Characterization of Simple Sequence Repeat (SSR) Markers Mined in Whole Grape Genomes
Abstract
:1. Introduction
2. Experiment Method and Content
2.1. Plant Materials
2.2. Data Sources
2.3. Identification of SSR Motifs
2.4. Distribution Density of SSRs and SNPs in the Genome
2.5. Functional Annotation of SSR Loci
2.6. Phylogenetic Analysis
3. Results
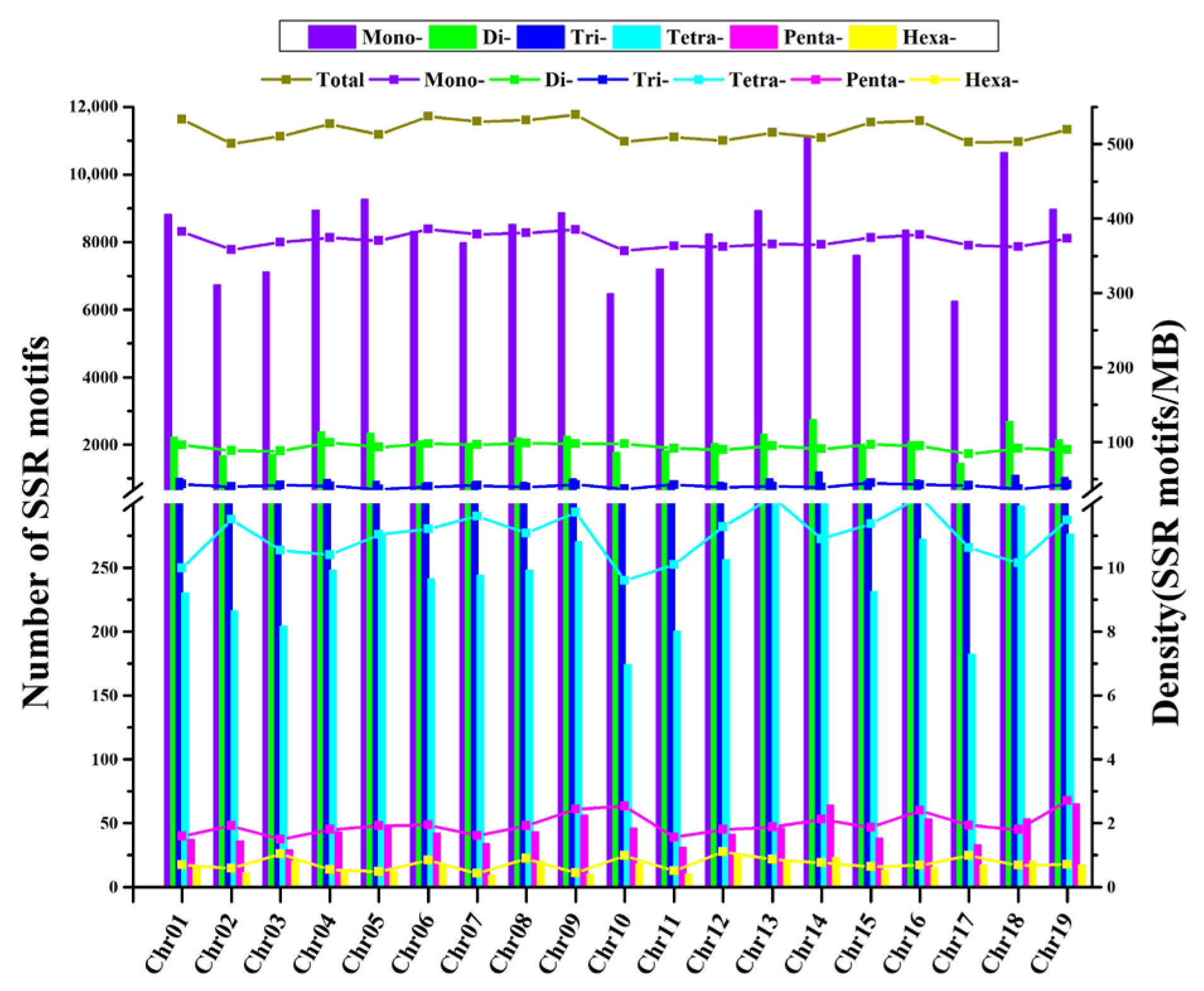
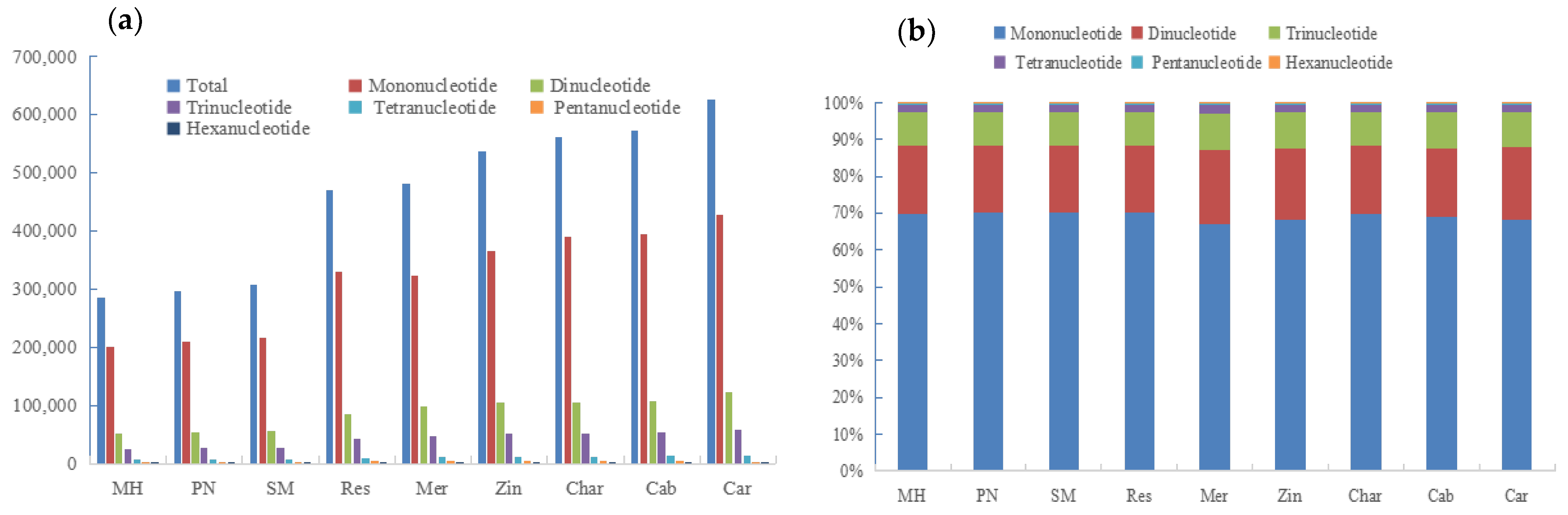
3.1. Number and Density of SSRs Identified in Nine Grape Genomes
3.2. Types of SSRs Repeat Motifs and Lengths in the Grape Genome
3.3. Length of Various SSR Loci and Repeating Times of the Different Repeat Motifs
3.4. Annotation of SSR Loci
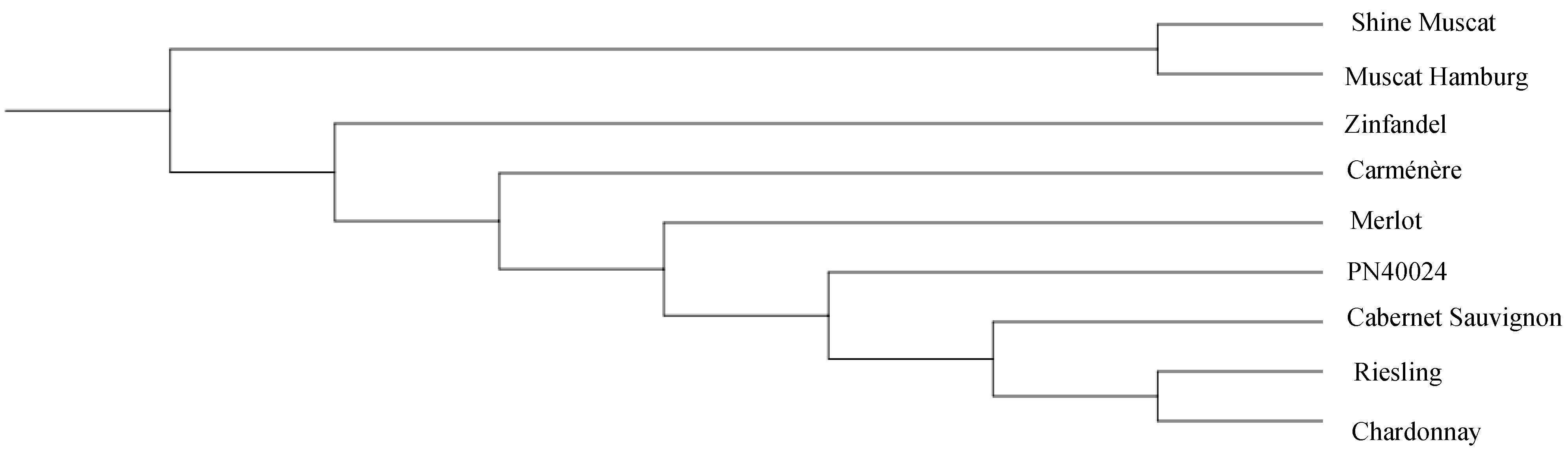
3.5. Comparison of the Phylogenetic Trees of Nine Grape Cultivars Constructed Using All the SSRs and SNPs
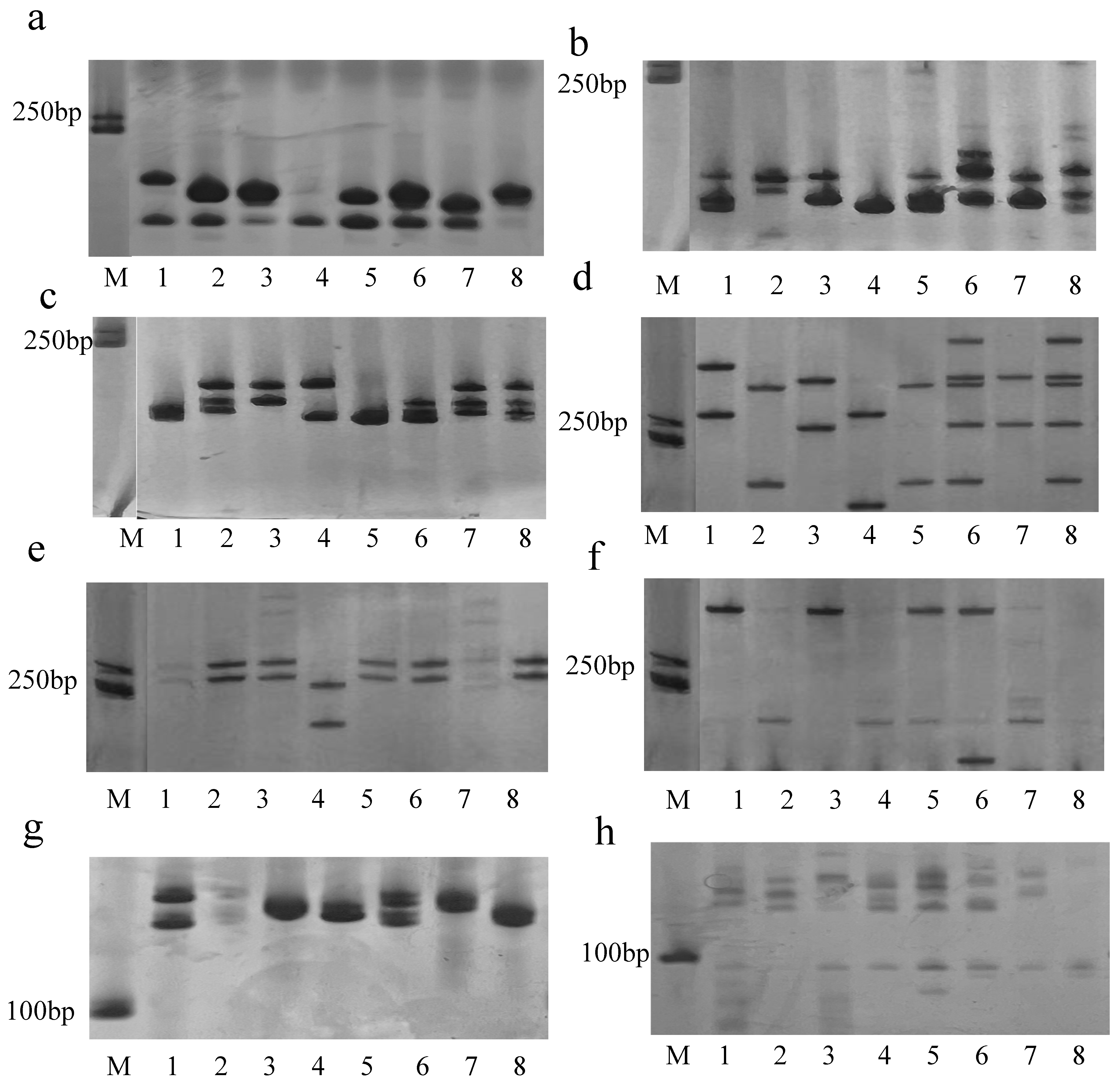
3.6. The Distribution and Numbers of ‘PN40024’ SSR Loci
4. Discussion
4.1. Relationship between the Grape Genome Sizes and the Distribution Density of SSR on Chromosomes
4.2. Conservation and Variety Specificity of the SSR Distribution Pattern of Grape Cultivars
4.3. Grape Genome Sequencing to Construct a Grape Variety Evolutionary Tree and Reveal Its Potential Genetic Relationship
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Varshney, R.K.; Graner, A.; Sorrells, M.E. Genic microsatellite markers in plants: Features and applications. Trends Biotechnol. 2005, 23, 48–55. [Google Scholar] [CrossRef] [PubMed]
- Grover, A.; Sharma, P.C. Development and use of molecular markers: Past and present. Crit. Rev. Biotechnol. 2016, 36, 290–302. [Google Scholar] [CrossRef] [PubMed]
- Kantartzi, S.K. Microsatellites: Methods and Protocols; Springer: New York, NY, USA, 2013; pp. 1–13. [Google Scholar]
- O’Hanlon, P.C.; Peakall, R.; Briese, D.T. A review of new PCR-based genetic markers and their utility to weed ecology. Weed Res. 2000, 40, 239–254. [Google Scholar] [CrossRef]
- Powell, W.; Machray, G.C.; Provan, J. Polymorphism revealed by simple sequence repeats. Trends Plant Sci. 1996, 1, 215–222. [Google Scholar] [CrossRef]
- Schlötterer, C. The evolution of molecular markers-just a matter of fashion? Nat. Rev. Genet. 2004, 5, 63–69. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, M.; Shrivastava, N.; Padh, H. Advances in molecular marker techniques and their applications in plant sciences. Plant Cell Rep. 2008, 27, 617–631. [Google Scholar] [CrossRef]
