Microsatellite Genome-Wide Database Development for the Commercial Blackhead Seabream (Acanthopagrus schlegelii)
Abstract
1. Introduction
2. Materials and Methods
2.1. Genome-Wide SSRs Analysis of the Blackhead Seabream
2.2. Genome Resequencing and Construction of Polymorphic SSR Database
2.3. Microsatellite Marker Analysis
2.4. Data Analysis
3. Results
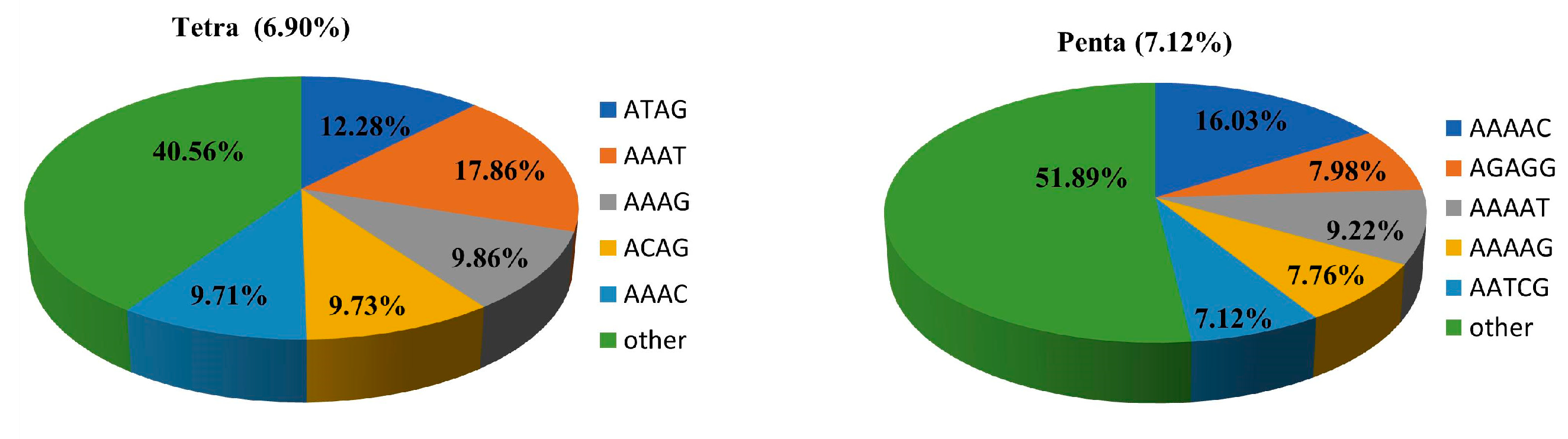
3.1. Identification of Genome-Wide SSRs
3.2. Identification of Genome-Wide Polymorphic SSRs
3.3. Identification of Polymorphic SSRs Using PCR Amplification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Acanthopagrus schlegelii in Fisheries Statistics Yearbook of Taiwan Area. Available online: https://fishdb.sinica.edu.tw/chi/yearrpt2.php?id=26 (accessed on 1 February 2023).
