Machine Learning-Assisted Approaches in Modernized Plant Breeding Programs
Abstract
1. Introduction
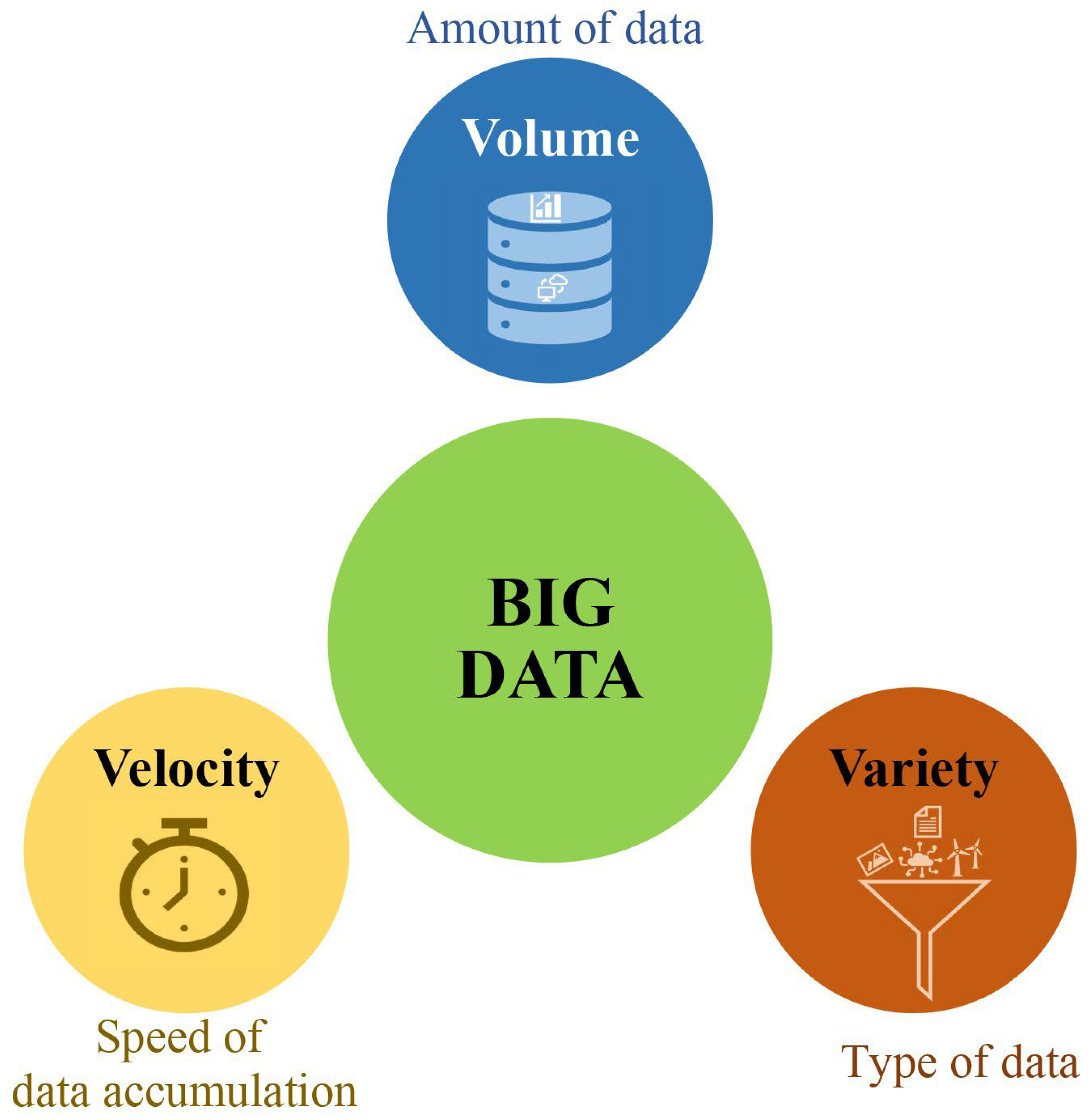
2. What Is Bigdata in Plant Breeding?
3. Artificial Intelligence at a Glance
4. Machine Learning: Basis and Function
5. Machine Learning: Model Evaluation
6. Common ML Algorithms in Plant Breeding
6.1. Support Vector Machine
6.2. Random Forest
6.3. Artificial Neural Network
6.4. Ensemble Learning
7. Data Integration Strategy
Recent Advances in Integration Strategies
8. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Farooq, A.; Farooq, N.; Akbar, H.; Hassan, Z.U.; Gheewala, S.H. A Critical Review of Climate Change Impact at a Global Scale on Cereal Crop Production. Agronomy 2023, 13, 162. [Google Scholar] [CrossRef]
- Coole, D. Should We Control World Population? John Wiley & Sons: Hoboken, NJ, USA, 2018. [Google Scholar]
- Dorling, D. World population prospects at the UN: Our numbers are not our problem? In The Struggle for Social Sustainability; Policy Press: New York, NY, USA, 2021; pp. 129–154. [Google Scholar]
- Yoosefzadeh-Najafabadi, M.; Rajcan, I.; Eskandari, M. Optimizing genomic selection in soybean: An important improvement in agricultural genomics. Heliyon 2022, 8, e11873. [Google Scholar] [CrossRef]
- Najafabadi, M.Y.; Soltani, F.; Noory, H.; Díaz-Pérez, J.C. Growth, yield and enzyme activity response of watermelon accessions exposed to irrigation water déficit. Int. J. Veg. Sci. 2018, 24, 323–337. [Google Scholar] [CrossRef]
- Xu, Y. Molecular Plant Breeding; CABI: Wallingford, UK, 2010. [Google Scholar]
- Poehlman, J.M. Breeding Field Crops; Springer Science & Business Media: Berlin/Heidelberg, Germany, 2013. [Google Scholar]
- Yoosefzadeh-Najafabadi, M.; Rajcan, I.; Vazin, M. High-throughput plant breeding approaches: Moving along with plant-based food demands for pet food industries. Front. Vet. Sci. 2022, 9, 1467. [Google Scholar] [CrossRef]
- Najafabadi, M.Y. Using Advanced Proximal Sensing and Genotyping Tools Combined with Bigdata Analysis Methods to Improve Soybean Yield. Ph.D. Thesis, University of Guelph, Guelph, ON, Canada, 2021. [Google Scholar]
- Mehta, S.; James, D.; Reddy, M. Omics Technologies for Abiotic Stress Tolerance in Plants: Current Status and Prospects. In Recent Approaches in Omics for Plant Resilience to Climate Change; Wani, S., Ed.; Springer: Cham, Switzerland, 2019; pp. 1–34. [Google Scholar]
- Li, Q.; Yan, J. Sustainable Agriculture in the Era of Omics: Knowledge-Driven Crop Breeding; Springer: Berlin/Heidelberg, Germany, 2020; Volume 21, pp. 1–5. [Google Scholar]
- Hesami, M.; Najafabadi, M.Y.; Adamek, K.; Torkamaneh, D.; Jones, A.M.P. Synergizing off-target predictions for in silico insights of CENH3 knockout in cannabis through CRISPR/CAS. Molecules 2021, 26, 2053. [Google Scholar] [CrossRef]
- Pramanik, D.; Shelake, R.M.; Kim, M.J.; Kim, J.-Y. CRISPR-Mediated Engineering across the Central Dogma in Plant Biology for Basic Research and Crop Improvement. Mol. Plant 2021, 14, 127–150. [Google Scholar] [CrossRef]
- Hesami, M.; Alizadeh, M.; Jones, A.M.P.; Torkamaneh, D. Machine learning: Its challenges and opportunities in plant system biology. Appl. Microbiol. Biotechnol. 2022, 106, 3507–3530. [Google Scholar] [CrossRef]
