R.; Ahlqvist, E.; Sandholm, N.; Deshmukh, H.; Rayner, N.W.; Abdalla, M.; Ladenvall, C.; Ziemek, D.; Fauman, E.; Robertson, N.R.; et al. A Genome-Wide Association Study of Diabetic Kidney Disease in Subjects With Type 2 Diabetes. Diabetes 2018, 67, 1414–1427. [Google Scholar] [CrossRef] [Green Version]
- Swan, E.J.; Salem, R.M.; Sandholm, N.; Tarnow, L.; Rossing, P.; Lajer, M.; Groop, P.H.; Maxwell, A.P.; Mcknight, A.J. The GENIE Consortium Genetic Risk Factors Affecting Mitochondrial Function Are Associated with Kidney Disease in People with Type 1 Diabetes. Diabet. Med. 2015, 32, 1104–1109. [Google Scholar] [CrossRef] [Green Version]
- Teumer, A.; Li, Y.; Ghasemi, S.; Prins, B.P.; Wuttke, M.; Hermle, T.; Giri, A.; Sieber, K.B.; Qiu, C.; Kirsten, H.; et al. Genome-Wide Association Meta-Analyses and Fine-Mapping Elucidate Pathways Influencing Albuminuria. Nat. Commun. 2019, 10, 4130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liao, L.N.; Li, T.C.; Li, C.I.; Liu, C.S.; Lin, W.Y.; Lin, C.H.; Yang, C.W.; Chen, C.C.; Chang, C.T.; Yang, Y.F.; et al. Genetic Risk Score for Risk Prediction of Diabetic Nephropathy in Han Chinese Type 2 Diabetes Patients. Sci. Rep. 2019, 9, 19897. [Google Scholar] [CrossRef] [Green Version]
- Smyth, L.J.; McKay, G.J.; Maxwell, A.P.; McKnight, A.J. DNA Hypermethylation and DNA Hypomethylation Is Present at Different Loci in Chronic Kidney Disease. Epigenetics 2013, 9, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Zhuo, C.; Feng, M.; Andrew, D.P.; John, M.L.; Lingxiao, Z.; Dustin, E.S.; Xiwei, W.; Jinhui, W.; Joshua, D.T.; Saul, G.; et al. Epigenomic Profiling Reveals an Association between Persistence of DNA Methylation and Metabolic Memory in the DCCT/EDIC Type 1 Diabetes Cohort. Proc. Natl. Acad. Sci. USA 2016, 113, E3002–E3011. [Google Scholar] [CrossRef] [Green Version]
- Sapienza, C.; Lee, J.; Powell, J.; Erinle, O.; Yafai, F.; Reichert, J.; Siraj, E.S.; Madaio, M. DNA Methylation Profling Identifes Epigenetic Differences between Diabetes Patients with ESRD and Diabetes Patients without Nephropathy. Epigenetics 2011, 6, 20–28. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, A.Y.; Tin, A.; Schlosser, P.; Ko, Y.A.; Qiu, C.; Yao, C.; Joehanes, R.; Grams, M.E.; Liang, L.; Gluck, C.A.; et al. Epigenome-Wide Association Studies Identify DNA Methylation Associated with Kidney Function. Nat. Commun. 2017, 8, 1286. [Google Scholar] [CrossRef] [Green Version]
- McKnight, A.J.; McKay, G.J.; Maxwell, A.P. Genetic and Epigenetic Risk Factors for Diabetic Kidney Disease. Adv. Chronic Kidney Dis. 2014, 21, 287–296. [Google Scholar] [CrossRef] [PubMed]
- Swan, E.J.; Maxwell, A.P.; Mcknight, A.J. Distinct Methylation Patterns in Genes That Affect Mitochondrial Function Are Associated with Kidney Disease in Blood-Derived DNA from Individuals with Type 1 Diabetes. Diabet. Med. 2015, 32, 1110–1115. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Miao, F.; Braffett, B.H.; Lachin, J.M.; Zhang, L.; Wu, X.; Roshandel, D.; Carless, M.; Li, X.A.; Tompkins, J.D.; et al. DNA Methylation Mediates HbA1c-Associated Complications Development in Type 1 Diabetes. Nat. Metab. 2020, 2, 744–762. [Google Scholar] [CrossRef]
- Smyth, L.J.; Maxwell, A.P.; Benson, K.A.; Kilner, J.; McKay, G.J.; McKnight, A.J. Validation of Differentially Methylated MicroRNAs Identified from an Epigenome-Wide Association Study; Sanger and next Generation Sequencing Approaches. BMC Res. Notes 2018, 11, 767. [Google Scholar] [CrossRef] [Green Version]
- Park, J.; Guan, Y.; Sheng, X.; Gluck, C.; Seasock, M.J.; Ari Hakimi, A.; Qiu, C.; Pullman, J.; Verma, A.; Li, H.; et al. Functional Methylome Analysis of Human Diabetic Kidney Disease. JCI Insight 2019, 4, e128886. [Google Scholar] [CrossRef] [Green Version]
- Smyth, L.J.; Kilner, J.; Nair, V.; Liu, H.; Brennan, E.; Kerr, K.; Sandholm, N.; Cole, J.; Dahlström, E.; Syreeni, A.; et al. Assessment of Differentially Methylated Loci in Individuals with End-Stage Kidney Disease Attributed to Diabetic Kidney Disease: An Exploratory Study. Clin. Epigenetics 2021, 13, 99. [Google Scholar] [CrossRef]
- Smyth, L.J.; Patterson, C.C.; Swan, E.J.; Maxwell, A.P.; McKnight, A.J. DNA Methylation Associated With Diabetic Kidney Disease in Blood-Derived DNA. Front. Cell Dev. Biol. 2020, 8, 561907. [Google Scholar] [CrossRef] [PubMed]
- Marumo, T.; Yagi, S.; Kawarazaki, W.; Nishimoto, M. Diabetes Induces Aberrant DNA Methylation in the Proximal Tubules of the Kidney. J. Am. Soc. Nephrol. 2015, 26, 2388–2397. [Google Scholar] [CrossRef] [Green Version]
- Kato, M.; Natarajan, R. Epigenetics and Epigenomics in Diabetic Kidney Disease and Metabolic Memory. Nat. Rev. Nephrol. 2019, 15, 327–345. [Google Scholar] [CrossRef]
- Shiels, P.G.; Painer, J.; Natterson-Horowitz, B.; Johnson, R.J.; Miranda, J.J.; Stenvinkel, P. Manipulating the Exposome to Enable Better Ageing. Biochem. J. 2021, 478, 2889–2898. [Google Scholar] [CrossRef] [PubMed]
- Mafra, D.; Borges, N.A.; Lindholm, B.; Shiels, P.G.; Evenepoel, P.; Stenvinkel, P. Food as Medicine: Targeting the Uraemic Phenotype in Chronic Kidney Disease. Nat. Rev. Nephrol. 2021, 17, 153–171. [Google Scholar] [CrossRef]
- Mohtat, D.; Susztak, K. Fine Tuning Gene Expression: The Epigenome. Semin. Nephrol. 2010, 30, 468–476. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moore, L.D.; Le, T.; Fan, G. DNA Methylation and Its Basic Function. Neuropsychopharmacology 2013, 38, 23–38. [Google Scholar] [CrossRef] [Green Version]
- Holliday, R.; Pugh, J.E. DNA Modification Mechanisms and Gene Activity during Development. Science 1975, 187, 226–232. [Google Scholar] [CrossRef]
- Compere, S.J.; Palmiter, R.D. DNA Methylation Control the Inducibility of the Mouse Metallothionein-I Gene in Lympohoid Cells. Cell 1981, 25, 233–240. [Google Scholar] [CrossRef]
- Allfrey, G.; Faulkner, R.; Mirsky, A.E. Acetylation and Methylation of Histones and Their Possible Role in Regulation of RNA Synthesis. Biochemistry 1964, 51, 786–794. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Sabari, B.R.; Garcia, B.A.; David Allis, C.; Zhao, Y. SnapShot: Histone Modifications. Cell 2014, 159, 458–458.e1. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Heyn, H.; Esteller, M. DNA Methylation Profiling in the Clinic: Applications and Challenges. Nat. Rev. Genet. 2012, 13, 679–692. [Google Scholar] [CrossRef]
