Nanopore-Based Direct RNA Sequencing of the Trypanosoma brucei Transcriptome Identifies Novel lncRNAs
Abstract
1. Introduction
2. Materials and Methods
2.1. Growth of Trypanosome Cells
2.2. Oligodeoxynucleotide Synthesis
2.3. RNA Isolation
2.4. Synthesis of cDNA and Quantitative Real-Time (qRT)-PCR
2.5. Preparation of DRS-Sequencing Libraries and Sequencing
2.6. Transcript Mapping
2.7. Identification of Full-Length Reads
2.8. Reference-Free Transcript Identification and Identification of lncRNA
2.9. Estimation of Transcript Abundance, DGE Analysis, and Statistics
3. Results and Discussion
3.1. RNA Sequencing Libraries—Quality Assessment
3.2. Identification of Full-Length Transcripts
3.3. Reference-Free Transcript Identification
3.4. Comparison with Annotated Exons
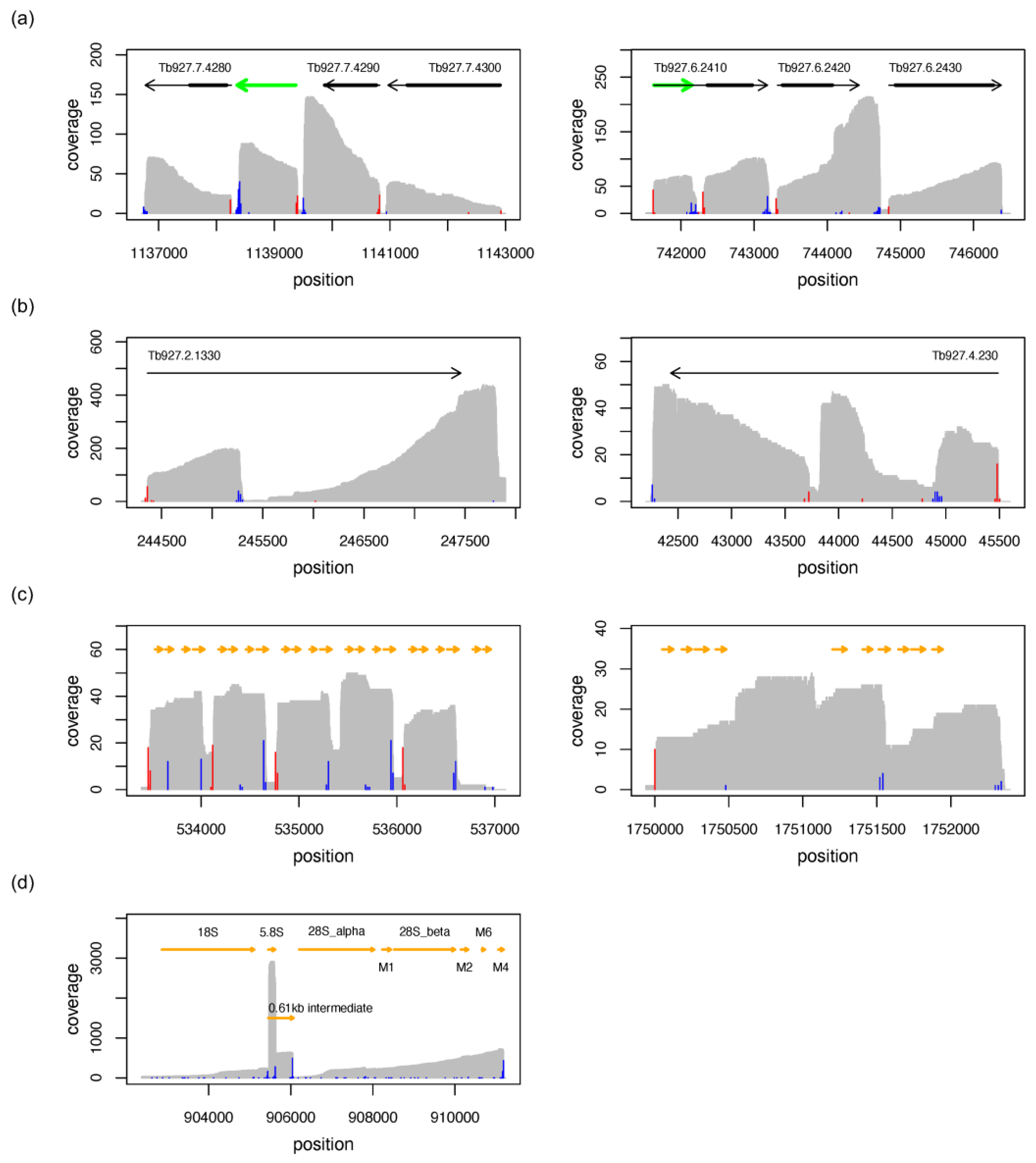
3.5. Long Noncoding RNAs
3.5.1. Characterisation of Long Noncoding RNAs
3.5.2. Novel lncRNAs
3.6. Differential Gene Expression (DGE) between Insect- and Bloodstream-Stage Trypanosomes
3.7. The Mitochondrial Transcriptome
3.8. RNA Editing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kaufer, A.; Stark, D.; Ellis, J. A review of the systematics, species identification and diagnostics of the Trypanosomatidae using the maxicircle kinetoplast DNA: From past to present. Int. J. Parasitol. 2020, 50, 449–460. [Google Scholar] [CrossRef] [PubMed]
- De Rycker, M.; Wyllie, S.; Horn, D.; Read, K.D.; Gilbert, I.H. Anti-trypanosomatid drug discovery: Progress and challenges. Nat. Rev. Microbiol. 2023, 21, 35–50. [Google Scholar] [CrossRef] [PubMed]
- Clayton, C. Regulation of gene expression in trypanosomatids: Living with polycistronic transcription. Open Biol. 2019, 9, 190072. [Google Scholar] [CrossRef] [PubMed]
- Kolev, N.G.; Ullu, E.; Tschudi, C. The emerging role of RNA-binding proteins in the life cycle of Trypanosoma brucei. Cell. Microbiol. 2014, 16, 482–489. [Google Scholar] [CrossRef] [PubMed]
- Clayton, C.; Shapira, M. Post-transcriptional regulation of gene expression in trypanosomes and leishmanias. Mol. Biochem. Parasitol. 2007, 156, 93–101. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S. Trans-splicing in trypanosomes: Machinery and its impact on the parasite transcriptome. Future Microbiol. 2011, 6, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Calvillo, S.; Florencio-Martínez, L.E.; Nepomuceno-Mejía, T. Nucleolar Structure and Function in Trypanosomatid Protozoa. Cells 2019, 8, 421. [Google Scholar] [CrossRef] [PubMed]
- Rajan, K.S.; Doniger, T.; Cohen-Chalamish, S.; Rengaraj, P.; Galili, B.; Aryal, S.; Unger, R.; Tschudi, C.; Michaeli, S. Developmentally Regulated Novel Non-coding Anti-sense Regulators of mRNA Translation in Trypanosoma brucei. iScience 2020, 23, 101780. [Google Scholar] [CrossRef]
- Kolev, N.G.; Rajan, K.S.; Tycowski, K.T.; Toh, J.Y.; Shi, H.; Lei, Y.; Michaeli, S.; Tschudi, C. The vault RNA of Trypanosoma brucei plays a role in the production of trans-spliced mRNA. J. Biol. Chem. 2019, 294, 15559–15574. [Google Scholar] [CrossRef]