- Zarouri, B.; Vargas, A.M.; Gaforio, L.; Aller, M.; de Andrés, M.T.; Cabezas, J.A. Whole-genome genotyping of grape using a panel of microsatellite multiplex PCRs. Tree Genet. Genomes 2015, 11, 17. [Google Scholar] [CrossRef] [Green Version]
- Singh, R.K.; Jena, S.N.; Khan, S.; Yadav, S.; Banarjee, N.; Raghuvanshi, S.; Bhardwaj, V.; Dattamajumder, S.K.; Kapur, R.; Solomon, S.; et al. Development, cross-species/genera transferability of novel EST-SSR markers and their utility in revealing population structure and genetic diversity in sugarcane. Gene 2013, 524, 309–329. [Google Scholar] [CrossRef]
- Haq, S.U.; Kumar, P.; Singh, R.K.; Verma, K.S.; Bhatt, R.; Sharma, M.; Kachhwaha, S.; Kothari, S.L. Assessment of functional EST-SSR markers (sugarcane) in cross-species transferability, genetic diversity among Poaceae plants, and bulk segregation analysis. Genet. Res. Int. 2016, 2016, 7052323. [Google Scholar]
- Upadhyay, A.; Kadam, U.S.; Chacko, P.; Aher, L.; Karibasappa, G.M. Microsatellite analysis to differentiate clones of thompson seedless grapevine. Indian J. Hortic. 2010, 67, 260. [Google Scholar]
- Upadhyay, A.; Kadam, U.S.; Priya, C.M.; Karibasappa, G.S. Microsatellite and RAPD analysis of grape (Vitis spp.) accessions and identification of duplicates/misnomers in germplasm collection. Indian J. Hortic. 2010, 67, 8. [Google Scholar]
- Pinto, L.R.; Oliveira, K.M.; Marconi, T.; Garcia, A.A.F.; Ulian, E.C.; DeSouza, A.P. Characterization of novel sugarcane expressed sequence tag microsatellites and their comparison with genomic SSRs. Plant Breed. 2006, 125, 378–384. [Google Scholar] [CrossRef]
- La Rota, M.; Kantety, R.V.; Yu, J.K.; Sorrells, M.E. Nonrandom distribution and frequencies of genomic and EST-derived microsatellite markers in rice, wheat, and barley. BMC Genom. 2005, 6, 23. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saha, M.C.; Mian, R.; Zwonitzer, J.C.; Chekhovskiy, K.; Hopkins, A.A. An SSR- and AFLP-based genetic linkage map of tall fescue (Festuca arundinacea Schreb.). Theor. Appl. Genet. 2005, 110, 323–336. [Google Scholar] [CrossRef]
- Lagercrantz, U.; Ellegren, H.; Andersson, L. The abundance of various polymorphic microsatellite motifs differs between plants and vertebrates. Nucl. Acids. Res. 1993, 21, 1111–1115. [Google Scholar] [CrossRef] [Green Version]
- Roa, A.C.; Chavarriaga-Aguirre, P.; Duque, M.C.; Maya, M.M.; Bonierbale, M.W.; Iglesias, C.; Tohme, J. Cross-species amplification of cassava (Manihot esculenta) (Euphorbiaceae) microsatellites: Allelic polymorphism and degree of relationship. Am. J. Bot. 2000, 87, 1647–1655. [Google Scholar] [CrossRef]