- Kelly, T.K.; De Carvalho, D.D.; Jones, P.A. Epigenetic Modifications as Therapeutic Targets. Nat. Biotechnol. 2010, 28, 1069–1078. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Zhang, Q.; Liu, S.; Chen, Y.; Li, R.; Lin, T.; Yu, C.; Zhang, H.; Huang, Z.; Zhao, X.; et al. DNA Methyltransferase 1 May Be a Therapy Target for Attenuating Diabetic Nephropathy and Podocyte Injury. Kidney Int. 2017, 92, 140–153. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Ge, C.; Tan, J.; Sun, Y.; Kuang, Q.; Dai, X.; Zhong, S.; Yi, C.; Hu, L.F.; Lou, D.S.; et al. Juglanin Protects against High Fat Diet-Induced Renal Injury by Suppressing Inflammation and Dyslipidemia via Regulating NF-ΚB/HDAC3 Signaling. Int. Immunopharmacol. 2021, 95, 107340. [Google Scholar] [CrossRef]
- Shan, Q.; Zheng, G.; Zhu, A.; Cao, L.; Lu, J.; Wu, D.; Zhang, Z.; Fan, S.; Sun, C.; Hu, B.; et al. Epigenetic Modification of MiR-10a Regulates Renal Damage by Targeting CREB1 in Type 2 Diabetes Mellitus. Toxicol. Appl. Pharmacol. 2016, 306, 134–143. [Google Scholar] [CrossRef]
- Zhang, L.; Cao, W. Histone Deacetylase 3 (HDAC3) as an Important Epigenetic Regulator of Kidney Diseases. J. Mol. Med. 2021, 100, 43–51. [Google Scholar] [CrossRef]
- Wilson, V.L.; Smith, R.A.; Mag, S.; Cutler, R.G. Genomic 5-Methyldeoxycytidine Decreases with Age. J. Biol. Chem. 1987, 262, 9948–9951. [Google Scholar] [CrossRef]
- Heyn, H.; Li, N.; Ferreira, H.J.; Moran, S.; Pisano, D.G.; Gomez, A.; Diez, J. Distinct DNA Methylomes of Newborns and Centenarians. PNAS 2012, 109, 10522–10527. [Google Scholar] [CrossRef] [Green Version]
- López-Otín, C.; Blasco, M.A.; Partridge, L.; Serrano, M.; Kroemer, G. The Hallmarks of Aging. Cell 2013, 153, 1194. [Google Scholar] [CrossRef] [Green Version]
- Horvath, S. DNA Methylation Age of Human Tissues and Cell Types DNA Methylation Age of Human Tissues and Cell Types. Genome Biol. 2013, 14, R15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bocklandt, S.; Lin, W.; Sehl, M.E.; Sánchez, F.J.; Sinsheimer, J.S.; Horvath, S.; Vilain, E. Epigenetic Predictor of Age. PLoS ONE 2011, 6, e14821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Weidner, C.I.; Lin, Q.; Koch, C.M.; Eisele, L.; Beier, F.; Ziegler, P.; Bauerschlag, D.O.; Jöckel, K.H.; Erbel, R.; Mühleisen, T.W.; et al. Aging of Blood Can Be Tracked by DNA Methylation Changes at Just Three CpG Sites. Genome Biol. 2014, 15, R24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, J.; Miao, K.; Wang, H.; Ding, H.; Wang, D.W. Association between Telomere Length and Type 2 Diabetes Mellitus: A Meta-Analysis. PLoS ONE 2013, 8, e79993. [Google Scholar] [CrossRef] [PubMed]
- Jeanclos, E.; Krolewski, A.; Skurnick, J.; Kimura, M.; Aviv, H.; Warram, J.H.; Aviv, A. Shortened Telomere Length in White Blood Cells of Patients with IDDM. Diabetes 1998, 47, 482–486. [Google Scholar] [CrossRef]
- Adaikalakoteswari, A.; Balasubramanyam, M.; Mohan, V. Telomere Shortening Occurs in Asian Indian Type 2 Diabetic Patients. Diabet. Med. 2005, 22, 1151–1156. [Google Scholar] [CrossRef] [Green Version]
- Testa, R.; Olivieri, F.; Sirolla, C.; Spazzafumo, L.; Rippo, M.R.; Marra, M.; Bonfigli, A.R.; Ceriello, A.; Antonicelli, R.; Franceschi, C.; et al. Leukocyte Telomere Length Is Associated with Complications of Type 2 Diabetes Mellitus. Diabet. Med. 2011, 28, 1388–1394. [Google Scholar] [CrossRef]
- White, W.E. Aging and Uremia: Is There Cellular and Molecular Crossover? World J. Nephrol. 2015, 4, 19–30. [Google Scholar] [CrossRef]
- Guo, J.; Zheng, H.J.; Zhang, W.; Lou, W.; Xia, C.; Han, X.T.; Huang, W.J.; Zhang, F.; Wang, Y.; Liu, W.J. Accelerated Kidney Aging in Diabetes Mellitus. Oxid. Med. Cell. Longev. 2020, 2020, 1234059. [Google Scholar] [CrossRef]
- Liu, J.; Yang, J.R.; Chen, X.M.; Cai, G.Y.; Lin, L.R.; He, Y.N. Impact of ER Stress-Regulated ATF4/P16 Signaling on the Premature Senescence of Renal Tubular Epithelial Cells in Diabetic Nephropathy. Am. J. Physiol. Cell Physiol. 2015, 308, C621–C630. [Google Scholar] [CrossRef] [Green Version]
- Verzola, D.; Gandolfo, M.T.; Gaetani, G.; Ferraris, A.; Mangerini, R.; Ferrario, F.; Villaggio, B.; Gianiorio, F.; Tosetti, F.; Weiss, U.; et al. Accelerated Senescence in the Kidneys of Patients with Type 2 Diabetic Nephropathy. Am. J. Physiol. Ren. Physiol. 2008, 295, F1563–F1573. [Google Scholar] [CrossRef] [Green Version]
- Shen, S.; Ji, C.; Wei, K. Cellular Senescence and Regulated Cell Death of Tubular Epithelial Cells in Diabetic Kidney Disease. Front. Endocrinol. 2022, 13, 924299. [Google Scholar] [CrossRef]
- Kooman, J.P.; Dekker, M.J.; Usvyat, L.A.; Kotanko, P.; van der Sande, F.M.; Schalkwijk, C.G.; Shiels, P.G.; Stenvinkel, P. Inflammation and Premature Aging in Advanced Chronic Kidney Disease. Am. J. Physiol. Ren. Physiol. 2017, 313, F938–F950. [Google Scholar] [CrossRef] [Green Version]
- Shiels, P.G.; McGuinness, D.; Eriksson, M.; Kooman, J.P.; Stenvinkel, P. The Role of Epigenetics in Renal Ageing. Nat. Rev. Nephrol. 2017, 13, 471–482. [Google Scholar] [CrossRef] [Green Version]
- Kooman, J.P.; Kotanko, P.; Schols, A.M.W.J.; Shiels, P.G.; Stenvinkel, P. Chronic Kidney Disease and Premature Ageing. Nat. Rev. Nephrol. 2014, 10, 732–742. [Google Scholar] [CrossRef]
- Serrano, M.; Blasco, M.A. Putting the Stress on Senescence. Curr. Opin. Cell Biol. 2001, 13, 748–753. [Google Scholar] [CrossRef]
- Mir, S.M.; Tehrani, S.S.; Goodarzi, G.; Jamalpoor, Z.; Asadi, J.; Khelghati, N.; Qujeq, D.; Maniati, M. Telomeres and Telomerase in Cardiovascular Diseases. Clin. Interv. Aging 2020, 15, 827–839. [Google Scholar] [CrossRef]
- Haycock, P.C.; Heydon, E.E.; Kaptoge, S.; Butterworth, A.S.; Thompson, A.; Willeit, P. Leucocyte Telomere Length and Risk of Cardiovascular Disease: Systematic Review and Meta- Analysis. BMJ 2014, 349, g4277. [Google Scholar] [CrossRef] [Green Version]
- Yeh, J.K.; Wang, C.Y. Telomeres and Telomerase in Cardiovascular Diseases. Genes 2016, 7, 58. [Google Scholar] [CrossRef] [Green Version]
- Spyridopoulos, I.; Von Zglinicki, T. Telomere Length Predicts Cardiovascular Disease: Measurement in Humans Is Unlikely to Be Useful until We Find out How and Why. BMJ 2014, 349, g4373. [Google Scholar] [CrossRef]
- De Vusser, K.; Pieters, N.; Janssen, B.; Lerut, E.; Kuypers, D.; Jochmans, I.; Monbaliu, D.; Pirenne, J.; Nawrot, T.; Naesens, M. Telomere Length, Cardiovascular Risk and Arteriosclerosis in Human Kidneys: An Observational Cohort Study. Aging 2015, 7, 766–775. [Google Scholar] [CrossRef] [Green Version]
- Kirchner, H.; Shaheen, F.; Kalscheuer, H.; Schmid, S.M.; Oster, H.; Lehnert, H. The Telomeric Complex and Metabolic Disease. Genes 2017, 8, 176. [Google Scholar] [CrossRef] [Green Version]