- Michaeli, S.; Podell, D.; Agabian, N.; Ullu, E. The 7SL RNA homologue of Trypanosoma brucei is closely related to mammalian 7SL RNA. Mol. Biochem. Parasitol. 1992, 51, 55–64. [Google Scholar] [CrossRef]
- Sandhu, R.; Sanford, S.; Basu, S.; Park, M.; Pandya, U.M.; Li, B.; Chakrabarti, K. A trans-spliced telomerase RNA dictates telomere synthesis in Trypanosoma brucei. Cell Res. 2013, 23, 537–551. [Google Scholar] [CrossRef]
- Fort, R.S.; Chavez, S.; Trinidad Barnech, J.M.; Oliveira-Rizzo, C.; Smircich, P.; Sotelo-Silveira, J.R.; Duhagon, M.A. Current Status of Regulatory Non-Coding RNAs Research in the Tritryp. Noncoding RNA 2022, 8, 54. [Google Scholar] [CrossRef] [PubMed]
- Koslowsky, D.; Sun, Y.; Hindenach, J.; Theisen, T.; Lucas, J. The insect-phase gRNA transcriptome in Trypanosoma brucei. Nucleic Acids Res. 2014, 42, 1873–1886. [Google Scholar] [CrossRef] [PubMed]
- Statello, L.; Guo, C.J.; Chen, L.L.; Huarte, M. Gene regulation by long non-coding RNAs and its biological functions. Nat. Rev. Mol. Cell Biol. 2021, 22, 96–118. [Google Scholar] [CrossRef]
- Kolev, N.G.; Franklin, J.B.; Carmi, S.; Shi, H.; Michaeli, S.; Tschudi, C. The transcriptome of the human pathogen Trypanosoma brucei at single-nucleotide resolution. PLoS Pathog. 2010, 6, e1001090. [Google Scholar] [CrossRef] [PubMed]
- Guegan, F.; Rajan, K.S.; Bento, F.; Pinto-Neves, D.; Sequeira, M.; Gumińska, N.; Mroczek, S.; Dziembowski, A.; Cohen-Chalamish, S.; Doniger, T.; et al. A long noncoding RNA promotes parasite differentiation in African trypanosomes. Sci. Adv. 2022, 8, eabn2706. [Google Scholar] [CrossRef]
- Minshall, N.; Git, A. Enzyme- and gene-specific biases in reverse transcription of RNA raise concerns for evaluating gene expression. Sci. Rep. 2020, 10, 8151. [Google Scholar] [CrossRef]
- Steijger, T.; Abril, J.F.; Engström, P.G.; Kokocinski, F.; RGASP Consortium; Hubbard, T.J.; Guigó, R.; Harrow, J.; Bertone, P. Assessment of transcript reconstruction methods for RNA-seq. Nat. Methods 2013, 10, 1177–1184. [Google Scholar] [CrossRef]
- Jan, C.H.; Friedman, R.C.; Ruby, J.G.; Bartel, D.P. Formation, regulation and evolution of Caenorhabditis elegans 3′UTRs. Nature 2011, 469, 97–101. [Google Scholar] [CrossRef]
- Houseley, J.; Tollervey, D. Apparent non-canonical trans-splicing is generated by reverse transcriptase in vitro. PLoS ONE 2010, 5, e12271. [Google Scholar] [CrossRef]
- Mourão, K.; Schurch, N.J.; Lucoszek, R.; Froussios, K.; MacKinnon, K.; Duc, C.; Simpson, G.; Barton, G.J. Detection and mitigation of spurious antisense expression with RoSA. F1000Research 2019, 8, 819. [Google Scholar] [CrossRef]
- Garalde, D.R.; Snell, E.A.; Jachimowicz, D.; Sipos, B.; Lloyd, J.H.; Bruce, M.; Pantic, N.; Admassu, T.; James, P.; Warland, A.; et al. Highly parallel direct RNA sequencing on an array of nanopores. Nat. Methods 2018, 15, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Cross, G.A. Identification, purification and properties of clone-specific glycoprotein antigens constituting the surface coat of Trypanosoma brucei. Parasitology 1975, 71, 393–417. [Google Scholar] [CrossRef] [PubMed]