- Nelson, R.; Wiesner-Hanks, T.; Wisser, R.; Balint-Kurti, P. Navigating complexity to breed disease-resistant crops. Nat. Rev. Genet. 2018, 19, 21–33. [Google Scholar] [CrossRef]
- Yoosefzadeh-Najafabadi, M.; Torabi, S.; Tulpan, D.; Rajcan, I.; Eskandari, M. Genome-wide association studies of soybean yield-related hyperspectral reflectance bands using machine learning-mediated data integration methods. Front. Plant Sci. 2021, 12, 777028. [Google Scholar] [CrossRef]
- Jafari, M.; Daneshvar, M.H.; Jafari, S.; Hesami, M. Machine Learning-Assisted In Vitro Rooting Optimization in Passiflora caerulea. Forests 2022, 13, 2020. [Google Scholar] [CrossRef]
- Zhang, N.; Gupta, A.; Chen, Z.; Ong, Y.-S. Evolutionary machine learning with minions: A case study in feature selection. IEEE Trans. Evol. Comput. 2021, 26, 130–144. [Google Scholar] [CrossRef]
- Libbrecht, M.W.; Noble, W.S. Machine learning applications in genetics and genomics. Nat. Rev. Genet. 2015, 16, 321–332. [Google Scholar] [CrossRef] [PubMed]
- Katal, A.; Wazid, M.; Goudar, R.H. Big data: Issues, challenges, tools and good practices. In Proceedings of the 2013 Sixth International Conference on Contemporary Computing (IC3), Noida, India, 8–10 August 2013; pp. 404–409. [Google Scholar]
- Dautov, R.; Distefano, S. Quantifying volume, velocity, and variety to support (Big) data-intensive application development. In Proceedings of the 2017 IEEE International Conference on Big Data (Big Data), Boston, MA, USA, 11–14 December 2017; pp. 2843–2852. [Google Scholar]
- Monino, J.-L.; Sedkaoui, S. Big Data, Open Data and Data Development; John Wiley & Sons: Hoboken, NJ, USA, 2016; Volume 3. [Google Scholar]
- Elgendy, N.; Elragal, A. Big data analytics: A literature review paper. In Proceedings of the Industrial Conference on Data Mining, St. Petersburg, Russia, 16–20 July 2014; pp. 214–227. [Google Scholar]
- Najafabadi, M.Y.; Rajcan, I. Six Decades of Soybean Breeding in Ontario, Canada: A Tradition of Innovation. Can. J. Plant Sci. 2022. [Google Scholar] [CrossRef]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Kim, K.D.; Kang, Y.; Kim, C. Application of genomic big data in plant breeding: Past, present, and future. Plants 2020, 9, 1454. [Google Scholar] [CrossRef]
- Niazian, M.; Niedbała, G. Machine learning for plant breeding and biotechnology. Agriculture 2020, 10, 436. [Google Scholar] [CrossRef]
- Xu, Y.; Zhang, X.; Li, H.; Zheng, H.; Zhang, J.; Olsen, M.S.; Varshney, R.K.; Prasanna, B.M.; Qian, Q. Smart breeding driven by big data, artificial intelligence and integrated genomic-enviromic prediction. Mol. Plant 2022. [Google Scholar] [CrossRef] [PubMed]
- Pearce, S.; Vazquez-Gross, H.; Herin, S.Y.; Hane, D.; Wang, Y.; Gu, Y.Q.; Dubcovsky, J. WheatExp: An RNA-seq expression database for polyploid wheat. BMC Plant Biol. 2015, 15, 1–8. [Google Scholar] [CrossRef]
- Gui, S.; Yang, L.; Li, J.; Luo, J.; Xu, X.; Yuan, J.; Chen, L.; Li, W.; Yang, X.; Wu, S. ZEAMAP, a comprehensive database adapted to the maize multi-omics era. IScience 2020, 23, 101241. [Google Scholar] [CrossRef]
- Wu, L.; Han, L.; Li, Q.; Wang, G.; Zhang, H.; Li, L. Using interactome big data to crack genetic mysteries and enhance future crop breeding. Mol. Plant 2021, 14, 77–94. [Google Scholar] [CrossRef]