- Gurung, R.L.; M, Y.; Liu, S.; Liu, J.J.; Lim, S.C. Short Leukocyte Telomere Length Predicts Albuminuria Progression in Individuals with Type 2 Diabetes. Kidney Int. Rep. 2018, 3, 592–601. [Google Scholar] [CrossRef] [Green Version]
- Ameh, O.I.; Okpechi, I.G.; Dandara, C.; Kengne, A.P. Association between Telomere Length, Chronic Kidney Disease, and Renal Traits: A Systematic Review. Omi. A J. Integr. Biol. 2017, 21, 143–155. [Google Scholar] [CrossRef]
- Fazzini, F.; Lamina, C.; Raschenberger, J.; Schultheiss, U.T.; Kotsis, F.; Schönherr, S.; Weissensteiner, H.; Forer, L.; Steinbrenner, I.; Meiselbach, H.; et al. Results from the German Chronic Kidney Disease (GCKD) Study Support Association of Relative Telomere Length with Mortality in a Large Cohort of Patients with Moderate Chronic Kidney Disease. Kidney Int. 2020, 98, 488–497. [Google Scholar] [CrossRef]
- Mazidi, M.; Rezaie, P.; Covic, A.; Malyszko, J.; Rysz, J.; Kengne, A.P.; Banach, M. Telomere Attrition, Kidney Function, and Prevalent Chronic Kidney Disease in the United States. Oncotarget 2017, 8, 80175–80181. [Google Scholar] [CrossRef] [Green Version]
- Carrero, J.J.; Shiels, P.G.; Stenvinkel, P. Telomere Biology Alterations as a Mortality Risk Factor in CKD. Am. J. Kidney Dis. 2008, 51, 1076–1077. [Google Scholar] [CrossRef]
- Carrero, J.J.; Stenvinkel, P.; Fellström, B.; Qureshi, A.R.; Lamb, K.; Heimbürger, O.; Bárány, P.; Radhakrishnan, K.; Lindholm, B.; Soveri, I.; et al. Telomere Attrition Is Associated with Inflammation, Low Fetuin-A Levels and High Mortality in Prevalent Haemodialysis Patients. J. Intern. Med. 2008, 263, 302–312. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Fogo, A.B. Cell Senescence in the Aging Kidney. J. Am. Soc. Nephrol. 2010, 21, 1436–1439. [Google Scholar] [CrossRef] [Green Version]
- Denic, A.; Glassock, R.J.; Rule, A.D. Structural and Functional Changes With the Aging Kidney. Adv. Chronic Kidney Dis. 2016, 23, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Eguchi, K.; Honig, L.S.; Lee, J.H.; Hoshide, S.; Kario, K. Short Telomere Length Is Associated with Renal Impairment in Japanese Subjects with Cardiovascular Risk. PLoS ONE 2017, 12, e0176138. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramírez, R.; Carracedo, J.; Soriano, S.; Jiménez, R.; Martín-Malo, A.; Rodríguez, M.; Blasco, M.; Aljama, P. Stress-Induced Premature Senescence in Mononuclear Cells from Patients on Long-Term Hemodialysis. Am. J. Kidney Dis. 2005, 45, 353–359. [Google Scholar] [CrossRef] [PubMed]
- van der Harst, P.; Wong, L.S.M.; de Boer, R.A.; Brouilette, S.W.; van der Steege, G.; Voors, A.A.; Hall, A.S.; Samani, N.J.; Wikstrand, J.; van Gilst, W.H.; et al. Possible Association Between Telomere Length and Renal Dysfunction in Patients With Chronic Heart Failure. Am. J. Cardiol. 2008, 102, 207–210. [Google Scholar] [CrossRef] [PubMed]
- Wong, L.S.M.; Van Der Harst, P.; De Boer, R.A.; Codd, V.; Huzen, J.; Samani, N.J.; Hillege, H.L.; Voors, A.A.; Van Gilst, W.H.; Jaarsma, T.; et al. Renal Dysfunction Is Associated with Shorter Telomere Length in Heart Failure. Clin. Res. Cardiol. 2009, 98, 629–634. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, W.G.; Wang, Y.; Hou, K.; Jia, L.P.; Ma, J.; Zhao, D.L.; Zhu, S.Y.; Bai, X.J.; Cai, G.Y.; Wang, Y.P.; et al. A Correlation Study of Telomere Length in Peripheral Blood Leukocytes and Kidney Function with Age. Mol. Med. Rep. 2015, 11, 4359–4364. [Google Scholar] [CrossRef] [Green Version]
- Betjes, M.G.H.; Langerak, A.W.; Van Der Spek, A.; De Wit, E.A.; Litjens, N.H.R. Premature Aging of Circulating T Cells in Patients with End-Stage Renal Disease. Kidney Int. 2011, 80, 208–217. [Google Scholar] [CrossRef] [Green Version]
- Melk, A.; Ramassar, V.; Helms, L.M.H.; Moore, R.; Rayner, D.; Solez, K.; Halloran, P.F. Telomere Shortening in Kidneys with Age. J. Am. Soc. Nephrol. 2000, 11, 444–453. [Google Scholar] [CrossRef]
- Cao, D.W.; Jiang, C.; Wan, C.; Zhang, M.; Zhang, Q.; Zhao, M.; Yang, B.; Zhu, D.; Han, X. Upregulation of MiR-126 Delays the Senescence of Human Glomerular Mesangial Cells Induced by High Glucose via Telomere-P53-P21-Rb Signaling Pathway. Curr. Med. Sci. 2018, 38, 758–764. [Google Scholar] [CrossRef]
- Tentolouris, N.; Nzietchueng, R.; Cattan, V.; Poitevin, G.; Lacolley, P.; Papazafiropoulou, A.; Perrea, D.; Katsilambros, N.; Benetos, A. White Blood Cells Telomere Length Is Shorter in Males with Type 2 Diabetes and Microalbuminuria. Diabetes Care 2007, 30, 2909–2915. [Google Scholar] [CrossRef] [Green Version]
- Cheng, F.; Luk, A.O.; Tam, C.H.T.; Fan, B.; Wu, H.; Yang, A.; Lau, E.S.H.; Ng, A.C.W.; Lim, C.K.P.; Lee, H.M.; et al. Shortened Relative Leukocyte Telomere Length Is Associated with Prevalent and Incident Cardiovascular Complications in Type 2 Diabetes: Analysis from the Hong Kong Diabetes Register. Diabetes Care 2020, 43, 2257–2265. [Google Scholar] [CrossRef]
- Akinnibosun, O.A.; Maier, M.C.; Eales, J.; Tomaszewski, M.; Charchar, F.J. Telomere Therapy for Chronic Kidney Disease. Epigenomics 2022, 14, 1039–1054. [Google Scholar] [CrossRef] [PubMed]
- Patnaik, M.M.; Kamath, P.S.; Simonetto, D.A. Hepatic Manifestations of Telomere Biology Disorders. J. Hepatol. 2018, 69, 736–743. [Google Scholar] [CrossRef]
- Khattar, E.; Tergaonkar, V. The Role of Telomeres and Telomere-Associated Proteins as Components of Interactome in Cell-Signaling Pathways. In Telomere—A Complex End of a Chromosome; Larramendy, M.L., Ed.; IntechOpen: London, UK, 2016; ISBN 978-953-51-2753-6. [Google Scholar]
- Vasan, R.S.; Demissie, S.; Kimura, M.; Cupples, L.A.; Rifai, N.; White, C.; Wang, T.J.; Gardner, J.P.; Cao, X.; Benjamin, E.J.; et al. Association of Leukocyte Telomere Length with Circulating Biomarkers of the Renin-Angiotensin-Aldosterone System: The Framingham Heart Study. Circulation 2008, 117, 1138–1144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Charlton, A.; Garzarella, J.; Jandeleit-Dahm, K.A.M.; Jha, J.C. Oxidative Stress and Inflammation in Renal and Cardiovascular Complications of Diabetes. Biology 2021, 10, 18. [Google Scholar] [CrossRef] [PubMed]
- Reichert, S.; Stier, A. Does Oxidative Stress Shorten Telomeres in Vivo? A Review. Biol. Lett. 2017, 13, 20170164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barnes, R.P.; Fouquerel, E.; Opresko, P.L. The Impact of Oxidative DNA Damage and Stress on Telomere Homeostasis. Mech. Ageing Dev. 2019, 177, 37–45. [Google Scholar] [CrossRef]
- Douglas, A.P.; Vance, D.R.; Kenny, E.M.; Morris, D.W.; Maxwell, A.P.; McKnight, A.J. Next-Generation Sequencing of the Mitochondrial Genome and Association with IgA Nephropathy in a Renal Transplant Population. Sci. Rep. 2014, 4, 7379. [Google Scholar] [CrossRef] [Green Version]
