Exploring the Lifetime Effect of Children on Wellbeing Using Two-Sample Mendelian Randomisation
Abstract
:1. Introduction
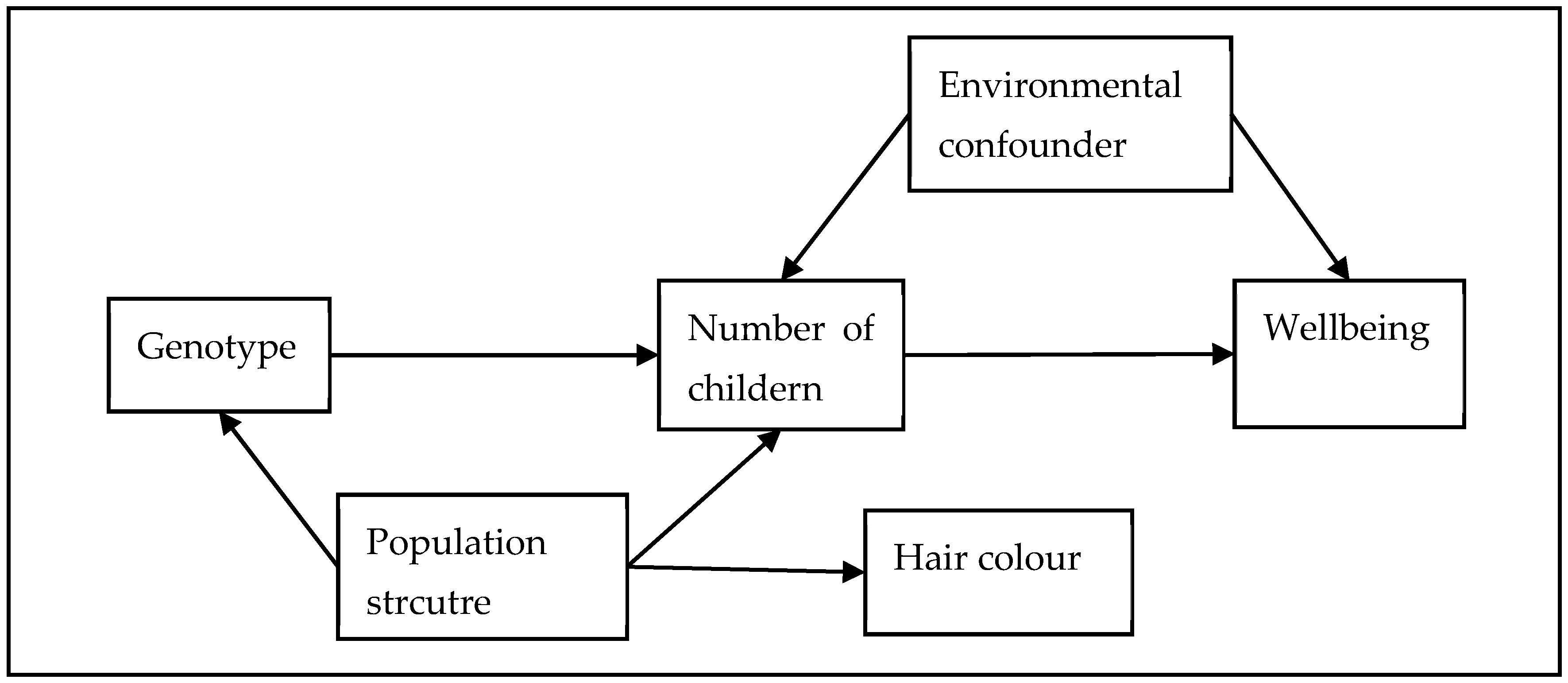
- Mendelian randomisation (MR) is a natural experiment that is theoretically robust for confounding and reverse causation.
- We were able to use two negative control analyses to explore the robustness of our study to two potential sources of residual confounding (populations structure and passive gene–environment correlation).