- Yamashita, Y.; Sanchez, G.; Kawai, K.; Tomano, S.; Fujita, H.; Umino, T. The role of the isolation of the marginal seas during the Pleistocene in the genetic structure of black sea bream Acanthopagrus schlegelii (Bleeker, 1854) in the coastal waters of Japan. PeerJ 2021, 9, e11001. [Google Scholar] [CrossRef] [PubMed]
- Chang, W.-C.; Lee, Y.-C.; Shih, C.-H.; Chu, T.-J.; Chang, P.-H. Population size and stocking contribution rates for marked and recaptured black porgy Acanthopagrus schlegelli, in Northwestern Taiwan, 2005–2008. Fish. Res. 2011, 109, 252–256. [Google Scholar]
- Chen, S.L.; Liu, Y.G.; Xu, M.Y.; Li, J. Isolation and characterization of polymorphic microsatellite loci from an EST-library of red sea bream (Chrysophrys major) and cross-species amplification. Mol. Ecol. Resour. 2005, 5, 215–217. [Google Scholar] [CrossRef]
- Song, W.; Li, Y.; Zhao, Y.; Liu, Y.; Niu, Y.; Pang, R.; Miao, G.; Liao, X.; Shao, C.; Gao, F.; et al. Construction of a high-density microsatellite genetic linkage map and mapping of sexual and growth-related traits in half-smooth tongue sole (Cynoglossus semilaevis). PLoS ONE 2012, 7, e52097. [Google Scholar] [CrossRef] [PubMed]
- Ji, X.S.; Chen, S.L.; Liao, X.L.; Yang, J.F.; Xu, T.J.; Ma, H.Y.; Tian, Y.S.; Jiang, Y.L.; Wu, P.F. Microsatellite-centromere mapping in Cynoglossus semilaevis using gynogenetic diploid families produced by the use of homologous and non-homologous sperm. J. Fish. Biol. 2009, 75, 422–434. [Google Scholar] [CrossRef]
- Dutta, N.; Singh, R.K.; Mohindra, V.; Pathak, A.; Kumar, R.; Sah, P.; Mandal, S.; Kaur, G.; Lal, K.K. Microsatellite marker set for genetic diversity assessment of primitive Chitala chitala (Hamilton, 1822) derived through SMRT sequencing technology. Mol. Biol. Rep. 2019, 46, 41–49. [Google Scholar] [CrossRef]
- Wang, X.; Liu, S.; Yang, Y.; Wu, L.; Huang, W.; Wu, R.; Li, G.; Zhang, H.; Meng, Z. Genetic evidence for the mating system and reproductive success of black sea bream (Acanthopagrus schlegelii). Ecol. Evol. 2020, 10, 4483–4494. [Google Scholar] [CrossRef]
- Guerrero-Cózar, I.; Perez-Garcia, C.; Benzekri, H.; Sánchez, J.J.; Seoane, P.; Cruz, F.; Gut, M.; Zamorano, M.J.; Claros, M.G.; Manchado, M. Development of whole-genome multiplex assays and construction of an integrated genetic map using SSR markers in Senegalese sole. Sci. Rep. 2020, 10, 21905. [Google Scholar] [CrossRef]
- Yang, Y.; Cao, X.; Feng, R.; Chen, S.; Zhang, Z.; Ren, W.; Xu, S. Isolation and characterization of thirteen polymorphic microsatellite loci from black porgy (Acanthopagrus schlegeli). J. Genet. 2014, 93, e97–e99. [Google Scholar] [CrossRef]
- Mao, X.Q.; Li, Z.B.; Yuan, Y.; Ning, Y.F.; Shangguan, J.B.; Huang, Y.S.; Yang, M.; Li, B.B. Isolation and characterization of eight novel microsatellite markers in Acanthopagrus schlegelii. Genet. Mol. Res. 2016, 15, 15027902. [Google Scholar] [CrossRef]