- Chow, C.-N.; Zheng, H.-Q.; Wu, N.-Y.; Chien, C.-H.; Huang, H.-D.; Lee, T.-Y.; Chiang-Hsieh, Y.-F.; Hou, P.-F.; Yang, T.-Y.; Chang, W.-C. PlantPAN 2.0: An update of plant promoter analysis navigator for reconstructing transcriptional regulatory networks in plants. Nucleic Acids Res. 2016, 44, D1154–D1160. [Google Scholar] [CrossRef]
- Sandelin, A.; Alkema, W.; Engström, P.; Wasserman, W.W.; Lenhard, B. JASPAR: An open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004, 32, D91–D94. [Google Scholar] [CrossRef]
- Riaño-Pachón, D.M.; Ruzicic, S.; Dreyer, I.; Mueller-Roeber, B. PlnTFDB: An integrative plant transcription factor database. BMC Bioinform. 2007, 8, 42. [Google Scholar] [CrossRef]
- Gu, H.; Zhu, P.; Jiao, Y.; Meng, Y.; Chen, M. PRIN: A predicted rice interactome network. BMC Bioinform. 2011, 12, 161. [Google Scholar] [CrossRef]
- Zhu, G.; Wu, A.; Xu, X.-J.; Xiao, P.-P.; Lu, L.; Liu, J.; Cao, Y.; Chen, L.; Wu, J.; Zhao, X.-M. PPIM: A protein-protein interaction database for maize. Plant Physiol. 2016, 170, 618–626. [Google Scholar] [CrossRef]
- Mueller, L.A.; Zhang, P.; Rhee, S.Y. AraCyc: A biochemical pathway database for Arabidopsis. Plant Physiol. 2003, 132, 453–460. [Google Scholar] [CrossRef]
- Kanehisa, M.; Araki, M.; Goto, S.; Hattori, M.; Hirakawa, M.; Itoh, M.; Katayama, T.; Kawashima, S.; Okuda, S.; Tokimatsu, T. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2007, 36, D480–D484. [Google Scholar] [CrossRef]
- Grafahrend-Belau, E.; Junker, B.H.; Schreiber, F. Plant metabolic pathways: Databases and pipeline for stoichiometric analysis. In Seed Development: OMICS Technologies Toward Improvement of Seed Quality and Crop Yield; Springer: Berlin/Heidelberg, Germany, 2012; pp. 345–366. [Google Scholar]
- Yoosefzadeh-Najafabadi, M.; Eskandari, M.; Torabi, S.; Torkamaneh, D.; Tulpan, D.; Rajcan, I. Machine-learning-based genome-wide association studies for uncovering QTL underlying soybean yield and its components. Int. J. Mol. Sci. 2022, 23, 5538. [Google Scholar] [CrossRef]
- Warwick, K. Artificial Intelligence: The Basics; Routledge: Oxfordshire, UK, 2013. [Google Scholar]
- Harfouche, A.L.; Nakhle, F.; Harfouche, A.H.; Sardella, O.G.; Dart, E.; Jacobson, D. A primer on artificial intelligence in plant digital phenomics: Embarking on the data to insights journey. Trends Plant Sci. 2022, 28, 154–184. [Google Scholar] [CrossRef]
- Pepe, M.; Hesami, M.; Small, F.; Jones, A.M.P. Comparative Analysis of Machine Learning and Evolutionary Optimization Algorithms for Precision Micropropagation of Cannabis sativa: Prediction and Validation of in vitro Shoot Growth and Development Based on the Optimization of Light and Carbohydrate Sources. Front. Plant Sci. 2021, 12, 757869. [Google Scholar] [CrossRef]
- Chandra, A.L.; Desai, S.V.; Guo, W.; Balasubramanian, V.N. Computer vision with deep learning for plant phenotyping in agriculture: A survey. arXiv 2020, arXiv:2006.11391. [Google Scholar]
- Pepe, M.; Hesami, M.; Jones, A.M.P. Machine Learning-Mediated Development and Optimization of Disinfection Protocol and Scarification Method for Improved In Vitro Germination of Cannabis Seeds. Plants 2021, 10, 2397. [Google Scholar] [CrossRef]