- Zhan, M.; Usman, I.M.; Sun, L.; Kanwar, Y.S. Disruption of Renal Tubular Mitochondrial Quality Control by Myo-Inositol Oxygenase in Diabetic Kidney Disease. J. Am. Soc. Nephrol. 2015, 26, 1304–1321. [Google Scholar] [CrossRef] [Green Version]
- Galvan, D.L.; Green, N.H.; Danesh, F.R. The Hallmarks of Mitochondrial Dysfunction in Chronic Kidney Disease. Kidney Int. 2017, 92, 1051–1057. [Google Scholar] [CrossRef]
- Zheng, Q.; Huang, J.; Wang, G. Mitochondria, Telomeres and Telomerase Subunits. Front. Cell Dev. Biol. 2019, 7, 274. [Google Scholar] [CrossRef] [Green Version]
- Codd, V.; Mangino, M.; Van Der Harst, P.; Braund, P.S.; Kaiser, M.; Beveridge, A.J.; Rafelt, S.; Moore, J.; Nelson, C.; Soranzo, N.; et al. Common Variants near TERC Are Associated with Mean Telomere Length. Nat. Genet. 2010, 42, 197–199. [Google Scholar] [CrossRef] [Green Version]
- Levy, D.; Neuhausen, S.L.; Hunt, S.C.; Kimura, M.; Hwang, S.J.; Chen, W.; Bis, J.C.; Fitzpatrick, A.L.; Smith, E.; Johnson, A.D.; et al. Genome-Wide Association Identifies OBFC1 as a Locus Involved in Human Leukocyte Telomere Biology. Proc. Natl. Acad. Sci. USA 2010, 107, 9293–9298. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Codd, V.; Nelson, C.P.; Albrecht, E.; Mangino, M.; Deelen, J.; Buxton, J.L.; Hottenga, J.J.; Fischer, K.; Esko, T.; Surakka, I.; et al. Identification of Seven Loci Affecting Mean Telomere Length and Their Association with Disease. Nat. Genet. 2013, 45, 422–427. [Google Scholar] [CrossRef] [Green Version]
- Do, S.K.; Yoo, S.S.; Choi, Y.Y.; Choi, J.E.; Jeon, H.S.; Lee, W.K.; Lee, S.Y.; Lee, J.; Cha, S.I.; Kim, C.H.; et al. Replication of the Results of Genome-Wide and Candidate Gene Association Studies on Telomere Length in a Korean Population. Korean J. Intern. Med. 2015, 30, 719–726. [Google Scholar] [CrossRef]
- Du, J.; Zhu, X.; Xie, C.; Dai, N.; Gu, Y.; Zhu, M.; Wang, C.; Gao, Y.; Pan, F.; Ren, C.; et al. Telomere Length, Genetic Variants and Gastric Cancer Risk in a Chinese Population. Carcinogenesis 2015, 36, 963–970. [Google Scholar] [CrossRef] [Green Version]
- Codd, V.; Wang, Q.; Allara, E.; Musicha, C.; Kaptoge, S.; Stoma, S.; Jiang, T.; Hamby, S.E.; Braund, P.S.; Bountziouka, V.; et al. Polygenic Basis and Biomedical Consequences of Telomere Length Variation. Nat. Genet. 2021, 53, 1425–1433. [Google Scholar] [CrossRef]
- van der Spek, A.; Warner, S.C.; Broer, L.; Nelson, C.P.; Vojinovic, D.; Ahmad, S.; Arp, P.P.; Brouwer, R.W.W.; Denniff, M.; van den Hout, M.C.G.N.; et al. Exome Sequencing Analysis Identifies Rare Variants in ATM and RPL8 That Are Associated With Shorter Telomere Length. Front. Genet. 2020, 11, 337. [Google Scholar] [CrossRef] [PubMed]
- Taub, M.A.; Conomos, M.P.; Keener, R.; Pankratz, N.; Reiner, A.P.; Mathias, R.A. Genetic Determinants of Telomere Length from 109,122 Ancestrally Diverse Whole-Genome Sequences in TOPMed. Cell Genomics 2022, 2, 100084. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Stoma, S.; Lotta, L.A.; Warner, S.; Albrecht, E.; Allione, A.; Arp, P.P.; Broer, L.; Buxton, J.L.; Da Silva Couto Alves, A.; et al. Genome-Wide Association Analysis in Humans Links Nucleotide Metabolism to Leukocyte Telomere Length. Am. J. Hum. Genet. 2020, 106, 389–404. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Liu, J.; Cheng, G.; Dai, M.; Liu, J.; Qi, Z.; Zhao, J.; Li, W.; Kong, F.; Liu, G.; et al. The Telomerase Gene Polymorphisms, but Not Telomere Length, Increase Susceptibility to Primary Glomerulonephritis/End Stage Renal Diseases in Females. J. Transl. Med. 2020, 18, 184. [Google Scholar] [CrossRef]
- Park, S.; Lee, S.; Kim, Y.; Cho, S.; Kim, K.; Kim, Y.C.; Han, S.S.; Lee, H.; Lee, J.P.; Joo, K.W.; et al. A Mendelian Randomization Study Found Causal Linkage between Telomere Attrition and Chronic Kidney Disease. Kidney Int. 2021, 100, 1063–1070. [Google Scholar] [CrossRef]
- Dorajoo, R.; Chang, X.; Gurung, R.L.; Li, Z.; Wang, L.; Wang, R.; Beckman, K.B.; Adams-Haduch, J.; M, Y.; Liu, S.; et al. Loci for Human Leukocyte Telomere Length in the Singaporean Chinese Population and Trans-Ethnic Genetic Studies. Nat. Commun. 2019, 10, 2491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gurung, R.L.; Dorajoo, R.; M, Y.; Wang, L.; Liu, S.; Liu, J.-J.; Shao, Y.M.; Chen, Y.; Sim, X.; Ang, K.; et al. Association of Leukocyte Telomere Length with Chronic Kidney Disease in East Asians with Type 2 Diabetes: A Mendelian Randomization Study. Clin. Kidney J. 2021, 14, 2371–2376. [Google Scholar] [CrossRef] [PubMed]
- Demanelis, K.; Jasmine, F.; Chen, L.S.; Chernoff, M.; Tong, L.; Delgado, D.; Zhang, C.; Shinkle, J.; Sabarinathan, M.; Lin, H.; et al. Determinants of Telomere Length across Human Tissues. Science 2020, 369, eaaz6876. [Google Scholar] [CrossRef] [PubMed]
- Haycock, P.C. Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study. JAMA Oncol. 2017, 3, 636–651. [Google Scholar] [CrossRef]
- Zhang, C.; Doherty, J.A.; Burgess, S.; Hung, R.J.; Lindström, S.; Kraft, P.; Gong, J.; Amos, C.I.; Sellers, T.A.; Monteiro, A.N.A.; et al. Genetic Determinants of Telomere Length and Risk of Common Cancers: A Mendelian Randomization Study. Hum. Mol. Genet. 2015, 24, 5356–5366. [Google Scholar] [CrossRef] [Green Version]
- Ojha, J.; Codd, V.; Nelson, C.P.; Samani, N.J.; Ivan, V.; Madsen, N.R.; Hansen, H.M.; De Smith, A.J.; Bracci, P.M.; Wiencke, K.; et al. Genetic Variation Associated with Longer Telomere Length Increases Risk of Chronic Lymphocytic Leukemia. Cancer Epidemiol. Biomarkers Prev. 2016, 25, 1043–1049. [Google Scholar] [CrossRef] [Green Version]
- Codd, V.; Denniff, M.; Swinfield, C.; Warner, S.C.; Papakonstantinou, M.; Sheth, S.; Nanus, D.E.; Budgeon, C.A.; Musicha, C.; Bountziouka, V.; et al. Measurement and Initial Characterization of Leukocyte Telomere Length in 474,074 Participants in UK Biobank. Nat. Aging 2022, 2, 170–179. [Google Scholar] [CrossRef]
- Fyhrquist, F.; Tiitu, A.; Saijonmaa, O.; Forsblom, C.; Groop, P.H. Telomere Length and Progression of Diabetic Nephropathy in Patients with Type 1 Diabetes. J. Intern. Med. 2010, 267, 278–286. [Google Scholar] [CrossRef]
- Raschenberger, J.; Kollerits, B.; Ritchie, J.; Lane, B.; Kalra, P.A.; Ritz, E.; Kronenberg, F. Association of Relative Telomere Length with Progression of Chronic Kidney Disease in Two Cohorts: Effect Modification by Smoking and Diabetes. Sci. Rep. 2015, 5, 11887. [Google Scholar] [CrossRef] [Green Version]