- Abuzayed, M.A.; Goktay, M.; Allmer, J.; Doganlar, S. Development of genomic simple sequence repeat markers in faba bean by next-generation sequencing. Plant Mol. Biol. Rep. 2017, 35, 61–71. [Google Scholar] [CrossRef]
- Gil, J.S.; Um, Y.; Kim, S.; Kim, O.K.; Koo, S.C.; Reddy, C.S.; Kim, S.C.; Hong, C.P.; Park, S.G.; Kim, H.B.; et al. Development of genome-wide SSR markers from Angelica gigas nakai using next generation sequencing. Genes 2017, 8, 238. [Google Scholar] [CrossRef]
- Jaillon, O.; Aury, J.M.; Noel, B.; Policriti, A.; Clepet, C.; Casagrande, A.; Choisne, N.; Aubourg, S.; Vitulo, N.; Jubin, C.; et al. French-Italian Public Consortium for Grapevine Genome Characterization. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature 2007, 449, 463–467. [Google Scholar]
- Shirasawa, K.; Hirakawa, H.; Azuma, A.; Taniguchi, F.; Yamamoto, T.; Sato, A.; Ghelfi, A.; Isobe, S.N. De novo whole-genome assembly in an interspecific hybrid table grape, ‘Shine Muscat’. DNA Res. 2022, 29, dsac040. [Google Scholar] [CrossRef]
- Zhao, J.; Huang, L.; Ren, X.; Pandey, M.K.; Wu, B.; Chen, Y.; Zhou, X.; Chen, W.; Xia, Y.; Li, Z.; et al. Genetic variation and association mapping of seed-related traits in cultivated peanut (Arachis hypogaea L.) using single-locus simple sequence repeat markers. Front. Plant Sci. 2017, 8, 2105. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gyawali, S.; Harrington, M.; Durkin, J.; Horner, K.; Parkin, I.A.; Hegedus, D.D.; Bekkaoui, D.; Buchwaldt, L. Microsatellite markers used for genome-wide association mapping of partial resistance to Sclerotinia sclerotiorum in a world collection of Brassica napus. Mol. Breed. 2016, 36, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zalapa, J.E.; Cuevas, H.; Zhu, H.; Steffan, S.; Senalik, D.; Zeldin, E.; McCown, B.; Harbut, R.; Simon, P. Using next-generation sequencing approaches to isolate simple sequence repeat (SSR) loci in the plant sciences. Am. J. Bot. 2012, 99, 193–208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, Y.W.; Wei, X.; Abbas, F.; Yu, Y.Y.; Yu, R.C.; Fan, Y.P. Genome-wide identification of simple sequence repeats and assessment of genetic diversity in Hedychium. J. Appl. Res. Med. Aroma 2021, 24, 100312. [Google Scholar] [CrossRef]
- Liao, Z.; Chen, M.; Guo, L.; Gong, Y.; Tang, F.; Sun, X.; Tang, K. Rapid isolation of high-quality total RNA from taxus and ginkgo. Prep. Biochem. Biotechnol. 2004, 34, 209–214. [Google Scholar] [CrossRef]