- We additionally used pleiotropy robust estimates (such as MR-PRESSO, MR-Egger, weighted median, and weighed mode) to explore if our result was affected by the direct effects of the genetic variants on the outcome not mediated by the exposure.
- Because we used summary data, we were unable to explore interactions or non-linear, time-varying, or time sensitive effects.
2. Methods
2.1. Study Design
2.2. Data Sources
2.2.1. UK Biobank (UKB)
2.2.2. Social Science Genetics Consortia (SSGAC)
2.2.3. Within Family Consortium (WFC)
2.3. Phenotyping
2.3.1. UKB
2.3.2. SSGAC
2.3.3. WFC
2.4. Statistical Analysis
2.4.1. Overview of the Analysis
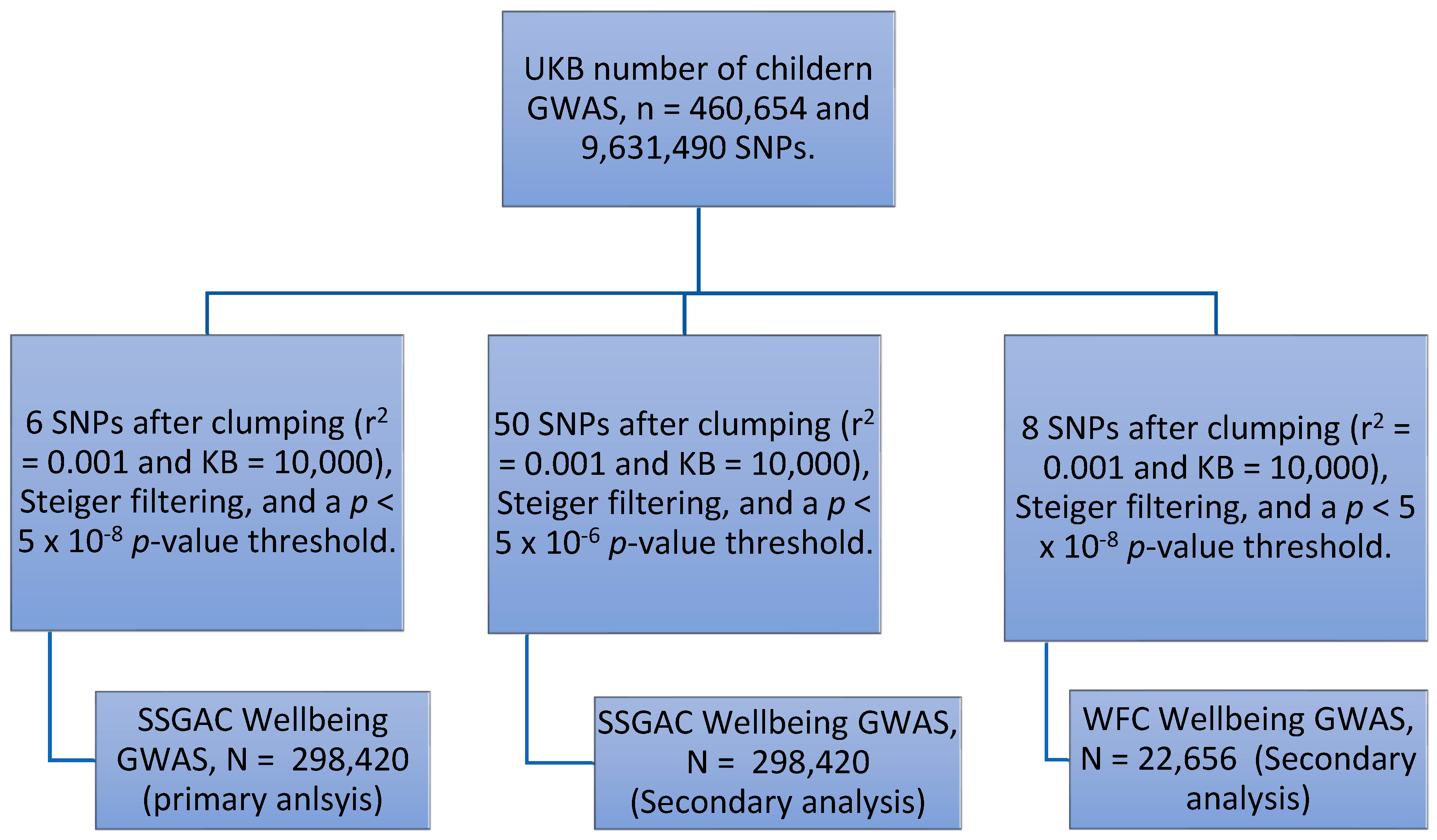
2.4.2. Instrument Construction
2.4.3. Statistical Methods
2.4.4. Assumptions of the Analysis
2.4.5. Assessment of Assumptions
2.5. Sensitivity and Additional Analyses
2.5.1. Negative Controls
2.5.2. WFC GWAS
2.5.3. Less stringent SNP Selection
2.5.4. Leave-One-Out Analysis
2.5.5. Bidirectional Analysis
3. Results
3.1. Descriptive Data
3.1.1. Number of Participants and SNPs in Each Stage
3.1.2. Two-Sample MR Specific Assumptions
3.2. Main Results
3.3. Assessment of Assumptions
3.3.1. Weak Instrument Bias and NOME
3.3.2. Heterogeneity and Exclusion Restriction Violations
3.4. Sensitivity and Additional Analyses
3.4.1. Pleiotropy Robust Estimators
3.4.2. Negative Controls
3.4.3. WFC Outcome
3.4.4. Less Stringent SNP Selection
3.4.5. Leave-One-Out Analysis and MR-PRESSO Outlier Test
3.4.6. Bidirectional MR
4. Discussion
4.1. Pre-Specified Interpretation
4.1.1. Pleiotropy
4.1.2. Residual Confounding
4.1.3. Low Power
4.2. Generalisability
4.3. Strengths and Limitations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Collins, C.; Glass, J. Effects of Work-Family Policies on Parenthood and Wellbeing. Handbook of Family Policy. 30 November 2018. Available online: https://www.elgaronline.com/view/edcoll/9781784719333/9781784719333.00035.xml (accessed on 18 March 2022).
- Herbst, C.M.; Ifcher, J. The increasing happiness of US parents. Rev. Econ. Househ. 2016, 14, 529–551. [Google Scholar] [CrossRef] [Green Version]
- Evenson, R.J.; Simon, R.W. Clarifying the Relationship Between Parenthood and Depression. J. Health Soc. Behav. 2005, 46, 341–358. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McLanahan, S.; Adams, J. Parenthood and Psychological Well-Being. Annu. Rev. Sociol. 1987, 13, 237–257. [Google Scholar] [CrossRef]