- Bansal, N.; Whooley, M.A.; Regan, M.; McCulloch, C.E.; Ix, J.H.; Epel, E.; Blackburn, E.; Lin, J.; Hsu, C.Y. Association between Kidney Function and Telomere Length: The Heart and Soul Study. Am. J. Nephrol. 2012, 36, 405–411. [Google Scholar] [CrossRef] [Green Version]
- Pykhtina, V.S.; Strazhesko, I.D.; Tkacheva, O.N.; Akasheva, D.U.; Dudinskaya, E.N.; Vygodin, V.A.; Plokhova, E.V.; Kruglikova, A.S.; Boitsov, S.A. Association of Renal Function, Telomere Length, and Markers of Chronic Inflammation in Patients without Chronic Kidney and Cardiovascular Diseases. Adv. Gerontol. 2016, 6, 217–223. [Google Scholar] [CrossRef]
- Liu, H.; Yang, Y.; Ge, Y.; Liu, J.; Zhao, Y. TERC Promotes Cellular Inflammatory Response Independent of Telomerase. Nucleic Acids Res. 2019, 47, 8084–8095. [Google Scholar] [CrossRef]
- Ding, D.; Xi, P.; Zhou, J.; Wang, M.; Cong, Y.S. Human Telomerase Reverse Transcriptase Regulates MMP Expression Independently of Telomerase Activity via NF-ΚB-Dependent Transcription. FASEB J. 2013, 27, 4375–4383. [Google Scholar] [CrossRef]
- Wang, M.; Xu, H.; Chong Lee Shin, O.L.S.; Li, L.; Gao, H.; Zhao, Z.; Zhu, F.; Zhu, H.; Liang, W.; Qian, K.; et al. Compound α-Keto Acid Tablet Supplementation Alleviates Chronic Kidney Disease Progression via Inhibition of the NF-KB and MAPK Pathways. J. Transl. Med. 2019, 17, 122. [Google Scholar] [CrossRef]
- Robin, J.D.; Ludlow, A.T.; Batten, K.; Magdinier, F.; Stadler, G.; Wagner, K.R.; Shay, J.W.; Wright, W.E. Telomere Position Effect: Regulation of Gene Expression with Progressive Telomere Shortening over Long Distances. Genes Dev. 2014, 28, 2464–2476. [Google Scholar] [CrossRef] [Green Version]
- McKnight, A.J.; O’Donoghue, D.; Peter Maxwell, A. Annotated Chromosome Maps for Renal Disease. Hum. Mutat. 2009, 30, 314–320. [Google Scholar] [CrossRef]
- Ma, C.; He, S.; Li, P.; Zhang, H.; Li, W.; Li, Y. Negative Association between Caloric Intake and Estimated Glomerular Filtration Rate in a Chinese Population: Mediation Models Involving Mitochondrial Function. Gerontology 2020, 66, 439–446. [Google Scholar] [CrossRef]
- Zhang, W.G.; Jia, L.; Ma, J.; Zhu, S.Y.; Nie, S.S.; Song, K.K.; Liu, X.M.; Zhang, Y.P.; Cao, D.; Yang, X.P.; et al. Peripheral Blood Leukocyte Telomere Length Is Associated with Age but Not Renal Function: A Cross-Sectional Follow-up Study. J. Nutr. Health Aging 2018, 22, 276–281. [Google Scholar] [CrossRef]
- Astrup, A.S.; Tarnow, L.; Jorsal, A.; Lajer, M.; Nzietchueng, R.; Benetos, A.; Rossing, P.; Parving, H.H. Telomere Length Predicts All-Cause Mortality in Patients with Type 1 Diabetes. Diabetologia 2010, 53, 45–48. [Google Scholar] [CrossRef] [Green Version]
- Dogan, F.; Forsyth, N.R. Telomerase Regulation: A Role for Epigenetics. Cancers 2021, 13, 1213. [Google Scholar] [CrossRef] [PubMed]
- Hill, C.; Avila-palencia, I.; Maxwell, A.P.; Hunter, R.F.; Mcknight, A.J. Harnessing the Full Potential of Multi-Omic Analyses to Advance the Study and Treatment of Chronic Kidney Disease. Front. Nephrol. 2022, 2, 923068. [Google Scholar] [CrossRef]
- Dessain, S.K.; Yu, H.Y.; Reddel, R.R.; Beijersbergen, R.L.; Weinberg, R.A. Methylation of the Human Telomerase Gene CpG Island. Cancer Res. 2000, 60, 537–541. [Google Scholar] [PubMed]
- Devereux, T.R.; Horikawa, I.; Anna, C.H.; Annab, L.A.; Afshari, C.A.; Barrett, J.C. DNA Methylation Analysis of the Promoter Region of the Human Telomerase Reverse Transcriptase (HTERT) Gene. Cancer Res. 1999, 59, 6087–6090. [Google Scholar]
- Barthel, F.P.; Wei, W.; Tang, M.; Martinez-ledesma, E.; Hu, X.; Lichtenberg, T.; Hu, J.; Zhang, J.; Zheng, S.; Roel, G.W. Systematic Analysis of Telomere Length and Somatic Alterations in 31 Cancer Types. Nat. Genet. 2017, 49, 349–357. [Google Scholar] [CrossRef]
- Renaud, S.; Loukinov, D.; Abdullaev, Z.; Guilleret, I.; Bosman, F.T. Dual Role of DNA Methylation inside and Outside of CTCF-Binding Regions in the Transcriptional Regulation of the Telomerase HTERT Gene. Nucleic Acids Res. 2007, 35, 1245–1256. [Google Scholar] [CrossRef] [Green Version]
- Zhao, X.; Tian, X.; Kajigaya, S.; Caroline, R.; Strickland, S.; Savani, B.N.; Mohan, S.; Feng, X.; Keyvanfar, K.; Dunavin, N.; et al. Epigenetic Landscape of the TERT Promoter: A Potential Biomarker for High Risk AML/MDS. Br. J. Haematol. 2016, 175, 427–439. [Google Scholar] [CrossRef] [Green Version]
- Shin, K.; Kang, M.K.; Dicterow, E.; Park, N. Hypermethylation of the HTERT Promoter Inhibits the Expression of Telomerase Activity in Normal Oral Fibroblasts and Senescent Normal Oral Keratinocytes. Br. J. Cancer 2003, 89, 1473–1478. [Google Scholar] [CrossRef] [Green Version]
- Guilleret, I.; Yan, P.; Grange, F.; Braunschweig, R.; Bosman, F.T.; Benhattar, J. Hypermethylation of the Human Telomerase Catalytic Subunit (HTERT) Gene Correlates with Telomerase Activity. Int. J. Cancer 2002, 101, 335–341. [Google Scholar] [CrossRef]
- Stern, J.L.; Paucek, R.D.; Huang, F.W.; Ghandi, M.; Nwumeh, R.; Costello, J.C.; Cech, T.R.; Stern, J.L.; Paucek, R.D.; Huang, F.W.; et al. Allele-Specific DNA Methylation and Its Interplay with Repressive Histone Marks at Promoter-Mutant TERT Genes. Cell Rep. 2017, 21, 3700–3707. [Google Scholar] [CrossRef] [Green Version]
- Tsirpanlis, G.; Chatzipanagiotou, S.; Boufidou, F.; Kordinas, V.; Alevyzaki, F.; Zoga, M.; Kyritsis, I.; Stamatelou, K.; Triantafyllis, G.; Nicolaou, C. Telomerase Activity Is Decreased in Peripheral Blood Mononuclear Cells of Hemodialysis Patients. Am. J. Nephrol. 2006, 26, 91–96. [Google Scholar] [CrossRef]
- Moreno, J.A.; Hamza, E.; Guerrero-Hue, M.; Rayego-Mateos, S.; García-Caballero, C.; Vallejo-Mudarra, M.; Metzinger, L.; Metzinger-Le Meuth, V. Non-Coding RNAs in Kidney Diseases: The Long and Short of Them. Int. J. Mol. Sci. 2021, 22, 6077. [Google Scholar] [CrossRef] [PubMed]
- Xiong, G.; Tang, W.; Zhang, D.; He, D.; Wei, G.; Atala, A.; Liang, X.J.; Bleyer, A.J.; Bleyer, M.E.; Yu, J.; et al. Impaired Regeneration Potential in Urinary Stem Cells Diagnosed from the Patients with Diabetic Nephropathy. Theranostics 2019, 9, 4221–4232. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Liu, Y.; Niu, X.; Yin, J.; Hu, B.; Guo, S.; Fan, Y.; Wang, Y.; Wang, N. Exosomes Secreted by Human Urine-Derived Stem Cells Could Prevent Kidney Complications from Type I Diabetes in Rats. Stem Cell Res. Ther. 2016, 7, 24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutkin, A.; Uziel, O.; Beery, E.; Nordenberg, J.; Pinchasi, M.; Goldvaser, H.; Henick, S.; Goldberg, M.; Lahav, M. Tumor Cells Derived Exosomes Contain HTERT MRNA and Transform Nonmalignant Fibroblasts into Telomerase Positive Cells. Oncotarget 2016, 7, 59173–59188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Goldvaser, H.; Gutkin, A.; Beery, E.; Edel, Y.; Nordenberg, J.; Wolach, O.; Rabizadeh, E.; Uziel, O.; Lahav, M. Characterisation of Blood-Derived Exosomal HTERT MRNA Secretion in Cancer Patients: A Potential Pan-Cancer Marker. Br. J. Cancer 2017, 117, 353–357. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, C.; Dellar, E.R.; Baena-Lopez, L.A. Caspases Help to Spread the Message via Extracellular Vesicles. FEBS J. 2022, 1–19. [Google Scholar] [CrossRef]