- Bhatt, D.; Patel, C.; Talsania, H.; Patel, J.; Vaghela, R.; Pandya, S.; Modi, K.; Ghayvat, H. CNN variants for computer vision: History, architecture, application, challenges and future scope. Electronics 2021, 10, 2470. [Google Scholar] [CrossRef]
- Voulodimos, A.; Doulamis, N.; Doulamis, A.; Protopapadakis, E. Deep learning for computer vision: A brief review. Comput. Intell. Neurosci. 2018, 2018, 7068349. [Google Scholar] [CrossRef]
- Kunze, L.; Hawes, N.; Duckett, T.; Hanheide, M.; Krajník, T. Artificial intelligence for long-term robot autonomy: A survey. IEEE Robot. Autom. Lett. 2018, 3, 4023–4030. [Google Scholar] [CrossRef]
- Murphy, R.R. Introduction to AI Robotics; MIT Press: Cambrige, MA, USA, 2019. [Google Scholar]
- Atefi, A.; Ge, Y.; Pitla, S.; Schnable, J. Robotic technologies for high-throughput plant phenotyping: Contemporary reviews and future perspectives. Front. Plant Sci. 2021, 12, 611940. [Google Scholar] [CrossRef] [PubMed]
- Bangert, W.; Kielhorn, A.; Rahe, F.; Albert, A.; Biber, P.; Grzonka, S.; Haug, S.; Michaels, A.; Mentrup, D.; Hänsel, M. Field-robot-based agriculture:“RemoteFarming. 1” and “BoniRob-Apps”. VDI-Berichte 2013, 2193, 2.1. [Google Scholar]
- Grimstad, L.; From, P.J. Thorvald II-a modular and re-configurable agricultural robot. IFAC-PapersOnLine 2017, 50, 4588–4593. [Google Scholar] [CrossRef]
- Falk, K.G.; Jubery, T.Z.; Mirnezami, S.V.; Parmley, K.A.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B.; Singh, A.K. Computer vision and machine learning enabled soybean root phenotyping pipeline. Plant Methods 2020, 16, 1–19. [Google Scholar] [CrossRef]
- Jafari, M.; Shahsavar, A. The application of artificial neural networks in modeling and predicting the effects of melatonin on morphological responses of citrus to drought stress. PLOS ONE 2020, 15, e0240427. [Google Scholar] [CrossRef] [PubMed]
- Hesami, M.; Jones, A.M.P. Application of artificial intelligence models and optimization algorithms in plant cell and tissue culture. Appl. Microbiol. Biotechnol. 2020, 104, 9449–9485. [Google Scholar] [CrossRef]
- Yoosefzadeh-Najafabadi, M.; Tulpan, D.; Eskandari, M. Using hybrid artificial intelligence and evolutionary optimization algorithms for estimating soybean yield and fresh biomass using hyperspectral vegetation indices. Remote Sens. 2021, 13, 2555. [Google Scholar] [CrossRef]
- Mahesh, B. Machine learning algorithms-a review. Int. J. Sci. Res. 2020, 9, 381–386. [Google Scholar]
- Lee, D.; Seo, H.; Jung, M.W. Neural basis of reinforcement learning and decision making. Annu. Rev. Neurosci. 2012, 35, 287. [Google Scholar] [CrossRef]
- Vabalas, A.; Gowen, E.; Poliakoff, E.; Casson, A.J. Machine learning algorithm validation with a limited sample size. PLoS ONE 2019, 14, e0224365. [Google Scholar] [CrossRef]
- Sakib, S.; Fouda, M.M.; Fadlullah, Z.M.; Nasser, N. Migrating intelligence from cloud to ultra-edge smart IoT sensor based on deep learning: An arrhythmia monitoring use-case. In Proceedings of the 2020 International Wireless Communications and Mobile Computing (IWCMC), Limassol, Cyprus, 15–19 June 2020; pp. 595–600. [Google Scholar]
- Derakhshan, B.; Mahdiraji, A.R.; Rabl, T.; Markl, V. Continuous Deployment of Machine Learning Pipelines. Open Proceed 2019, 36, 397–408. [Google Scholar] [CrossRef]