- Hughes, M. Parenthood and Psychological Well-Being Among the Formerly Married: Are Children the Primary Source of Psychological Distress? J. Fam. Issues 1989, 10, 463–481. [Google Scholar] [CrossRef]
- Keizer, R.; Dykstra, P.A.; Poortman, A.R. The transition to parenthood and well-being: The impact of partner status and work hour transitions. J. Fam. Psychol. 2010, 24, 429–438. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nomaguchi, K.; Milkie, M.A. Parenthood and Well-Being: A Decade in Review. J. Marriage Fam. 2020, 82, 198–223. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T. Parenthood and Happiness: A Review of Folk Theories Versus Empirical Evidence. Soc. Indic. Res. 2012, 108, 29–64. [Google Scholar] [CrossRef] [Green Version]
- Nelson, S.K.; Kushlev, K.; English, T.; Dunn, E.W.; Lyubomirsky, S. In Defense of Parenthood: Children Are Associated With More Joy Than Misery. Psychol. Sci. 2013, 24, 3–10. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horton, R.S. Parenthood, subjective well-being, and the moderating effects of parent narcissism. J. Individ. Differ. 2021, 42, 57–63. [Google Scholar] [CrossRef]
- Nelson, S.K.; Kushlev, K.; Lyubomirsky, S. The pains and pleasures of parenting: When, why, and how is parenthood associated with more or less well-being? Psychol. Bull. 2014, 140, 846–895. [Google Scholar] [CrossRef] [Green Version]
- Nelson-Coffey, S.K.; Killingsworth, M.; Layous, K.; Cole, S.W.; Lyubomirsky, S. Parenthood Is Associated With Greater Well-Being for Fathers Than Mothers. Pers. Soc. Psychol. Bull. 2019, 45, 1378–1390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brandel, M.; Melchiorri, E.; Ruini, C. The Dynamics of Eudaimonic Well-Being in the Transition to Parenthood: Differences Between Fathers and Mothers. J. Fam. Issues 2018, 39, 2572–2589. [Google Scholar] [CrossRef]
- Umberson, D.; Gove, W.R. Parenthood and Psychological Well-Being: Theory, Measurement, and Stage in the Family Life Course. J. Fam. Issues 1989, 10, 440–462. [Google Scholar] [CrossRef]
- Deaton, A.; Stone, A.A. Evaluative and hedonic wellbeing among those with and without children at home. Proc. Natl. Acad. Sci. USA 2014, 111, 1328–1333. [Google Scholar] [CrossRef] [Green Version]
- Radó, M.K. Tracking the Effects of Parenthood on Subjective Well-Being: Evidence from Hungary. J. Happiness Stud. 2020, 21, 2069–2094. [Google Scholar] [CrossRef] [Green Version]
- Yu, Q.; Zhang, J.; Zhang, L.; Zhang, Q.; Guo, Y.; Jin, S.; Chen, J. Who Gains More? The Relationship Between Parenthood and Well-Being. Evol. Psychol. 2019, 17, 1474704919860467. [Google Scholar] [CrossRef] [Green Version]