- Chin, C.S.; Peluso, P.; Sedlazeck, F.J.; Nattestad, M.; Concepcion, G.T.; Clum, A.; Dunn, C.; O’Malley, R.; Figueroa-Balderas, R.; Morales-Cruz, A.; et al. Phased diploid genome assembly with single-molecule real-time sequencing. Nat. Methods 2016, 13, 1050–1054. [Google Scholar] [CrossRef] [Green Version]
- Minio, A.; Cochetel, N.; Vondras, A.; Massonnet, M.; Cantu, D. Assembly of complete diploid phased chromosomes from draft genome sequences. bioRxiv 2021, 1–8. [Google Scholar] [CrossRef]
- Minio, A.; Massonnet, M.; Figueroa-Balderas, R.; Vondras, A.M.; Blanco-Ulate, B.; Cantu, D. Iso-seq allows genome-independent transcriptome profiling of grape berry development. G3 Genes Genom. Genet. 2019, 9, 755–767. [Google Scholar] [CrossRef] [Green Version]
- Minio, A.; Massonnet, M.; Figueroa-Balderas, R.; Castro, A.; Cantu, D. Diploid genome assembly of the wine grape Carménère. G3 Genes Genom. Genet. 2019, 9, 1331–1337. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.F.; Minio, A.; Massonnet, M.; Solares, E.; Lv, Y.D.; Beridze, T.; Cauto, D.; Gaut, B.S. The population genetics of structural variants in grapevine domestication. Nat. Plants 2019, 5, 965–979. [Google Scholar] [CrossRef]
- Massonnet, M.; Cochetel, N.; Minio, A.; Vondras, A.M.; Lin, J.; Muyle, A.; Garcia, J.F.; Zhou, Y.; Delledonne, M.; Riaz, S.; et al. The genetic basis of sex determination in grapes. Nat. Commun. 2020, 11, 2902. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Massonnet, M.; Minio, A.; Patel, S.; Llaca, V.; Karn, A.; Gouker, F.; Cadle-Davidson, L.; Reisch, B.; Fennell, A.; et al. Multiple independent recombinations led to hermaphroditism in grapevine. Proc. Natl. Acad. Sci. USA 2021, 118, e2023548118. [Google Scholar] [CrossRef] [PubMed]
- Vondras, A.M.; Minio, A.; Blanco-Ulate, B.; Figueroa-Balderas, R.; Penn, M.A.; Zhou, Y.; Seymour, D.; Ye, Z.; Liang, D.; Espinoza, L.K.; et al. The genomic diversification of grapevine clones. BMC Genom. 2019, 20, 972. [Google Scholar] [CrossRef] [Green Version]
- Vitulo, N.; Forcato, C.; Carpinelli, E.C.; Telatin, A.; Campagna, D.; D’Angelo, M.; Zimbello, R.; Corso, M.; Vannozzi, A.; Bonghi, C.; et al. A deep survey of alternative splicing in grape reveals changes in the splicing machinery related to tissue, stress condition and genotype. BMC Plant Biol. 2014, 14, 99. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Canaguier, A.; Grimplet, J.; Di Gaspero, G.; Scalabrin, S.; Duchêne, E.; Choisne, N.; Mohellibi, N.; Guichard, C.; Rombauts, S.; Le Clainche, I.; et al. A new version of the grapevine reference genome assembly (12X.v2) and of its annotation (VCost.v3). Genom. Data 2017, 14, 56–62. [Google Scholar] [CrossRef]