- Liu, Y.G.; Liu, L.X.; Wu, Z.X.; Lin, H.; Li, B.F.; Sun, X.Q. Isolation and characterization of polymorphic microsatellite loci in black sea bream (Acanthopagrus schlegeli) by cross-species amplification with six species of the Sparidae family. Aquat. Living Resour. 2007, 20, 257–262. [Google Scholar] [CrossRef]
- Wu, R.X.; Xiao, Y.; Niu, S.F.; Zhai, Y.; Zhang, H.R.; Chen, W.Y.; Li, X. Development of microsatellite markers for Acanthopagrus schlegelii based on SLAF-seq technology and generality in the family Sparidae. Oceanol. Limnol. Sin. 2019, 50, 365–377. [Google Scholar] [CrossRef]
- Gymrek, M.; Golan, D.; Rosset, S.; Erlich, Y. lobSTR: A short tandem repeat profiler for personal genomes. Genome Res. 2012, 22, 1154–1162. [Google Scholar] [CrossRef]
- King, J.L.; Wendt, F.R.; Sun, J.; Budowle, B. STRait Razor v2s: Advancing sequence-based STR allele reporting and beyond to other marker systems. Forensic Sci. Int. Genet. 2017, 29, 21–28. [Google Scholar] [CrossRef]
- Ganschow, S.; Silvery, J.; Kalinowski, J.; Tiemann, C. toaSTR: A web application for forensic STR genotyping by massively parallel sequencing. Forensic Sci. Int. Genet. 2018, 37, 21–28. [Google Scholar] [CrossRef]
- Willems, T.; Zielinski, D.; Yuan, J.; Gordon, A.; Gymrek, M.; Erlich, Y. Genome-wide profiling of heritable and de novo STR variations. Nat. Methods 2017, 14, 590–592. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhang, K.; Chen, S.; Zhang, Z.; Zhang, J.; You, X.; Bian, C.; Xu, J.; Jia, C.; Qiang, J.; et al. Draft genome of the protandrous Chinese black porgy, Acanthopagrus schlegelii. Gigascience 2018, 7, giy012. [Google Scholar] [CrossRef]
- Du, L.; Zhang, C.; Liu, Q.; Zhang, X.; Yue, B.; Hancock, J. Krait: An ultrafast tool for genome-wide survey of microsatellites and primer design. Bioinformatics 2018, 34, 681–683. [Google Scholar] [CrossRef]
- Zane, L.; Bargelloni, L.; Patarnello, T. Strategies for microsatellite isolation: A review. Mol. Ecol. 2002, 11, 1–16. [Google Scholar] [CrossRef]
- Schuelke, M. An economic method for the fluorescent labeling of PCR fragments. Nat. Biotechnol. 2000, 18, 233–234. [Google Scholar] [CrossRef]
- Holland, M.M.; Parson, W. GeneMarker® HID: A reliable software tool for the analysis of forensic STR data. J. Forensic Sci. 2011, 56, 29–35. [Google Scholar] [CrossRef]
- Peakall, R.; Smouse, P.E. GenAlEx 6.5: Genetic analysis in Excel. Population genetic software for teaching and research—An update. Bioinformatics 2012, 28, 2537–2539. [Google Scholar] [CrossRef]
- Oosterhout, C.V.; Hutchinson, W.F.; Wills, D.P.M.; Shipley, P. micro-checker: Software for identifying and correcting genotyping errors in microsatellite data. Mol. Ecol. Resour. 2010, 4, 535–538. [Google Scholar] [CrossRef]
- Liu, K.; Muse, S.V. PowerMarker: An integrated analysis environment for genetic marker analysis. Bioinformatics 2005, 21, 2128–2129. [Google Scholar] [CrossRef]