- Dellar, E.R.; Hill, C.; Melling, G.E.; Carter, D.R.; Baena-Lopez, L.A. Unpacking Extracellular Vesicles: RNA Cargo Loading and Function. J. Extracell. Biol. 2022, 1, e40. [Google Scholar] [CrossRef]
- Hong, J.; Yun, C. Telomere Gene Therapy: Polarizing Therapeutic. Cells 2019, 8, 1–17. [Google Scholar] [CrossRef] [Green Version]
- Liu, J.; Ge, Y.; Wu, S.; Ma, D.; Xu, W.; Zhang, Y.; Yang, Y. Association between Antidiabetic Agents Use and Leukocyte Telomere Shortening Rates in Patients with Type 2 Diabetes. Aging (Albany NY) 2019, 11, 741–755. [Google Scholar] [CrossRef]
- Ma, D.; Yu, Y.; Yu, X.; Zhang, M.; Yan, Y. The Changes of Leukocyte Telomere Length and Telomerase Activity after Sitagliptin Intervention in Newly Diagnosed Type 2 Diabetes. Diabetes. Metab. Res. Rev. 2015, 31, 256–261. [Google Scholar] [CrossRef]
- De Jesus, B.B.; Vera, E.; Schneeberger, K.; Tejera, A.M.; Ayuso, E.; Bosch, F.; Blasco, M.A. Telomerase Gene Therapy in Adult and Old Mice Delays Aging and Increases Longevity without Increasing Cancer. EMBO Mol. Med. 2012, 4, 691–704. [Google Scholar] [CrossRef]
- Jaskelioff, M.; Muller, F.L.; Paik, J.H.; Thomas, E.; Jiang, S.; Adams, A.C.; Sahin, E.; Kost-Alimova, M.; Protopopov, A.; Cadiñanos, J.; et al. Telomerase Reactivation Reverses Tissue Degeneration in Aged Telomerase-Deficient Mice. Nature 2011, 469, 102–107. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Townsley, D.M.; Dumitriu, B.; Liu, D.; Biancotto, A.; Weinstein, B.; Chen, C.; Hardy, N.; Mihalek, A.D.; Lingala, S.; Kim, Y.J.; et al. Danazol Treatment for Telomere Diseases. N. Engl. J. Med. 2016, 374, 1922–1931. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, A.; Arimondo, P.B.; Rots, M.G.; Jeronimo, C.; Berdasco, M. The Timeline of Epigenetic Drug Discovery: From Reality to Dreams. Clin. Epigenetics 2019, 11, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fontecha-Barriuso, M.; Martin-Sanchez, D.; Ruiz-Andres, O.; Poveda, J.; Sanchez-Niño, M.D.; Valiño-Rivas, L.; Ruiz-Ortega, M.; Ortiz, A.; Sanz, A.B. Targeting Epigenetic DNA and Histone Modifications to Treat Kidney Disease. Nephrol. Dial. Transplant. 2018, 33, 1875–1886. [Google Scholar] [CrossRef] [PubMed]
- Kushwaha, K.; Garg, S.S.; Gupta, J. Targeting Epigenetic Regulators for Treating Diabetic Nephropathy. Biochimie 2022, 202, 146–158. [Google Scholar] [CrossRef]
- Kang, J.G.; Park, J.S.; Ko, J.H.; Kim, Y.S. Regulation of Gene Expression by Altered Promoter Methylation Using a CRISPR/Cas9-Mediated Epigenetic Editing System. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Osteikoetxea, X.; Silva, A.; Lázaro-Ibáñez, E.; Salmond, N.; Shatnyeva, O.; Stein, J.; Schick, J.; Wren, S.; Lindgren, J.; Firth, M.; et al. Engineered Cas9 Extracellular Vesicles as a Novel Gene Editing Tool. J. Extracell. Vesicles 2022, 11, e12225. [Google Scholar] [CrossRef]
- Tang, T.T.; Wang, B.; Lv, L.L.; Dong, Z.; Liu, B.C. Extracellular Vesicles for Renal Therapeutics: State of the Art and Future Perspective. J. Control. Release 2022, 349, 32–50. [Google Scholar] [CrossRef]
| Paper | Author, Year [Reference] | Key Findings | Method Summary | Relevance |
|---|---|---|---|---|
| Novel genetic determinants of telomere length from a multi-ethnic analysis of 109,122 whole genome sequences in TOPMed | Taub et al., 2022 [132] | 59 novel variants associated with telomere length were identified. One SNP (rs1008438 in the HSPA1A gene) was significantly associated with risk of renal manifestations in T1D. | Whole genome sequencing (WGS) of whole blood for 109,122 individuals. TL was estimated from WGS data via the TelSeq methods. Tests for novelty were performed by checking LD with previously conducted GWAS and discarding those that had LD > 0.7 with previously described loci. PheWAS were conducted within the UK Biobank and Vanderbilt University biobank. | T1D, DKD |
| Polygenic basis and biomedical consequences of telomere length variation | Codd et al., 2021 [130] | Identified 193 novel variants significantly associated with leukocyte TL. No causal association between genetically estimated TL and CKD/T1D/T2D (p = 0.819, 0.845 and 0.163, respectively). CKD and T2D were significantly associated with experimentally determined leukocyte telomere length (p = 9.4527 × 10−17 and 0.000316, respectively). | Leukocyte TL measurements from the UK Biobank (n = 474,074), generated via qPCR [142]. By removing nonconditionally independent, pleiotropic and correlated variants, an instrument with 130 variants was created. MR was conducted on 93 biomedical traits and 123 disease outcomes from the UK Biobank, including CKD, T1D and T2D. For these three, the data sets contained: CKD (14,485 cases/437,060 controls); T1D (4227 cases/437,060 control); T2D (36,324 cases/437,060 control) | CKD, T1D, T2D |
| A Mendelian randomization study found causal linkage between telomere attrition and chronic kidney disease | Park et al., 2021 [135] | Significant causal association supporting TL shortening with increased CKD risk. IVW method (1.20 OR; 95% CI, 1.08–1.33; p < 0.001). All implemented MR sensitivity analyses did not affect significance. The only non-significant causal estimate was the MR-Egger regression analysis (1.10 OR; 95% CI, 0.92–1.54; p < 0.19) performed after SNPs with strong associations with other phenotypes (n = 13) were excluded. Reverse-direction MR for kidney functions effect on telomere attrition yielded significant causal estimates for all analyses excluding both the MR-Egger regressions performed. The MR-Egger intercept (p = 0.04) indicates the presence of directional pleiotropy in the reverse-direction MR. | A genetic instrument of 46 SNPs associated with leukocyte TL was used [133]. The SNPs were tested for genome-wide associations with confounders (hypertension, diabetes mellitus, cholesterol lowering medications, blood lipid profiles, smoking, or obesity). Summary level MR performed using European ancestry outcome data from CKDGen Consortium (n = 480,698, CKD cases = 41,395). Polygenic score analysis was performed using the 46 SNP instrument on UKBiobank data (Individuals with cystatin C/creatinine-eGFR data = 321,024, CKD cases = 8118). Reverse causation was investigated using a second instrument with 140 SNPs created from CKDGen GWAS data for European ancestry eGFR. This instrument was then used on UK Biobank data for individuals with TL data available (n = 326,075). | CKD |