- Margolis, R.; Myrskylä, M. A Global Perspective on Happiness and Fertility. Popul. Dev. Rev. 2011, 37, 29–56. [Google Scholar] [CrossRef] [PubMed]
- Stanca, L. Suffer the little children: Measuring the effects of parenthood on well-being worldwide. J. Econ. Behav. Organ. 2012, 81, 742–750. [Google Scholar] [CrossRef] [Green Version]
- Novoa, C.; Bustos, C.; Bühring, V.; Oliva, K.; Páez, D.; Vergara-Barra, P.; Cova, F. Subjective Well-Being and Parenthood in Chile. Int. J. Environ. Res. Public Health 2021, 18, 7408. [Google Scholar] [CrossRef] [PubMed]
- Neuberger, F.S.; Preisner, K. Parenthood and Quality of Life in Old Age: The Role of Individual Resources, the Welfare State and the Economy. Soc. Indic. Res. 2018, 138, 353–372. [Google Scholar] [CrossRef]
- Becker, C.; Kirchmaier, I.; Trautmann, S.T. Marriage, parenthood and social network: Subjective well-being and mental health in old age. PLoS ONE 2019, 14, e0218704. [Google Scholar] [CrossRef] [Green Version]
- Hansen, T.; Slagsvold, B.; Moum, T. Childlessness and Psychological Well-Being in Midlife and Old Age: An Examination of Parental Status Effects Across a Range of Outcomes. Soc. Indic. Res. 2009, 94, 343–362. [Google Scholar] [CrossRef]
- Munafò, M.R.; Davey Smith, G. Robust research needs many lines of evidence. Nature 2018, 553, 399–401. [Google Scholar] [CrossRef] [Green Version]
- Davey Smith, G.; Holmes, M.V.; Davies, N.M.; Ebrahim, S. Mendel’s laws, Mendelian randomization and causal inference in observational data: Substantive and nomenclatural issues. Eur. J. Epidemiol. 2020, 35, 99–111. [Google Scholar] [CrossRef] [Green Version]
- Gage, S.H.; Smith, G.D.; Ware, J.J.; Flint, J.; Munafò, M.R. G = E: What GWAS Can Tell Us about the Environment. PLoS Genet. 2016, 12, e1005765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sanderson, E.; Glymour, M.M.; Holmes, M.V.; Kang, H.; Morrison, J.; Munafò, M.R.; Palmer, T.; Schooling, C.M.; Wallace, C.; Zhao, Q.; et al. Mendelian randomization. Nat. Rev. Methods Prim. 2022, 2, 6. [Google Scholar] [CrossRef]
- Bennett, D.A.; Holmes, M.V. Mendelian randomisation in cardiovascular research: An introduction for clinicians. Heart 2017, 103, 1400–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gill, D.; Georgakis, M.K.; Walker, V.M.; Schmidt, A.F.; Gkatzionis, A.; Freitag, D.F.; Finan, C.; Hingorani, A.D.; Howson, J.M.M.; Burgess, S.; et al. Mendelian randomization for studying the effects of perturbing drug targets. Wellcome Open Res. 2021. Available online: https://wellcomeopenresearch.org/articles/6-16 (accessed on 18 March 2022). [CrossRef]