- Hirumi, H.; Hirumi, K. Continuous cultivation of Trypanosoma brucei bloodstream forms in a medium containing a low concentration of serum protein without feeder cell layers. J. Parasitol. 1989, 75, 985–989. [Google Scholar] [CrossRef] [PubMed]
- Brun, R.; Schönenberger, M. Cultivation and in vitro cloning or procyclic culture forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979, 36, 289–292. [Google Scholar]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Wick, R.R.; Judd, L.M.; Holt, K.E. Performance of neural network basecalling tools for Oxford Nanopore sequencing. Genome Biol. 2019, 20, 129. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 2009, 25, 1754–1760. [Google Scholar] [CrossRef]
- Li, H. Minimap2: Pairwise alignment for nucleotide sequences. Bioinformatics 2018, 34, 3094–3100. [Google Scholar] [CrossRef]
- Naguleswaran, A.; Fernandes, P.; Bevkal, S.; Rehmann, R.; Nicholson, P.; Roditi, I. Developmental changes and metabolic reprogramming during establishment of infection and progression of Trypanosoma brucei brucei through its insect host. PLoS Negl. Trop. Dis. 2021, 15, e0009504. [Google Scholar] [CrossRef] [PubMed]
- Loman, N.J.; Quick, J.; Simpson, J.T. A complete bacterial genome assembled de novo using only nanopore sequencing data. Nat. Methods 2015, 12, 733–735. [Google Scholar] [CrossRef] [PubMed]
- Workman, R.E.; Tang, A.D.; Tang, P.S.; Jain, M.; Tyson, J.R.; Razaghi, R.; Zuzarte, P.C.; Gilpatrick, T.; Payne, A.; Quick, J.; et al. Nanopore native RNA sequencing of a human poly(A) transcriptome. Nat. Methods 2019, 16, 1297–1305, Erratum in: Nat. Methods 2020, 7, 114. [Google Scholar] [CrossRef] [PubMed]
- Quinlan, A.R.; Hall, I.M. BEDTools: A flexible suite of utilities for comparing genomic features. Bioinformatics 2010, 26, 841–842. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinform. 2014, 47, 11.12.1–11.12.34. [Google Scholar] [CrossRef] [PubMed]
- Kang, Y.J.; Yang, D.C.; Kong, L.; Hou, M.; Meng, Y.Q.; Wei, L.; Gao, G. CPC2: A fast and accurate coding potential calculator based on sequence intrinsic features. Nucleic Acids Res. 2017, 45, W12–W16. [Google Scholar] [CrossRef] [PubMed]
- Han, S.; Liang, Y.; Ma, Q.; Xu, Y.; Zhang, Y.; Du, W.; Wang, C.; Li, Y. LncFinder: An integrated platform for long non-coding RNA identification utilizing sequence intrinsic composition, structural information and physicochemical property. Brief Bioinform. 2019, 20, 2009–2027. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. The R package Rsubread is easier, faster, cheaper and better for alignment and quantification of RNA sequencing reads. Nucleic Acids Res. 2019, 47, e47. [Google Scholar] [CrossRef] [PubMed]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Jain, M.; Abu-Shumays, R.; Olsen, H.E.; Akeson, M. Advances in nanopore direct RNA sequencing. Nat. Methods 2022, 19, 1160–1164. [Google Scholar] [CrossRef]