- Sawata, Y.; Matsukawa, T.; Doi, S.; Tsunoda, T.; Arikawa, N.; Matsunaga, N.; Ohnuki, K.; Shirasawa, S.; Kotake, Y. A novel compound, ferulic acid-bound resveratrol, induces the tumor suppressor gene p15 and inhibits the three-dimensional proliferation of colorectal cancer cells. Mol. Cell. Biochem. 2019, 462, 25–31. [Google Scholar] [CrossRef]
- Shi, J.Q.; Huang, S.M.; Fu, D.H.; Yu, J.Y.; Wang, X.F.; Hua, W.; Liu, S.Y.; Liu, G.H. Evolutionary dynamics of microsatellite distribution in plants: Insight from the comparison of sequenced Brassica, Arabidopsis and other angiosperm species. PLoS ONE 2013, 8, e59988. [Google Scholar] [CrossRef] [Green Version]
- Rozen, S.; Skaletsky, H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000, n132, 365–386. [Google Scholar]
- Chory, J.; Baldwin, A.S. Nondenaturing Polyacrylamide Gel Electrophoresis: Preparation and Analysis of DNA. Curr. Protoc. Mol. Biol. 1994, 25, 2.7.1–2.7.8. [Google Scholar] [CrossRef]
- Anderson, J.A.; Churchill, G.A.; Autrique, J.E.; Tanksley, S.D.; Sorrells, M.E. Optimizing parental selection for genetic linkage maps. Genome 1993, 36, 181–186. [Google Scholar] [CrossRef]
- Krzywinski, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [Green Version]
- Lee, T.H.; Guo, H.; Wang, X.; Kim, C.; Paterson, A.H. SNPhylo: A pipeline to construct a phylogenetic tree from huge SNP data. BMC Genom. 2014, 15, 162. [Google Scholar] [CrossRef] [Green Version]
- Willis, L.G.; Winston, M.L.; Honda, B.M. Phylogenetic relationships in the honeybee (genus Apis) as determined by the sequence of the cytochrome oxidase II region of mitochondrial DNA. Mol. Phylogenetics Evol. 1992, 1, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Varshney, R.K.; Nayak, S.N.; May, G.D.; Jackson, S.A. Next-generation sequencing technologies and their implications for crop genetics and breeding. Trends Biotechnol. 2009, 27, 522–530. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Guo, L.; Song, P.; Luan, F.; Hu, J.; Sun, X.; Yang, L. Development of genome-wide SSR markers in melon with their cross-species transferability analysis and utilization in genetic diversity study. Mol. Breed. 2016, 36, 153. [Google Scholar] [CrossRef]