- Wootton, R.E.; Jones, H.J.; Sallis, H.M. Mendelian randomisation for psychiatry: How does it work, and what can it tell us? Mol. Psychiatry 2021, 27, 53–57. [Google Scholar] [CrossRef] [PubMed]
- McMartin, A.; Conley, D. Commentary: Mendelian randomization and education–Challenges remain. Int. J. Epidemiol. 2020, 49, 1193–1206. [Google Scholar] [CrossRef]
- Jaffee, S.; Price, T. Gene–environment correlations: A review of the evidence and implications for prevention of mental illness. Mol. Psychiatry 2007, 12, 432–442. [Google Scholar] [CrossRef] [Green Version]
- Dawkins, R. The Extended Phenotype: The Long Reach of the Gene, reprint ed.; OUP Oxford: Oxford, UK, 2016; 496p. [Google Scholar]
- Darwin, C. On the Origin of Species: By Means of Natural Selection or the Preservation of Favoured Races in the Struggle for Life, 1st ed.; Bynum, W., Ed.; Penguin Classics: London, UK, 2009; 576p. [Google Scholar]
- The Selfish Gene (Audio Download): Richard Dawkins, Richard Dawkins, Lalla Ward, Audible Studios: Amazon.co.uk: Audible Books & Originals. Available online: https://www.amazon.co.uk/The-Selfish-Gene/dp/B004UAZL5U/ref=sr_1_1?keywords=the+selfish+gene&qid=1647643192&sprefix=the+selfish+g%2Caps%2C119&sr=8-1 (accessed on 18 March 2022).
- Collins, R. What makes UK Biobank special. Lancet 2012, 379, 1173–1174. [Google Scholar] [CrossRef] [PubMed]
- Elsworth, B.; Lyon, M.; Alexander, T.; Liu, Y.; Matthews, P.; Hallett, J.; Bates, P.; Palmer, T.; Haberland, V.; Smith, G.D.; et al. The MRC IEU OpenGWAS data infrastructure. bioRxiv 2020. Available online: https://www.biorxiv.org/content/10.1101/2020.08.10.244293v1 (accessed on 30 March 2022).
- Okbay, A.; Baselmans, B.M.L.; De Neve, J.E.; Turley, P.; Nivard, M.G.; Fontana, M.A.; Meddens, S.F.W.; Linnér, R.K.; Rietveld, C.A.; Derringer, J.; et al. Genetic variants associated with subjective well-being, depressive symptoms, and neuroticism identified through genome-wide analyses. Nat. Genet. 2016, 48, 624–633. [Google Scholar] [CrossRef] [Green Version]
- Howe, L.J.; Nivard, M.G.; Morris, T.T.; Hansen, A.F.; Rasheed, H.; Cho, Y.; Chittoor, G.; Lind, P.A.; Palviainen, T.; van der Zee, M.D.; et al. Within-sibship GWAS improve estimates of direct genetic effects. bioRxiv 2021. Available online: https://www.biorxiv.org/content/10.1101/2021.03.05.433935v1 (accessed on 12 March 2022).