- Jeong, D.S.; Gonzalez, E.B.; Morishima, K.; Arai, K.; Umino, T. Parentage assignment of stocked black sea bream Acanthopagrus schlegelii in Hiroshima Bay using microsatellite DNA markers. Fish. Sci. 2010, 73, 823–830. [Google Scholar] [CrossRef]
- Botstein, D.; White, R.L.; Skolnick, M.; Davis, R.W. Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 1980, 32, 314–331. [Google Scholar]
- Xu, J.J.; Zheng, X.; Zhang, X.Y.; Wang, T.; Yin, S.W. Analysis of Distribution Characteristics of Microsatellites in Four Genomes of Puffer Fish. Genom. Appl. Biol. 2021, 40, 1441–1451. [Google Scholar] [CrossRef]
- Tian, H.F.; Hu, Q.M.; Li, Z. Genome-wide identification of simple sequence repeats and development of polymorphic SSR markers in swamp eel (Monopterus albus). Sci. Prog. 2021, 104, 368504211035597. [Google Scholar] [CrossRef]
- Schorderet, D.F.; Gartler, S.M. Analysis of CpG suppression in methylated and nonmethylated species. Proc. Natl. Acad. Sci. USA 1992, 89, 957–961. [Google Scholar] [CrossRef]
- Han, X.; Ling, Q.; Li, C.; Wang, G.; Xu, Z.; Lu, G. Characterization of pikeperch (Sander lucioperca) transcriptome and development of SSR markers. Biochem. Syst. Ecol. 2016, 66, 188–195. [Google Scholar] [CrossRef]
- Sigang, F.; Hao, H.; Yong, L.; Pengfei, W.; Chao, Z.; Lulu, Y.; Xiuting, Q.; Qiu, L. Genome-wide identification of microsatellite and development of polymorphic SSR markers for spotted sea bass (Lateolabrax maculatus). Aquac. Rep. 2021, 20, 100677. [Google Scholar] [CrossRef]
- Maduna, S.N.; Vivian-Smith, A.; Jónsdóttir, Ó.D.B.; Imsland, A.K.D.; Klütsch, C.F.C.; Nyman, T.; Eiken, H.G.; Hagen, S.B. Genome- and transcriptome-derived microsatellite loci in lumpfish Cyclopterus lumpus: Molecular tools for aquaculture, conservation and fisheries management. Sci. Rep. 2020, 10, 559. [Google Scholar] [CrossRef]
- Jiang, Q.; Li, Q.; Yu, H.; Kong, L. Genome-wide analysis of simple sequence repeats in marine animals—A comparative approach. Mar. Biotechnol. 2014, 16, 604–619. [Google Scholar] [CrossRef]
- Serapion, J.; Kucuktas, H.; Feng, J.; Liu, Z. Bioinformatic mining of type I microsatellites from expressed sequence tags of channel catfish (Ictalurus punctatus). Mar. Biotechnol. 2004, 6, 364–377. [Google Scholar] [CrossRef]
- Ma, Y.; Lou, F.; Yin, X.; Cong, B.; Liu, S.; Zhao, L.; Zheng, L. Whole-genome survey and phylogenetic analysis of Gadus macrocephalus. Biosci. Rep. 2022, 42, BSR20221037. [Google Scholar] [CrossRef]
- Bhattarai, G.; Shi, A.; Kandel, D.R.; Solís-Gracia, N.; da Silva, J.A.; Avila, C.A. Genome-wide simple sequence repeats (SSR) markers discovered from whole-genome sequence comparisons of multiple spinach accessions. Sci. Rep. 2021, 11, 9999. [Google Scholar] [CrossRef]
- Wang, H.; Gao, S.; Liu, Y.; Wang, P.; Zhang, Z.; Chen, D. A pipeline for effectively developing highly polymorphic simple sequence repeats markers based on multi-sample genomic data. Ecol. Evol. 2022, 12, e8705. [Google Scholar] [CrossRef]