- Yoosefzadeh-Najafabadi, M.; Heidari, A.; Rajcan, I. AllInOne. 2022. Available online: https://github.com/MohsenYN/AllInOne/wiki (accessed on 15 February 2023).
- Yoosefzadeh-Najafabadi, M.; Earl, H.J.; Tulpan, D.; Sulik, J.; Eskandari, M. Application of machine learning algorithms in plant breeding: Predicting yield from hyperspectral reflectance in soybean. Front. Plant Sci. 2021, 11, 624273. [Google Scholar] [CrossRef]
- Beyene, Y.; Semagn, K.; Mugo, S.; Tarekegne, A.; Babu, R.; Meisel, B.; Sehabiague, P.; Makumbi, D.; Magorokosho, C.; Oikeh, S. Genetic gains in grain yield through genomic selection in eight bi-parental maize populations under drought stress. Crop Sci. 2015, 55, 154–163. [Google Scholar] [CrossRef]
- Fu, W.; Nair, V.; Menzies, T. Why is differential evolution better than grid search for tuning defect predictors? arXiv 2016, arXiv:1609.02613. [Google Scholar]
- Wang, X.; Jin, Y.; Schmitt, S. Recent Advances in Bayesian Optimization. arXiv 2022, arXiv:2206.03301v2. [Google Scholar] [CrossRef]
- Harfouche, A.L.; Jacobson, D.A.; Kainer, D.; Romero, J.C.; Harfouche, A.H.; Mugnozza, G.S.; Moshelion, M.; Tuskan, G.A.; Keurentjes, J.J.; Altman, A. Accelerating climate resilient plant breeding by applying next-generation artificial intelligence. Trends Biotechnol. 2019, 37, 1217–1235. [Google Scholar] [CrossRef] [PubMed]
- Laptev, N.; Amizadeh, S.; Flint, I. Generic and scalable framework for automated time-series anomaly detection. In Proceedings of the 21th ACM SIGKDD international conference on knowledge discovery and data mining, Sydney, Australia, 10–13 August 2015; pp. 1939–1947. [Google Scholar]
- Agarwal, N.; Sondhi, A.; Chopra, K.; Singh, G. Transfer learning: Survey and classification. Smart Innovations in Communication and Computational Sciences; Springer: Singapore, 2021; pp. 145–155. [Google Scholar] [CrossRef]
- Turner, C.R.; Fuggetta, A.; Lavazza, L.; Wolf, A.L. A conceptual basis for feature engineering. J. Syst. Softw. 1999, 49, 3–15. [Google Scholar] [CrossRef]
- Huang, X.; Wu, L.; Ye, Y. A review on dimensionality reduction techniques. Int. J. Pattern Recognit. Artif. Intell. 2019, 33, 1950017. [Google Scholar] [CrossRef]
- Christ, M.; Braun, N.; Neuffer, J.; Kempa-Liehr, A.W. Time series feature extraction on basis of scalable hypothesis tests (tsfresh–a python package). Neurocomputing 2018, 307, 72–77. [Google Scholar] [CrossRef]
- Browne, M.W. Cross-validation methods. J. Math. Psychol. 2000, 44, 108–132. [Google Scholar] [CrossRef] [PubMed]
- Johnson, R.W. An introduction to the bootstrap. Teach. Stat. 2001, 23, 49–54. [Google Scholar] [CrossRef]
- Cawley, G.C.; Talbot, N.L. Efficient leave-one-out cross-validation of kernel fisher discriminant classifiers. Pattern Recognit. 2003, 36, 2585–2592. [Google Scholar] [CrossRef]
- Refaeilzadeh, P.; Tang, L.; Liu, H. Cross-validation. Encycl. Database Syst. 2009, 5, 532–538. [Google Scholar]
- Rao, R.B.; Fung, G.; Rosales, R. On the dangers of cross-validation. An experimental evaluation. In Proceedings of the 2008 SIAM International Conference on Data Mining, Atlanta, Georgia, 24–26 April 2008; pp. 588–596. [Google Scholar]
- Das, K.; Jiang, J.; Rao, J. Mean squared error of empirical predictor. Ann. Stat. 2004, 32, 818–840. [Google Scholar] [CrossRef]