| Association of leukocyte telomere length with chronic kidney disease in East Asians with type 2 diabetes: a Mendelian randomization study | Gurung et al., 2021 [137] | Genetically determined shorter TL was associated with increased CKD risk in patients with T2D (meta-IVW adjusted odds ratio = 1.51, 95% CI 1.12–2.12, p = 0.007). Similar results were obtained following sensitivity analysis. MR-Egger analysis suggested no evidence of horizontal pleiotropy. | MR analysis was performed using 16 leukocyte TL SNPs [136], investigating CKD as the outcome, defined as an eGFR of <60mL/min/1.73m2 (1628 cases/3140 controls). Participants were from the Singapore Study of Macro-angiopathy and Micro-vascular Reactivity in T2D and Diabetic Nephropathy cohorts. | T2D, DKD |
| Results from the German Chronic Kidney Disease (GCKD) study support association of relative telomere length with mortality in a large cohort of patients with moderate chronic kidney disease | Fazzini et al., 2020 [98] | RTL appeared positively associated with eGFR (p < 0.001) and Urine Albumin-Creatine ratio (p < 0.001); however this association did not remain after age and sex adjustment. Each 0.1 RTL unit decrease was associated with a 16% increase in all-cause mortality, even after age and sex adjustment. Patients in the lowest RTL quartile had a 75% higher risk for all-cause mortality than those in the highest quartile. | Relative TL was measured using qPCR within a cohort of 4955 patients from the GCKD study. Participants were divided into quartiles based on RTL and numbers of participants with confounders were presented for each quartile (smoking status, DM, prevalent CVD, sex, BMI). Average values for markers of kidney disease, BP and blood cholesterol were presented for each quartile. | CKD |
| The telomerase gene polymorphisms, but not telomere length, increase susceptibility to primary glomerulonephritis/end-stage kidney disease in females | Sun et al., 2020 [134] | No significant difference between TL between cases and controls. In females, a slightly shorter TL was observed in patients versus controls, but this was non-significant (p = 0.590). They instead identified genetic variants in telomere-related genes that contributed to disease susceptibility/progression. | 515 healthy controls and 769 primary glomerulonephritis(GN)/CKD/ESKD patients from a Han Chinese population. Genomic DNA was extracted from peripheral blood. Leukocyte TL measured via qPCR. LTL was assessed in 327 controls and 592 patients. | CKD |
| Genome-wide Association Analysis in Humans Links Nucleotide Metabolism to Leukocyte Telomere Length | Li et al., 2020 [133] | MR analysis did not yield significant causal estimates for TL and CKD/T1D/T2D. The MR-Egger intercepts for all three indicated that directional pleiotropy was not present. | Meta-analysis of 78,592 individuals from the ENGAGE, EPIC, CVD and InterAct studies. Leukocyte TL measurements made via qPCR. A 52 SNP genetic instrument for telomere attrition was generated and used to conduct an MR investigation on 122 disease outcomes from the UK Biobank. CKD cases = 5536. T1D cases = 3469. T2D cases = 20,575. | T1D, T2D, CKD |
| Negative Association between Caloric Intake and Estimated Glomerular Filtration Rate in a Chinese Population: Mediation Models Involving Mitochondrial Function | Ma et al., 2020 [152] | Leukocyte TL was not significantly associated with eGFR (r = 0.056, p = 0.260) or urinary microalbumin to creatinine ratio (UACR) (r = 0.069, p = 0.168), with these associations adjusted for age.Harnessing a multiple linear regression model, these associations were also not significant (eGFR: β = 0.672 (–0.629 to 1.973), p = 0.310; UACR: β = 0.075 (–0.035 to 0.185), p = 0.183). | 599 participants with different types of glucose tolerance were recruited from a Chinese rural cohort. Leukocyte TL (from peripheral blood) was determined via qPCR. Their multiple linear regression model was adjusted for age, gender, BMI, waist circumference, low-density lipoprotein cholesterol, triglycerides, abnormal glucose tolerance (including diabetes and prediabetes) and hypertension. In addition, when eGFR was a dependent variable, UACR was adjusted for; when UACR was a dependent variable, eGFR was adjusted for. | Renal function |
| Short Leukocyte Telomere Length Predicts Albuminuria Progression in Individuals With Type 2 Diabetes | Gurung et al., 2018 [96] | Leukocyte TL independently predicted the progression of albuminuria in T2D with preserved renal filtration function (eGFR > 60 mL/min/1.73 m2 and UACR < 300 mg/mg). The TL and albuminuria progression association was independent of risk factors, such as hypertension, hyperglycaemia, long diabetes duration, dyslipidaemia, and existing kidney function impairment. | A cohort of 691 Asian individuals with T2D who had preserved glomerular filtration rates. Leukocyte TL was measured via qPCR. | T2D, DKD |
| Peripheral blood leukocyte telomere length is associated with age but not renal function: A cross-sectional follow-up study | Zhang et al., 2018 [153] | Leukocyte TRF length was positively associated with eGFR (r = 0.182, 0.122, 0.290, and 0.254 depending on the specific eGFR calculation used, p < 0.01), but negatively correlated with serum cystatin C (r = −0.180, p < 0.01). The association with serum cystatin C was lost after adjusting for age. No association was observed between TRF length change and renal function. | Utilised a Han Chinese heathy population (n = 471). Telomere restriction fragment (TRF) length of genomic DNA was determined via a Southern blotting method. This study investigated Peripheral blood leukocyte telomere length. 3-year follow up TRF length data were available for 80 participants. | Renal function |