- Valle-Silva, G.; Frontanilla, T.S.; Ayala, J.; Donadi, E.A.; Simões, A.L.; Castelli, E.C.; Mendes-Junior, C.T. Analysis and comparison of the STR genotypes called with HipSTR, STRait Razor and toaSTR by using next generation sequencing data in a Brazilian population sample. Forensic Sci. Int. Genet. 2022, 58, 102676. [Google Scholar] [CrossRef]
- Rocca, M.S.; Ferrarini, M.; Msaki, A.; Vinanzi, C.; Ghezzi, M.; De Rocco Ponce, M.; Foresta, C.; Ferlin, A. Comparison of NGS panel and Sanger sequencing for genotyping CAG repeats in the AR gene. Mol. Genet. Genom. Med. 2020, 8, e1207. [Google Scholar] [CrossRef]
- Halman, A.; Oshlack, A. Accuracy of short tandem repeats genotyping tools in whole exome sequencing data. F1000Research 2020, 9, 200. [Google Scholar] [CrossRef]
- Frontanilla, T.S.; Valle-Silva, G.; Ayala, J.; Mendes-Junior, C.T. Open-Access Worldwide Population STR Database Constructed Using High-Coverage Massively Parallel Sequencing Data Obtained from the Genomes Project. Genes 2022, 13, 2205. [Google Scholar] [CrossRef] [PubMed]
- Han, J.; Munro, J.E.; Kocoski, A.; Barry, A.E.; Bahlo, M. Population-level genome-wide STR discovery and validation for population structure and genetic diversity assessment of Plasmodium species. PLoS Genet. 2022, 18, e1009604. [Google Scholar] [CrossRef] [PubMed]



| Alleles | Di- | Tri- | Tetra- | Total |
|---|---|---|---|---|
| 3 | 11,626 | 3339 | 2075 | 17,040 (15.61)% |
| 4, 5 | 19,917 | 4791 | 2524 | 27,232 (24.95%) |
| ≥6 | 56,264 | 5407 | 3188 | 64,859 (59.43%) |
| Total | 87,807 (80.16%) | 13,537 (12.40%) | 7787 (7.14%) | 109,131 (100%) |
| Loci | Primer Left Sequence | Primer Right Sequence | Ta | Product Size (bp) | Motif | Na | Ho | He | PIC | P-HWE |
|---|---|---|---|---|---|---|---|---|---|---|
| acs36 | CCAAACCTGAGCACGACA | TAACACCTACACCCATCCCT | 53 | 214–238 | (GT)10 | 6 | 0.7857 | 0.8006 | 0.7550 | 0.5013 |
| acs146 | CATTTCTGGGCAAACAAGC | GGGTGGATAATCACACAGGG | 53 | 252–270 | (CA)6 | 5 | 0.7692 | 0.7217 | 0.6608 | 0.4491 |
| acs168 | GCATCTGGCAGCGTTTATC | TAGATACAGGGTTTTATTGTTTCC | 53 | 233–263 | (TG)10 | 5 | 0.4483 | 0.6346 | 0.6176 | 0.2949 |
| acs12 | TTCTCTTGGGCTTAGGATG | GTGACAGAGGTAAACTGAGGT | 50 | 179–183 | (GT)7 | 2 | 0.1333 | 0.1266 | 0.1167 | 0.6956 |
| acs145 | TTAGGTCAGGTCGGAGCACG | CAAAACGGGGTCATCAGAGC | 54 | 196–220 | (AC)20 | 8 | 0.9000 | 0.8452 | 0.8096 | 0.2175 |
| acs133 | AATGGTCGCTTCTCCTCC | TCACTGTCTTACTTGTATTGACG | 53 | 214–241 | (CTC)7 | 7 | 0.8000 | 0.7977 | 0.7513 | 0.4107 |
| acs201 | CCTATGTATGCCTGTCACTC | TGTGTAACTCAATATGATAGAAC | 52 | 226–240 | (GT)9 | 6 | 0.7586 | 0.6824 | 0.6352 | 0.4065 |
| acs173 | CCAGAGAGAGAAGTGAGGGGA | TGCAGGGCAGCTCTAACCT | 53 | 248–251 | (TGA)6 | 2 | 0.0667 | 0.0655 | 0.0624 | 0.8502 |
| acs28 | ACAGTGAGAGGCTTTGGTGG | ATGGGTTTGTCAGGATGAGC | 52 | 189–195 | (GT)7 | 3 | 0.5333 | 0.6305 | 0.5478 | 0.2352 |
| acse37 | ACACTAAGACAACTCAAGCAAC | CACCATTGTTTGTTTGAGC | 51 | 228–234 | (CAA)4 | 2 | 0.2000 | 0.1831 | 0.1638 | 0.5428 |