- Wang, Q.; Fang, L.; Chen, J.; Hu, Y.; Si, Z.; Wang, S.; Chang, L.; Guo, W.; Zhang, T. Genome-wide mining, characterization, and development of microsatellite markers in Gossypium species. Sci. Rep. 2015, 5, 10638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Deng, Y.; Tan, J.; Hu, S.; Yu, J.; Xue, Q. A genome-wide microsatellite polymorphism database for the indica and japonica rice. DNA Res. 2007, 14, 37–45. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Song, P.; Koo, D.H.; Guo, L.; Li, Y.; Sun, S.; Weng, Y.; Yang, L. Genome wide characterization of simple sequence repeats in watermelon genome and their application in comparative mapping and genetic diversity analysis. BMC Genom. 2016, 17, 557. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, J.F.; Zhang, J.Y.; Jiang, M.Y.; Wang, W.R.; Zhou, X.R. Development of genome-wide SSR markers in rapeseed by next generation sequencing. Gene 2021, 798, 145798. [Google Scholar] [CrossRef]
- Cui, J.; Cheng, J.; Nong, D.; Peng, J.; Hu, Y.; He, W.; Zhou, Q.; Dhillon, N.P.S.; Hu, K. Genome-wide analysis of simple sequence repeats in bitter gourd (Momordica charantia). Front. Plant Sci. 2017, 8, 1103. [Google Scholar] [CrossRef] [Green Version]
- Cheng, J.; Zhao, Z.; Li, B.; Qin, C.; Wu, Z.; Trejo-Saavedra, D.L.; Luo, X.; Cui, J.; Rivera-Bustamante, R.F.; Li, S.; et al. A comprehensive characterization of simple sequence repeats in pepper genomes provides valuable resources for marker development in Capsicum. Sci. Rep. 2016, 6, 18919. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, P.; Wang, M.; Feng, K.; Cui, L.; Tong, W.; Song, W.; Nie, X. Genome-wide characterization of microsatellites in Triticeae species: Abundance, distribution and evolution. Sci. Rep. 2016, 6, 32224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ellegren, H. Microsatellites: Simple sequences with complex evolution. Nat. Rev. Genet. 2004, 5, 435–445. [Google Scholar] [CrossRef] [PubMed]
- Morgante, M.; Hanafey, M.; Powell, W. Microsatellites are preferentially associated with nonrepetitive DNA in plant genomes. Nat. Genet. 2002, 30, 194–200. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.C.; Korol, A.B.; Fahima, T.; Beiles, A.; Nevo, E. Microsatellites: Genomic distribution, putative functions and mutational mechanisms: A review. Mol. Ecol. 2002, 11, 2453–2465. [Google Scholar] [CrossRef]
- Zhang, L.; Peng, J.; Li, X.; Cui, C.; Sun, J.; Yang, G. Characterization of genome-wide microsatellites of Saccharina japonica based on a preliminary assembly of Illumina sequencing reads. J. Ocean Univ. China 2016, 15, 523–532. [Google Scholar] [CrossRef]