- Davies, N.M.; Howe, L.J.; Brumpton, B.; Havdahl, A.; Evans, D.M.; Davey Smith, G. Within family Mendelian randomization studies. Hum. Mol. Genet. 2019, 28, R170–R179. [Google Scholar] [CrossRef] [Green Version]
- Bigdeli, T.B.; Lee, D.; Riley, B.P.; Vladimirov, V.; Fanous, A.H.; Kendler, K.S.; Bacanu, S.A. FIQT: A simple, powerful method to accurately estimate effect sizes in genome scans. bioRxiv 2015, 019299. Available online: https://www.biorxiv.org/content/10.1101/019299v1 (accessed on 12 March 2022).
- Slob, E.A.W.; Burgess, S. A comparison of robust Mendelian randomization methods using summary data. Genet. Epidemiol. 2020, 44, 313–329. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bowden, J.; Del Greco, M.F.; Minelli, C.; Davey Smith, G.; Sheehan, N.A.; Thompson, J.R. Assessing the suitability of summary data for two-sample Mendelian randomization analyses using MR-Egger regression: The role of the I2 statistic. Int. J. Epidemiol. 2016, 45, 1961–1974. [Google Scholar]
- Bowden, J.; Davey Smith, G.; Burgess, S. Mendelian randomization with invalid instruments: Effect estimation and bias detection through Egger regression. Int. J. Epidemiol. 2015, 44, 512–525. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Q.; Wang, J.; Hemani, G.; Bowden, J.; Small, D.S. Statistical inference in two-sample summary-data Mendelian randomization using robust adjusted profile score. Ann. Stat. 2020, 48, 1742–1769. Available online: http://arxiv.org/abs/1801.09652 (accessed on 17 April 2019). [CrossRef]
- Verbanck, M.; Chen, C.Y.; Neale, B.; Do, R. Detection of widespread horizontal pleiotropy in causal relationships inferred from Mendelian randomization between complex traits and diseases. Nat. Genet. 2018, 50, 693–698. [Google Scholar] [CrossRef] [PubMed]
- Hartwig, F.P.; Wang, L.; Smith, G.D.; Davies, N.M. Average causal effect estimation via instrumental variables: The no simultaneous heterogeneity assumption. arXiv 2021. Available online: http://arxiv.org/abs/2010.10017 (accessed on 28 June 2022). [CrossRef] [PubMed]
- Burgess, S.; Davey Smith, G.; Davies, N.M.; Dudbridge, F.; Gill, D.; Glymour, M.M.; Hartwig, F.P.; Holmes, M.V.; Minelli, C.; Relton, C.L.; et al. Guidelines for performing Mendelian randomization investigations. Wellcome Open Res. 2020, 4, 186. [Google Scholar] [CrossRef] [PubMed]
- Davies, N.M.; Holmes, M.V.; Smith, G.D. Reading Mendelian randomisation studies: A guide, glossary, and checklist for clinicians. BMJ 2018, 362, k601. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Woolf, B.; Di Cara, N.; Moreno-Stokoe, C.; Skrivankova, V.; Drax, K.; Higgins, J.P.T.; Hemani, G.; Munafò, M.R.; Smith, G.D.; Yarmolinsky, J.; et al. Investigating the transparency of reporting in two-sample summary data Mendelian randomization studies using the MR-Base platform. Int. J. Epidemiol. 2022, 51, dyac074. [Google Scholar] [CrossRef] [PubMed]
- Burgess, S.; Davies, N.M.; Thompson, S.G. Instrumental Variable Analysis with a Nonlinear Exposure–Outcome Relationship. Epidemiology 2014, 25, 877–885. [Google Scholar] [CrossRef] [Green Version]