| acse121 | GCCGAAATAAACCAGATGATG | TTGTGACGGGAATGAGAGAC | 52 | 210–238 | (CA)10 | 6 | 0.5667 | 0.7610 | 0.7056 | 0.0011 * |
| acs199 | GCGGAAGCCTTTGATTGATG | CAGTTGGAGCATCGGAGCAG | 53 | 186–216 | (GT)15 | 6 | 0.5600 | 0.7461 | 0.6870 | 0.0000 * |
| acs107 | CTGGAATGTTACTGGAGCC | CCACAAATACACTACAATAGAAA | 50 | 190–210 | (AC)22 | 4 | 0.6000 | 0.5949 | 0.5319 | 0.6639 |
| acse85 | TTGCTGACCCATTGTGAAGT | AATAACTCCTCGCACCAGACT | 52 | 178–205 | (GCTGT)3 | 7 | 0.7931 | 0.8469 | 0.8100 | 0.9946 |
| acse86 | GTGCTGAGGGTCTTCGTTT | CTCCCACTGAGTCCAGGTAA | 52 | 179–185 | (AC)10 | 2 | 0.4000 | 0.4520 | 0.3457 | 0.5839 |
| acse54 | CAGCAGATCGGATCAGAAC | TGCGAGAACATCACAATAAAC | 52 | 175–201 | (AC)24 | 7 | 0.3214 | 0.8084 | 0.7694 | 0.0000 * |
| acs22 | GCTGCCAGGGTGCAGTTTCG | TCTGTCCATCCCGCCCGTCT | 54 | 182–197 | (GAA)4 | 4 | 0.3103 | 0.6528 | 0.5822 | 0.0001 * |
| acs16 | GACATTTTACAGCAGACAACC | TTCTTTATCCACAGAGCAGG | 50 | 245–275 | (CA)18 | 9 | 0.5357 | 0.6318 | 0.5976 | 0.2055 |
| acse34 | GTGGTTTACAAGGTGTGGCT | TGTTATTGTGTCACCCTGTCC | 50 | 164–178 | (GT)9 | 6 | 0.8333 | 0.7299 | 0.6816 | 0.0596 |
| acs151 | AGCAGACTGACAAGAGCACCT | AGATAGGTGGGCTGCGAG | 54 | 234–238 | (CA)6 | 2 | 0.0714 | 0.0701 | 0.0665 | 0.8446 |
| acse93 | GACAGCAACAATAATCATAA | CGTTTCACTGCCATCTG | 50 | 286–304 | (CA)8 | 6 | 0.2667 | 0.7684 | 0.7139 | 0.0000 * |
| acs4 | CGTACATTCTGTCAAAGCCA | AAAGGAGAAGTGCCGAGC | 52 | 202–238 | (CA)15 | 10 | 0.5862 | 0.8536 | 0.8214 | 0.0042 * |
| acs75 | TGTTATCCGACCTCAATCTC | TCTCACCATCACCTCCCA | 51 | 227–245 | (AC)13 | 8 | 0.8889 | 0.8637 | 0.8298 | 0.7215 |
| acse101 | AGGTATGTGGCTTGGTGTTG | TTGGCTCCATAGCACATTG | 52 | 230–260 | (GT)9 | 6 | 0.6552 | 0.7901 | 0.7412 | 0.1989 |
| acse138 | TGTGAGAAAGGAAGAATGAG | TGCTGGGAAACTACAATAAC | 50 | 185–205 | (GA)12 | 7 | 0.7333 | 0.7463 | 0.6910 | 0.5044 |
| acse141 | GGGCACGAGCAGTGATGTAG | CCATGTGTCTGCTATTTGCAAT | 50 | 250–280 | (TG)9 | 8 | 0.6667 | 0.7983 | 0.7585 | 0.1691 |
| acs3 | TCGTGGAGAAAGGAAGAG | CAGAAGGAGACATTTGAACT | 50 | 183–201 | (GT)18 | 7 | 0.8077 | 0.8205 | 0.7790 | 0.5272 |
| acs78 | ATCCAGTGGCAACAGAGGTC | TGACCCATCCCCAATCCC | 53 | 215–219 | (GT)11 | 2 | 0.2000 | 0.1831 | 0.1638 | 0.5428 |
| acse165 | TACGCTTCAAACGGAGACTT | GGGAACATTATTTGATTGGC | 53 | 220–232 | (GT)7 | 7 | 0.2610 | 0.2830 | 0.2708 | 0.3550 |
| acse76 | ACAACAATGCTACCAACA | AAACAAAACCTTTCCACT | 50 | 220–292 | (GAA)15 | 11 | 0.7500 | 0.8416 | 0.8080 | 0.3155 |
| AS1 | GTCCTTGTTGTAATGAGGGTTT | TCTTCATGTGTGGGGGCT | 60 | 159–183 | (AC)17 | 11 | 0.5313 | 0.8011 | 0.7638 | 0.0029 * |
| AS2 | CGTGTTTATGTGGGTGTG | ACACGGACAAAGTCACAGT | 53 | 146–184 | (AG)20 | 15 | 0.7813 | 0.9122 | 0.8896 | 0.1611 |
| AS3 | TGGACCGTATTGAAACCTGT | CAACATCAAGGCAGCAGAGT | 59 | 153–163 | (TG)10 | 4 | 0.4063 | 0.3914 | 0.3516 | 0.9632 |
| AS4 | CAACTACCTGTTCTCTGTGTCCT | CAAAATGTGCCATGACGAC | 55 | 272–294 | (TG)18 | 12 | 0.5625 | 0.9107 | 0.8877 | 0.0000 * |