- Han, B.; Wang, C.; Tang, Z.; Ren, Y.; Li, Y.; Zhang, D.; Dong, Y.; Zhao, X. Genome-wide analysis of microsatellite markers based on sequenced database in Chinese spring wheat (Triticum aestivum L.). PLoS ONE 2015, 10, e0141540. [Google Scholar] [CrossRef] [Green Version]
- Bowers, J.E.; Dangl, G.S.; Meredith, C.P. Development and characterization of additional microsatellite DNA markers for grape. Am. J. Enol. Viticult. 1999, 50, 243–246. [Google Scholar] [CrossRef]
- Röckel, F.; Moock, C.; Schwander, F.; Maul, E.; Töpfer, R.; Hausmann, L. A 69 kbp Deletion at the Berry Color Locus Is Responsible for Berry Color Recovery in Vitis vinifera L. Cultivar ‘Riesling Rot’. Int. J. Mol. Sci. 2022, 23, 3708. [Google Scholar] [CrossRef]


| Cultivars | SSRs Number | GC (%) | Genome Length (Mb) | Frequency | |
|---|---|---|---|---|---|
| Per Mb | One Every (Kb) | ||||
| ‘Cabernet Sauvignon’ | 572,976 | 34.61 | 591.0 | 969.50 | 1.03 |
| ‘Carménère’ | 627,429 | 34.45 | 622.8 | 1007.43 | 0.99 |
| ‘Chardonnay’ | 561,635 | 34.34 | 606.0 | 926.79 | 1.08 |
| ‘Merlot’ | 481,491 | 34.56 | 606.1 | 794.41 | 1.26 |
| ‘Muscat Hamburg’ | 286,192 | 34.61 | 457.0 | 626.24 | 1.60 |
| ‘PN40024’ | 297,659 | 34.55 | 486.0 | 612.07 | 1.63 |
| ‘Riesling’ | 470,670 | 34.43 | 741.8 | 634.50 | 1.58 |
| ‘Shine Muscat’ | 308,365 | 34.49 | 484.9 | 635.94 | 1.57 |
| ‘Zinfandel’ | 537,606 | 34.42 | 591.2 | 909.35 | 1.10 |
| Repeat Type | Repeat Motif | Number | Proportion (%) | Frequency |
|---|---|---|---|---|
| Mononucleotide | A/T | 196,103–419,278 | 65.334–68.659 | 0.4579–0.5442 |
| C/G | 4014–9509 | 1.341–1.778 | 0.0134–0.0186 | |
| Dinucleotide | AT/AT | 34,612–81,602 | 11.937–13.387 | 0.1666–0.1797 |
| AG/CT | 13,551–30,551 | 4.5270–5.163 | 0.529–0.571 | |
| AC/GT | 4403–10,218 | 1.522–1.727 | 0.0167–0.0178 | |
| CG/CG | 21–50 | 0.0070–0.0100 | 0.001 | |
| Trinucleotide | AAT/ATT | 19,253–44,482 | 6.702–7.434 | 0.0888–0.0953 |
| AAG/CTT | 2884–6769 | 0.9700–1.1111 | 0.0113–0.0124 | |
| ACC/GGT | 702–1474 | 0.2300–0.2620 | 0.0025–0.0029 | |
| AAC/GTT | 631–1488 | 0.2120–0.2500 | 0.0024–0.0029 | |
| AGG/CCT | 509–1110 | 0.1610–0.1890 | 0.0019–0.0020 | |
| ACT/AGT | 179–418 | 0.0620–0.0770 | 0.0008–0.0010 | |
| CCG/CGG | 70–170 | 0.0240–0.0310 | 0.0002–0.0003 | |
| ACG/CGT | 62–143 | 0.0200–0.0240 | 0.0002 | |
| Tetranucleotide | AAAT/ATTT | 4156–9370 | 1.396–1.545 | 0.0187–0.0200 |
| AATT/AATT | 475–1100 | 0.1600–0.1810 | 0.0021–0.0022 | |