- Loh, P.-R.; Tucker, G.; Bulik-Sullivan, B.K.; Vilhjálmsson, B.J.; Finucane, H.K.; Salem, R.M.; Chasman, D.; Ridker, P.M.; Neale, B.M.; Berger, B.; et al. Efficient Bayesian mixed-model analysis increases association power in large cohorts. Nat. Genet. 2015, 47, 284–290. [Google Scholar] [CrossRef]
- Bulik-Sullivan, B.K.; Loh, P.R.; Finucane, H.K.; Ripke, S.; Yang, J.; Schizophrenia Working Group of the Psychiatric Genomics Consortium; Patterson, N.; Daly, M.J.; Price, A.L.; Neale, B.M. LD Score regression distinguishes confounding from polygenicity in genome-wide association studies. Nat. Genet. 2015, 47, 291–295. [Google Scholar] [CrossRef] [Green Version]
- Lesko, C.R.; Buchanan, A.L.; Westreich, D.; Edwards, J.K.; Hudgens, M.G.; Cole, S.R. Generalizing study results: A potential outcomes perspective. Epidemiology 2017, 28, 553–561. [Google Scholar] [CrossRef]
- Dahabreh, I.J.; Hernán, M.A. Extending inferences from a randomized trial to a target population. Eur. J. Epidemiol. 2019, 34, 719–722. [Google Scholar] [CrossRef] [PubMed]
- Sanderson, E.; Richardson, T.G.; Hemani, G.; Davey Smith, G. The use of negative control outcomes in Mendelian randomization to detect potential population stratification. Int. J. Epidemiol. 2021, 50, 1350–1361. [Google Scholar] [CrossRef] [PubMed]
- Skrivankova, V.W.; Richmond, R.C.; Woolf, B.A.R.; Davies, N.M.; A Swanson, S.; VanderWeele, T.J.; Timpson, N.J.; Higgins, J.P.T.; Dimou, N.; Langenberg, C.; et al. Strengthening the reporting of observational studies in epidemiology using mendelian randomisation (STROBE-MR): Explanation and elaboration. BMJ 2021, 375, n2233. [Google Scholar] [CrossRef] [PubMed]
- Woolf, B.; Sallis, H.; Munafo, M.; Gill, D. MRSamePopTest: Introducing a simple falsification test for the Two-Sample Mendelian randomisation ‘same population’ assumption. OSF Prepr. 2022. Available online: https://osf.io/gvt87/ (accessed on 21 June 2022).
- Staley, J.R.; Blackshaw, J.; Kamat, M.A.; Ellis, S.; Surendran, P.; Sun, B.B.; Paul, D.S.; Freitag, D.; Burgess, S.; Danesh, J.; et al. PhenoScanner: A database of human genotype-phenotype associations. Bioinformatics 2016, 32, 3207–3209. [Google Scholar] [CrossRef] [Green Version]
- Giannelis, A.; Palmos, A.; Hagenaars, S.P.; Breen, G.; Lewis, C.M.; Mutz, J. Examining the association between family status and depression in the UK Biobank. J. Affect. Disord. 2021, 279, 585–598. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, C.; Mutz, J.; Lewis, C.M.; Serretti, A. Stratification of individuals with lifetime depression and low wellbeing in the UK Biobank. J. Affect. Disord. 2022, 314, 281–292. [Google Scholar] [CrossRef]
- Labrecque, J.A.; Swanson, S.A. Interpretation and Potential Biases of Mendelian Randomization Estimates With Time-Varying Exposures. Am. J. Epidemiol. 2019, 188, 231–238. [Google Scholar] [CrossRef]
- Sanderson, E.; Richardson, T.G.; Morris, T.T.; Tilling, K.; Smith, G.D. Estimation of causal effects of a time-varying exposure at multiple time points through Multivariable Mendelian randomization. medRxiv 2022. Available online: https://www.medrxiv.org/content/10.1101/2022.01.04.22268740v1 (accessed on 11 May 2022). [CrossRef]