- Chai, T.; Draxler, R.R. Root mean square error (RMSE) or mean absolute error (MAE). Geosci. Model Dev. Discuss. 2014, 7, 1525–1534. [Google Scholar]
- Recchia, A. R-squared measures for two-level hierarchical linear models using SAS. J. Stat. Softw. 2010, 32, 1–9. [Google Scholar] [CrossRef]
- Vrieze, S.I. Model selection and psychological theory: A discussion of the differences between the Akaike information criterion (AIC) and the Bayesian information criterion (BIC). Psychol. Methods 2012, 17, 228. [Google Scholar] [CrossRef] [PubMed]
- Huang, J.; Ling, C.X. Using AUC and accuracy in evaluating learning algorithms. IEEE Trans. Knowl. Data Eng. 2005, 17, 299–310. [Google Scholar] [CrossRef]
- Galdi, P.; Tagliaferri, R. Data mining: Accuracy and error measures for classification and prediction. Encycl. Bioinform. Comput. Biol. 2018, 1, 431–436. [Google Scholar]
- Akin, M.; Eyduran, S.P.; Eyduran, E.; Reed, B.M. Analysis of macro nutrient related growth responses using multivariate adaptive regression splines. Plant Cell Tissue Organ Cult. 2020, 140, 661–670. [Google Scholar] [CrossRef]
- Pisner, D.A.; Schnyer, D.M. Support vector machine. In Machine Learning; Elsevier: Amsterdam, The Netherlands, 2020; pp. 101–121. [Google Scholar]
- Griffel, L.; Delparte, D.; Edwards, J. Using Support Vector Machines classification to differentiate spectral signatures of potato plants infected with Potato Virus Y. Comput. Electron. Agric. 2018, 153, 318–324. [Google Scholar] [CrossRef]
- Zhao, W.; Lai, X.; Liu, D.; Zhang, Z.; Ma, P.; Wang, Q.; Zhang, Z.; Pan, Y. Applications of support vector machine in genomic prediction in pig and maize populations. Front. Genet. 2020, 11, 598318. [Google Scholar] [CrossRef]
- Shafiee, S.; Lied, L.M.; Burud, I.; Dieseth, J.A.; Alsheikh, M.; Lillemo, M. Sequential forward selection and support vector regression in comparison to LASSO regression for spring wheat yield prediction based on UAV imagery. Comput. Electron. Agric. 2021, 183, 106036. [Google Scholar] [CrossRef]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Qi, Y. Random forest for bioinformatics. In Ensemble Machine Learning; Springer: Berlin/Heidelberg, Germany, 2012; pp. 307–323. [Google Scholar]
- Parmley, K.; Nagasubramanian, K.; Sarkar, S.; Ganapathysubramanian, B.; Singh, A.K. Development of optimized phenomic predictors for efficient plant breeding decisions using phenomic-assisted selection in soybean. Plant Phenomics 2019, 2019, 5809404. [Google Scholar] [CrossRef]
- Akin, M.; Hand, C.; Eyduran, E.; Reed, B.M. Predicting minor nutrient requirements of hazelnut shoot cultures using regression trees. Plant Cell Tissue Organ Cult. 2018, 132, 545–559. [Google Scholar] [CrossRef]
- Kovalchuk, I.Y.; Mukhitdinova, Z.; Turdiyev, T.; Madiyeva, G.; Akin, M.; Eyduran, E.; Reed, B.M. Modeling some mineral nutrient requirements for micropropagated wild apricot shoot cultures. Plant Cell Tissue Organ Cult. 2017, 129, 325–335. [Google Scholar] [CrossRef]
- Shi, T.; Horvath, S. Unsupervised learning with random forest predictors. J. Comput. Graph. Stat. 2006, 15, 118–138. [Google Scholar] [CrossRef]
- Zou, J.; Han, Y.; So, S.-S. Overview of artificial neural networks. Artif. Neural Netw. 2008, 458, 14–22. [Google Scholar]