- Krause, M.; Niazi, A.M.; Labun, K.; Torres Cleuren, Y.N.; Müller, F.S.; Valen, E. tailfindr: Alignment-free poly(A) length measurement for Oxford Nanopore RNA and DNA sequencing. RNA 2019, 25, 1229–1241. [Google Scholar] [CrossRef] [PubMed]
- Maier, K.C.; Gressel, S.; Cramer, P.; Schwalb, B. Native molecule sequencing by nano-ID reveals synthesis and stability of RNA isoforms. Genome Res. 2020, 30, 1332–1344. [Google Scholar] [CrossRef] [PubMed]
- Dawe, H.R.; Shaw, M.K.; Farr, H.; Gull, K. The hydrocephalus inducing gene product, Hydin, positions axonemal central pair microtubules. BMC Biol. 2007, 5, 33. [Google Scholar] [CrossRef] [PubMed]
- Fickett, J.W.; Tung, C.S. Assessment of protein coding measures. Nucleic Acids Res. 1992, 20, 6441–6450. [Google Scholar] [CrossRef] [PubMed]
- Kong, L.; Zhang, Y.; Ye, Z.Q.; Liu, X.Q.; Zhao, S.Q.; Wei, L.; Gao, G. CPC: Assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Res. 2007, 35, W345–W349. [Google Scholar] [CrossRef] [PubMed]
- Ersfeld, K.; Melville, S.E.; Gull, K. Nuclear and genome organization of Trypanosoma brucei. Parasitol. Today 1999, 15, 58–63. [Google Scholar] [CrossRef]
- Veitch, N.J.; Johnson, P.C.; Trivedi, U.; Terry, S.; Wildridge, D.; MacLeod, A. Digital gene expression analysis of two life cycle stages of the human-infective parasite, Trypanosoma brucei gambiense reveals differentially expressed clusters of co-regulated genes. BMC Genom. 2010, 11, 124. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Siegel, T.N.; Hekstra, D.R.; Wang, X.; Dewell, S.; Cross, G.A. Genome-wide analysis of mRNA abundance in two life-cycle stages of Trypanosoma brucei and identification of splicing and polyadenylation sites. Nucleic Acids Res. 2010, 38, 4946–4957. [Google Scholar] [CrossRef]
- Christiano, R.; Kolev, N.G.; Shi, H.; Ullu, E.; Walther, T.C.; Tschudi, C. The proteome and transcriptome of the infectious metacyclic form of Trypanosoma brucei define quiescent cells primed for mammalian invasion. Mol. Microbiol. 2017, 106, 74–92. [Google Scholar] [CrossRef]
- Aird, D.; Ross, M.G.; Chen, W.S.; Danielsson, M.; Fennell, T.; Russ, C.; Jaffe, D.B.; Nusbaum, C.; Gnirke, A. Analyzing and minimizing PCR amplification bias in Illumina sequencing libraries. Genome Biol. 2011, 12, R18. [Google Scholar] [CrossRef]
- Parekh, S.; Ziegenhain, C.; Vieth, B.; Enard, W.; Hellmann, I. The impact of amplification on differential expression analyses by RNA-seq. Sci. Rep. 2016, 6, 25533. [Google Scholar] [CrossRef] [PubMed]
- Byrne, A.; Beaudin, A.E.; Olsen, H.E.; Jain, M.; Cole, C.; Palmer, T.; DuBois, R.M.; Forsberg, E.C.; Akeson, M.; Vollmers, C. Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells. Nat. Commun. 2017, 8, 16027. [Google Scholar] [CrossRef] [PubMed]
- Jenjaroenpun, P.; Wongsurawat, T.; Pereira, R.; Patumcharoenpol, P.; Ussery, D.W.; Nielsen, J.; Nookaew, I. Complete genomic and transcriptional landscape analysis using third-generation sequencing: A case study of Saccharomyces cerevisiae CEN.PK113-7D. Nucleic Acids Res. 2018, 46, e38. [Google Scholar] [CrossRef] [PubMed]