| Telomere attrition, kidney function, and prevalent chronic kidney disease in the United States | Mazidi et al., 2017 [99] | TL was negatively associated with urea albumin and ACR and positively associated with serum creatinine and eGFR (p < 0.001). In adjusted models, the association only remained significant for eGFR. Logistic regression between TL quartiles and chance of CKD did not reveal significant associations. | National Health and Nutrition Examination Survey (NHANES) cohort (n = 10,568). Univariable and multivariable (age, sex, race, smoking, fasting blood glucose, systolic and diastolic blood pressure, body mass index, and C-reactive protein) regression analyses were carried out. Note that diabetes and blood glucose were used as covariates. TL was measured via qPCR on whole blood-derived genomic DNA. | CKD |
| Association Between Telomere Length and Risk of Cancer and Non-Neoplastic Diseases: A Mendelian Randomization Study | Haycock et al., 2017 [139] | No significant association between genetically increased TL and CKD risk (0.94 OR; 95% CI, 0.77–1.16; p < 0.59) or T2D (1.00 OR; 95% CI, 0.84–1.20; p < 0.98). A statistically significant association between increased TL and lower T1D risk was reported (0.71 OR; 95% CI, 0.51–0.98; p < 0.04). | 16 SNPs selected as genetic proxies for telomere length, derived from original GWAS reports and the NHGRI-EBI GWAS catalogue. Outcome summary data obtained for 83 diseases and 46 risk factors. CKD data were obtained from CKDGen (5807 cases/56,430 controls), with only 13 of the instrumental SNPs present in the outcome dataset. T1D dataset was obtained from T1DBase (7514 cases/9045 controls), with 13 SNPs present in the dataset. T2D data were obtained from DIAGRAM Consortium (10,415 cases/53,655 controls), with 12 SNPs present in dataset. | T1D, T2D, CKD |
| Association of renal function, telomere length, and markers of chronic inflammation in patients without chronic kidney and cardiovascular diseases | Pykhtina et al., 2016 [146] | Significant associations were found between TL and increased albuminuria levels (p = 0.023), CRP (p = 0.047) and fibrinogen (p = 0.001) even after adjustment for age and gender. No associations were found between TL and eGFR, urea levels or serum creatinine. | A cohort of 253 individuals (aged 25–85) with no chronic non-infectious diseases (cardiovascular diseases linked to atherosclerosis; arterial hypertension (AH) III degree; diabetes; CKD (glomerular filtration rate (GFR) < 60 mL/min/1.73 m2 or GFR ≥ 60 mL/min/1.73 m2 with albuminuria ≥ 30 mg/24 h), chronic and acute infectious diseases, oncological diagnoses, pregnancy, or lactation period. Measurements were performed on numerous variables (serum creatinine levels, urinary albumin level, serum fibrinogen level, blood CRP level). Note that eGFR was not measured, but derived from the MDRD equation. TL was measured via qPCR. | Renal Function |
| Association of relative telomere length with progression of chronic kidney disease in two cohorts: Effect modification by smoking and diabetes | Raschenberger et al., 2015 [144] | Shorter TL was a predictor of more rapid CKD progression in patients with diabetes, determining that each 0.1 unit decrease in telomere length was significantly associated with an increased hazard ratio for CKD progression of 16%. | One of the two cohorts included in this study contained patients with diabetes. A non-dialysis-dependent CKD cohort of a predominantly white population in Greater Manchester (n = 889). 33% of the patients had diabetes mellitus. TL measured via qPCR on whole blood-derived genomic DNA. | Diabetes (T1D and T2D), CKD, DKD |
| Association between kidney function and telomere length: The heart and soul study | Bansal et al., 2012 [145] | When age was included as a confounder, lower creatinine-derived eGFR, was associated with shorter telomere length at baseline (β = 9.1, 95% CI 1.2–16.9, p < 0.05) and predicted more rapid telomere shortening (10.8, 95% CI 4.3–17.3, p < 0.05) over 5 years. Once results were adjusted for age, the association was no longer statistically significant. Serum creatinine, urine creatinine clearance, cystatin C, eGFRcys, urine albumin to creatinine ratio were not significantly associated with TL. | The Heart and Soul study cohort of heart disease patients (n = 1024). Only 608 subjects had TL measured both at baseline and at 6 years. TL was measured via qPCR. | CKD, coronary heart disease |
| Telomere length and progression of diabetic nephropathy in patients with type 1 diabetes | Fyhrquist et al., 2010 [143] | TL was not significantly different between those with T1D and healthy controls, nor between healthy controls and T1D patients with normoalbuminuria (normal albumin excretion), microalbuminuria (moderate increase in albumin excretion) or macroalbuminuria (highly elevated albumin excretion). However, a higher proportion of short telomeres was an independent predictor of DKD progression (HR = 1.115, [1.039–1.195], p = 0.0023), alongside HbA1c and smoking. | Leukocyte TL was measured using a Southern blot technique, harnessing blood samples from 132 patients with T1D (Finnish Diabetic Nephropathy Study) and 44 healthy controls. | T1D, DKD |
| Telomere length predicts all-cause mortality in patients with type 1 diabetes | Astrup et al., 2010 [154] | Telomere length did not differ between patients with or without DKD. Telomere length was significantly inversely correlated to age, systolic blood pressure and duration of diabetes (p < 0.01). | TL was measured in 157 patients with DKD and 116 patients with persistent normoalbuminuria (Steno Diabetes Center cohort). Telomere length was measured via Southern blot from DNA samples extracted from white blood cells. | T1D, DKD |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hill, C.; Duffy, S.; Coulter, T.; Maxwell, A.P.; McKnight, A.J. Harnessing Genomic Analysis to Explore the Role of Telomeres in the Pathogenesis and Progression of Diabetic Kidney Disease. Genes 2023, 14, 609. https://doi.org/10.3390/genes14030609
Hill C, Duffy S, Coulter T, Maxwell AP, McKnight AJ. Harnessing Genomic Analysis to Explore the Role of Telomeres in the Pathogenesis and Progression of Diabetic Kidney Disease. Genes. 2023; 14(3):609. https://doi.org/10.3390/genes14030609
Chicago/Turabian StyleHill, Claire, Seamus Duffy, Tiernan Coulter, Alexander Peter Maxwell, and Amy Jayne McKnight. 2023. "Harnessing Genomic Analysis to Explore the Role of Telomeres in the Pathogenesis and Progression of Diabetic Kidney Disease" Genes 14, no. 3: 609. https://doi.org/10.3390/genes14030609
APA StyleHill, C., Duffy, S., Coulter, T., Maxwell, A. P., & McKnight, A. J. (2023). Harnessing Genomic Analysis to Explore the Role of Telomeres in the Pathogenesis and Progression of Diabetic Kidney Disease. Genes, 14(3), 609. https://doi.org/10.3390/genes14030609