- Morris, T.T.; Heron, J.; Sanderson, E.C.M.; Davey Smith, G.; Didelez, V.; Tilling, K. Interpretation of Mendelian randomization using a single measure of an exposure that varies over time. Int. J. Epidemiol. 2022, 51, dyac136. [Google Scholar] [CrossRef] [PubMed]
- van Smeden, M.; Lash, T.L.; Groenwold, R.H.H. Reflection on modern methods: Five myths about measurement error in epidemiological research. Int. J. Epidemiol. 2020, 49, 338–347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Angrist, J.D.; Pischke, J.S. Mostly Harmless Econometrics: An Empiricist’s Companion, illustrated ed.; Princeton University Press: Princeton, NJ, USA, 2009; 392p. [Google Scholar]
- Minelli, C.; Del Greco, M.F.; van der Plaat, D.A.; Bowden, J.; Sheehan, N.A.; Thompson, J. The use of two-sample methods for Mendelian randomization analyses on single large datasets. Int. J. Epidemiol. 2021, 50, 1651–1659. [Google Scholar] [CrossRef] [PubMed]
- Ferron, J.; Rendina-Gobioff, G. Interrupted Time Series Design. In Encyclopedia of Statistics in Behavioral Science; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2005; Available online: https://onlinelibrary.wiley.com/doi/abs/10.1002/0470013192.bsa312 (accessed on 11 May 2022).
- Ruth Mitchell, E. MRC IEU UK Biobank GWAS Pipeline Version 2. 2019. Available online: https://data.bris.ac.uk/data/dataset/pnoat8cxo0u52p6ynfaekeigi (accessed on 12 March 2022).
- Hemani, G.; Zheng, J.; Elsworth, B.; Wade, K.H.; Haberland, V.; Baird, D.; Laurin, C.; Burgess, S.; Bowden, J.; Langdon, R.; et al. The MR-Base platform supports systematic causal inference across the human phenome. eLife 2018, 7, e34408. [Google Scholar] [CrossRef] [PubMed]
- Schwarzer, G.; Carpenter, J.R.; Rücker, G. Meta-Analysis with R; Springer International Publishing: Cham, Switzerland, 2015; Available online: http://link.springer.com/10.1007/978-3-319-21416-0 (accessed on 14 April 2022).
- Textor, J.; van der Zander, B.; Gilthorpe, M.S.; Liśkiewicz, M.; Ellison, G.T. Robust causal inference using directed acyclic graphs: The R package ‘dagitty’. Int. J. Epidemiol. 2016, 45, 1887–1894. [Google Scholar] [CrossRef] [PubMed] [Green Version]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Woolf, B.; Sallis, H.M.; Munafò, M.R. Exploring the Lifetime Effect of Children on Wellbeing Using Two-Sample Mendelian Randomisation. Genes 2023, 14, 716. https://doi.org/10.3390/genes14030716
Woolf B, Sallis HM, Munafò MR. Exploring the Lifetime Effect of Children on Wellbeing Using Two-Sample Mendelian Randomisation. Genes. 2023; 14(3):716. https://doi.org/10.3390/genes14030716
Chicago/Turabian StyleWoolf, Benjamin, Hannah M. Sallis, and Marcus R. Munafò. 2023. "Exploring the Lifetime Effect of Children on Wellbeing Using Two-Sample Mendelian Randomisation" Genes 14, no. 3: 716. https://doi.org/10.3390/genes14030716
APA StyleWoolf, B., Sallis, H. M., & Munafò, M. R. (2023). Exploring the Lifetime Effect of Children on Wellbeing Using Two-Sample Mendelian Randomisation. Genes, 14(3), 716. https://doi.org/10.3390/genes14030716