- Gleeson, J.; Leger, A.; Prawer, Y.D.J.; Lane, T.A.; Harrison, P.J.; Haerty, W.; Clark, M.B. Accurate expression quantification from nanopore direct RNA sequencing with NanoCount. Nucleic Acids Res. 2022, 50, e19. [Google Scholar] [CrossRef]
- Bringaud, F.; Biteau, N.; Melville, S.E.; Hez, S.; El-Sayed, N.M.; Leech, V.; Berriman, M.; Hall, N.; Donelson, J.E.; Baltz, T. A new, expressed multigene family containing a hot spot for insertion of retroelements is associated with polymorphic subtelomeric regions of Trypanosoma brucei. Eukaryot. Cell 2002, 1, 137–151, Erratum in: Eukaryot. Cell 2002, 1, 305. [Google Scholar] [CrossRef]
- Bringaud, F.; Biteau, N.; Zuiderwijk, E.; Berriman, M.; El-Sayed, N.M.; Ghedin, E.; Melville, S.E.; Hall, N.; Baltz, T. The ingi and RIME non-LTR retrotransposons are not randomly distributed in the genome of Trypanosoma brucei. Mol. Biol. Evol. 2004, 21, 520–528. [Google Scholar] [CrossRef]
- Sement, F.M.; Suematsu, T.; Zhang, L.; Yu, T.; Huang, L.; Aphasizheva, I.; Aphasizhev, R. Transcription initiation defines kinetoplast RNA boundaries. Proc. Natl. Acad. Sci. USA 2018, 115, E10323–E10332. [Google Scholar] [CrossRef]
- Adler, B.K.; Harris, M.E.; Bertrand, K.I.; Hajduk, S.L. Modification of Trypanosoma brucei mitochondrial rRNA by posttranscriptional 3′ polyuridine tail formation. Mol. Cell. Biol. 1991, 11, 5878–5884. [Google Scholar]
- Aphasizhev, R.; Aphasizheva, I. Mitochondrial RNA processing in trypanosomes. Res. Microbiol. 2011, 162, 655–663. [Google Scholar] [CrossRef]
- Aphasizheva, I.; Alfonzo, J.; Carnes, J.; Cestari, I.; Cruz-Reyes, J.; Göringer, H.U.; Hajduk, S.; Lukeš, J.; Madison-Antenucci, S.; Maslov, D.A.; et al. Lexis and Grammar of Mitochondrial RNA Processing in Trypanosomes. Trends Parasitol. 2020, 36, 337–355. [Google Scholar] [CrossRef]
- Opperdoes, F.R.; Michels, P.A. Complex I of Trypanosomatidae: Does it exist? Trends Parasitol. 2008, 24, 310–317. [Google Scholar] [CrossRef] [PubMed]








Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kruse, E.; Göringer, H.U. Nanopore-Based Direct RNA Sequencing of the Trypanosoma brucei Transcriptome Identifies Novel lncRNAs. Genes 2023, 14, 610. https://doi.org/10.3390/genes14030610
Kruse E, Göringer HU. Nanopore-Based Direct RNA Sequencing of the Trypanosoma brucei Transcriptome Identifies Novel lncRNAs. Genes. 2023; 14(3):610. https://doi.org/10.3390/genes14030610
Chicago/Turabian StyleKruse, Elisabeth, and H. Ulrich Göringer. 2023. "Nanopore-Based Direct RNA Sequencing of the Trypanosoma brucei Transcriptome Identifies Novel lncRNAs" Genes 14, no. 3: 610. https://doi.org/10.3390/genes14030610
APA StyleKruse, E., & Göringer, H. U. (2023). Nanopore-Based Direct RNA Sequencing of the Trypanosoma brucei Transcriptome Identifies Novel lncRNAs. Genes, 14(3), 610. https://doi.org/10.3390/genes14030610