| AS5 | TCTGTGAGAATGAGACGGAGTA | TGAGGTTTGCGAATGTGG | 57 | 271–293 | (AC)13 | 11 | 0.4375 | 0.7386 | 0.7081 | 0.0000 * |
| AS8 | CAGTCTCCAGAGTGCGATAAC | AAGAAACAGGTGTCTCGTGAA | 57 | 244–258 | (GT)8 | 6 | 0.8125 | 0.7490 | 0.6960 | 0.5973 |
| AS9 | GCCGCAACTAACAAAGAG | ACATACAGAATACATAAACGACAG | 55 | 198–216 | (GT)14 | 8 | 0.3330 | 0.7460 | 0.7045 | 0.0000 * |
| AS10 | ACACACTGGTCATTCGGTAA | TAGCGATAAAGAGGCACAAC | 57 | 270–298 | (GT)14 | 13 | 0.8438 | 0.9023 | 0.8779 | 0.0000 * |
| AS14 | AGAAAGGGCGGTCTGATG | GATTGACACCATCCCAGAACT | 59 | 214–246 | (GT)14 | 14 | 0.5333 | 0.6531 | 0.6301 | 0.0000 * |
| AS15 | TTGATTACTGTTCTCGTCTT | ATACAATACAAAACGATTACAA | 53 | 205–215 | (TG)13 | 6 | 0.8000 | 0.7062 | 0.6644 | 0.0669 |
| AS19 | AGCAGGAGATTCATGTGAGTC | ACTGCCCATGCTAACTGTTA | 57 | 224–238 | (TG)18 | 9 | 0.6875 | 0.8681 | 0.8374 | 0.0899 |
| AS20 | TTTATGTAACAAAGTGGATTTC | CAGTGCTGAGTTGGTACAAT | 53 | 225–231 | (AC)12 | 4 | 0.3667 | 0.3486 | 0.3206 | 0.1141 |
| AS25 | GTTTTCAATGTAAAGTCGGTCA | AATCGCCCAATCTGTAAGTG | 57 | 252–280 | (CA)17 | 12 | 0.3750 | 0.8309 | 0.7996 | 0.0000 * |
| AS26 | AGCAGATGGTCGATGTTG | TTTGTTAGAGCGAAGAAGTG | 53 | 137–143 | (AC)8 | 4 | 0.5625 | 0.6424 | 0.5666 | 0.1208 |
| AS27 | CACAATGAGTTGCTGGAGGT | TTCACAGTCCCAAAGCACA | 59 | 228–256 | (CA)12 | 11 | 0.7333 | 0.8311 | 0.7977 | 0.5160 |
| AS28 | ACCGTGGATGTGTTTCGTCA | GCTTGGTAGTATTCAGGCTCTCAG | 60 | 157–169 | (CA)11 | 8 | 0.6563 | 0.7128 | 0.6696 | 0.0136 * |
| AS29 | TTTATTGGAACACAGGGATT | GTTTCATTTTGACCATTTACC | 57 | 195–211 | (AC)17 | 8 | 0.8000 | 0.7768 | 0.7269 | 0.5920 |
| AS31 | CAGCGGCGTCTCACCTTG | GCCCGTTCAGCCTTCTCA | 60 | 253–265 | (CA)10 | 5 | 0.4063 | 0.5432 | 0.4757 | 0.2522 |
| AS34 | GCTACAGCCGCCCCACT | GGATTCAGGAAGATTGATTTGC | 60 | 205–217 | (AGAA)6 | 3 | 0.6667 | 0.6266 | 0.5391 | 0.6575 |
| AS35 | GGCTTAGGTGGAGCGTTT | GAGCGTGTTCCCAGTCATTA | 57 | 225–253 | (AGAA)8 | 8 | 0.7000 | 0.8260 | 0.7877 | 0.8708 |
| AS36 | GCAGCCTGAGCCTGGAGATA | TTAGACCGTAGAGTGTCACCGAAG | 60 | 262–286 | (GAAA)4 | 6 | 0.5450 | 0.6280 | 0.5707 | 0.0000 * |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Luo, X.; Zhang, L.; Chen, S. Microsatellite Genome-Wide Database Development for the Commercial Blackhead Seabream (Acanthopagrus schlegelii). Genes 2023, 14, 620. https://doi.org/10.3390/genes14030620
Luo X, Zhang L, Chen S. Microsatellite Genome-Wide Database Development for the Commercial Blackhead Seabream (Acanthopagrus schlegelii). Genes. 2023; 14(3):620. https://doi.org/10.3390/genes14030620
Chicago/Turabian StyleLuo, Xinhui, Lichun Zhang, and Songlin Chen. 2023. "Microsatellite Genome-Wide Database Development for the Commercial Blackhead Seabream (Acanthopagrus schlegelii)" Genes 14, no. 3: 620. https://doi.org/10.3390/genes14030620
APA StyleLuo, X., Zhang, L., & Chen, S. (2023). Microsatellite Genome-Wide Database Development for the Commercial Blackhead Seabream (Acanthopagrus schlegelii). Genes, 14(3), 620. https://doi.org/10.3390/genes14030620