- Agatonovic-Kustrin, S.; Beresford, R. Basic concepts of artificial neural network (ANN) modeling and its application in pharmaceutical research. J. Pharm. Biomed. Anal. 2000, 22, 717–727. [Google Scholar] [CrossRef]
- Silva, G.N.; Tomaz, R.S.; Sant’Anna, I.d.C.; Nascimento, M.; Bhering, L.L.; Cruz, C.D. Neural networks for predicting breeding values and genetic gains. Sci. Agric. 2014, 71, 494–498. [Google Scholar] [CrossRef]
- Brasileiro, B.P.; Marinho, C.D.; Costa, P.M.d.A.; Cruz, C.D.; Peternelli, L.A.; Barbosa, M.H.P. Selection in sugarcane families with artificial neural networks. Crop Breed. Appl. Biotechnol. 2015, 15, 72–78. [Google Scholar] [CrossRef]
- Sant’Anna, I.; Tomaz, R.S.; Silva, G.N.; Nascimento, M.; Bhering, L.L.; Cruz, C.D. Superiority of artificial neural networks for a genetic classification procedure. Genet. Mol. Res. 2015, 14, 9898–9906. [Google Scholar] [CrossRef]
- Dongare, A.; Kharde, R.; Kachare, A.D. Introduction to artificial neural network. Int. J. Eng. Innov. Technol. 2012, 2, 189–194. [Google Scholar]
- Hesami, M.; Alizadeh, M.; Naderi, R.; Tohidfar, M. Forecasting and optimizing Agrobacterium-mediated genetic transformation via ensemble model- fruit fly optimization algorithm: A data mining approach using chrysanthemum databases. PLoS ONE 2020, 15, e0239901. [Google Scholar] [CrossRef]
- Hesami, M.; Naderi, R.; Tohidfar, M. Introducing a hybrid artificial intelligence method for high-throughput modeling and optimizing plant tissue culture processes: The establishment of a new embryogenesis medium for chrysanthemum, as a case study. Appl. Microbiol. Biotechnol. 2020, 104, 10249–10263. [Google Scholar] [CrossRef] [PubMed]
- Dietterich, T.G. Ensemble learning. In The Handbook of Brain Theory and Neural Networks; MIT Press: Cambrige, MA, USA, 2002; Volume 2, pp. 110–125. [Google Scholar]
- Polikar, R. Ensemble learning. In Ensemble Machine Learning; Springer: Berlin/Heidelberg, Germany, 2012; pp. 1–34. [Google Scholar]
- Zhou, Z.-H. Ensemble learning. In Machine Learning; Springer: Berlin/Heidelberg, Germany, 2021; pp. 181–210. [Google Scholar]
- Sewell, M. Ensemble learning. RN 2008, 11, 1–34. [Google Scholar]
- Wu, T.; Zhang, W.; Jiao, X.; Guo, W.; Hamoud, Y.A. Evaluation of stacking and blending ensemble learning methods for estimating daily reference evapotranspiration. Comput. Electron. Agric. 2021, 184, 106039. [Google Scholar] [CrossRef]
- Yu, T.; Zhang, W.; Han, J.; Li, F.; Wang, Z.; Cao, C. An ensemble learning approach for predicting phenotypes from genotypes. In Proceedings of the 2021 20th International Conference on Ubiquitous Computing and Communications (IUCC/CIT/DSCI/SmartCNS), London, UK, 20 December 2021; pp. 382–389. [Google Scholar]
- Yoosefzadeh-Najafabadi, M.; Eskandari, M.; Belzile, F.; Torkamaneh, D. Genome-wide association study statistical models: A review. Genome-Wide Assoc. Stud. 2022, 1, 43–62. [Google Scholar]
- Yoosefzadeh-Najafabadi, M.; Tulpan, D.; Eskandari, M. Application of machine learning and genetic optimization algorithms for modeling and optimizing soybean yield using its component traits. PLoS ONE 2021, 16, e0250665. [Google Scholar] [CrossRef] [PubMed]
- Fabres, P.J.; Collins, C.; Cavagnaro, T.R.; Rodríguez López, C.M. A concise review on multi-omics data integration for terroir analysis in Vitis vinifera. Front. Plant Sci. 2017, 8, 1065. [Google Scholar] [CrossRef] [PubMed]