| AAAG/CTTT | 429–969 | 0.1450–0.1710 | 0.0021–0.0022 | |
| ACAT/ATGT | 321–743 | 0.1100–0.1300 | 0.0018–0.0020 | |
| AAAC/GTTT | 198–437 | 0.0640–0.0750 | 0.0008–0.0010 | |
| AGAT/ATCT | 146–368 | 0.0490–0.0600 | 0.0008–0.0009 | |
| AGGG/CCCT | 88–209 | 0.0260–0.0360 | 0.0004–0.0005 | |
| Pentanucleotide | AAAAT/ATTTT | 530–1169 | 0.1680–0.2090 | 0.0027–0.0034 |
| AAATT/AATTT | 48–112 | 0.0120–0.0180 | 0.0002–0.0003 | |
| AATAT/ATATT | 41–98 | 0.0140–0.0180 | 0.0003 | |
| AAAAC/GTTTT | 37–84 | 0.0100–0.0160 | 0.0001–0.0002 | |
| CCCCG/CGGGG | 30–53 | 0.0070–0.0120 | 0.0001–0.0002 | |
| AAAGG/CCTTT | 15–37 | 0.0040–0.0070 | 0.0001 | |
| Hexanucleotide | AAAAAT/ATTTTT | 77–170 | 0.0220–0.0320 | 0.0004–0.0006 |
| AAAAAG/CTTTTT | 75–174 | 0.0190–0.0300 | 0.0004–0.0006 | |
| AGAGGG/CCCTCT | 22–58 | 0.0060–0.0100 | 0.0001–0.0002 | |
| AAAAAC/GTTTTT | 13–32 | 0.0040–0.0060 | 0.0001 | |
| ACTCCC/AGTGGG | 7–28 | 0.0020–0.0050 | 0.0000–0.0001 | |
| AAAAGG/CCTTTT | 5–20 | 0.0020–0.0030 | 0.0000–0.0001 |
| Mono- | Di- | Tri- | Tetra- | Penta- | Hexa- | ||
|---|---|---|---|---|---|---|---|
| Total number | 227,078 | 56,388 | 25,230 | 6679 | 1090 | 409 | |
| Between genes | 160,947 | 39,104 | 17,937 | 4973 | 790 | 250 | |
| Within genes | Exon | 281 | 186 | 1363 | 11 | 3 | 46 |
| Others | 44,466 | 11,970 | 3670 | 1107 | 199 | 83 | |
| Others | 21,384 | 5128 | 2260 | 588 | 98 | 30 | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pei, D.; Song, S.; Kang, J.; Zhang, C.; Wang, J.; Dong, T.; Ge, M.; Pervaiz, T.; Zhang, P.; Fang, J. Characterization of Simple Sequence Repeat (SSR) Markers Mined in Whole Grape Genomes. Genes 2023, 14, 663. https://doi.org/10.3390/genes14030663
Pei D, Song S, Kang J, Zhang C, Wang J, Dong T, Ge M, Pervaiz T, Zhang P, Fang J. Characterization of Simple Sequence Repeat (SSR) Markers Mined in Whole Grape Genomes. Genes. 2023; 14(3):663. https://doi.org/10.3390/genes14030663
Chicago/Turabian StylePei, Dan, Siyan Song, Jun Kang, Chuan Zhang, Jing Wang, Tianyu Dong, Mengqing Ge, Tariq Pervaiz, Peian Zhang, and Jinggui Fang. 2023. "Characterization of Simple Sequence Repeat (SSR) Markers Mined in Whole Grape Genomes" Genes 14, no. 3: 663. https://doi.org/10.3390/genes14030663
APA StylePei, D., Song, S., Kang, J., Zhang, C., Wang, J., Dong, T., Ge, M., Pervaiz, T., Zhang, P., & Fang, J. (2023). Characterization of Simple Sequence Repeat (SSR) Markers Mined in Whole Grape Genomes. Genes, 14(3), 663. https://doi.org/10.3390/genes14030663