- Acerbi, F.; Sassanelli, C.; Taisch, M. A conceptual data model promoting data-driven circular manufacturing. Oper. Manag. Res. 2022, 15, 1–20. [Google Scholar]
- Cavill, R.; Jennen, D.; Kleinjans, J.; Briedé, J.J. Transcriptomic and metabolomic data integration. Brief. Bioinform. 2016, 17, 891–901. [Google Scholar] [CrossRef]
- Espíndola, D.B.; Fumagalli, L.; Garetti, M.; Pereira, C.E.; Botelho, S.S.; Henriques, R.V. A model-based approach for data integration to improve maintenance management by mixed reality. Comput. Ind. 2013, 64, 376–391. [Google Scholar] [CrossRef]
- ElKarami, B.; Alkhateeb, A.; Qattous, H.; Alshomali, L.; Shahrrava, B. Multi-omics Data Integration Model Based on UMAP Embedding and Convolutional Neural Network. Cancer Inform. 2022, 21, 11769351221124205. [Google Scholar] [CrossRef]
- Fei, S.; Hassan, M.A.; Xiao, Y.; Su, X.; Chen, Z.; Cheng, Q.; Duan, F.; Chen, R.; Ma, Y. UAV-based multi-sensor data fusion and machine learning algorithm for yield prediction in wheat. Precis. Agric. 2022, 24, 187–212. [Google Scholar] [CrossRef] [PubMed]
- Moeinizade, S.; Pham, H.; Han, Y.; Dobbels, A.; Hu, G. An applied deep learning approach for estimating soybean relative maturity from UAV imagery to aid plant breeding decisions. Mach. Learn. Appl. 2022, 7, 100233. [Google Scholar] [CrossRef]
- Chawade, A.; van Ham, J.; Blomquist, H.; Bagge, O.; Alexandersson, E.; Ortiz, R. High-throughput field-phenotyping tools for plant breeding and precision agriculture. Agronomy 2019, 9, 258. [Google Scholar] [CrossRef]
- Sandhu, K.S.; Lozada, D.N.; Zhang, Z.; Pumphrey, M.O.; Carter, A.H. Deep learning for predicting complex traits in spring wheat breeding program. Front. Plant Sci. 2021, 11, 613325. [Google Scholar] [CrossRef]
- Szymczak, S.; Biernacka, J.M.; Cordell, H.J.; González-Recio, O.; König, I.R.; Zhang, H.; Sun, Y.V. Machine learning in genome-wide association studies. Genet. Epidemiol. 2009, 33, S51–S57. [Google Scholar] [CrossRef]
- Asif, H.; Alliey-Rodriguez, N.; Keedy, S.; Tamminga, C.A.; Sweeney, J.A.; Pearlson, G.; Clementz, B.A.; Keshavan, M.S.; Buckley, P.; Liu, C. GWAS significance thresholds for deep phenotyping studies can depend upon minor allele frequencies and sample size. Mol. Psychiatry 2020, 26, 1–8. [Google Scholar] [CrossRef]
- Arshadi, N.; Chang, B.; Kustra, R. Predictive modeling in case-control single-nucleotide polymorphism studies in the presence of population stratification: A case study using Genetic Analysis Workshop 16 Problem 1 dataset. BMC Proc. 2009, 3, S60. [Google Scholar] [CrossRef]
- Grömping, U. Variable importance assessment in regression: Linear regression versus random forest. Am. Stat. 2009, 63, 308–319. [Google Scholar] [CrossRef]
- Ziliak, S. P values and the search for significance. Nat. Methods 2017, 14, 3–4. [Google Scholar]
- Di Leo, G.; Sardanelli, F. Statistical significance: p value, 0.05 threshold, and applications to radiomics—reasons for a conservative approach. Eur. Radiol. Exp. 2020, 4, 1–8. [Google Scholar] [CrossRef]
- Tang, R.; Sinnwell, J.P.; Li, J.; Rider, D.N.; de Andrade, M.; Biernacka, J.M. Identification of genes and haplotypes that predict rheumatoid arthritis using random forests. BMC Proc. 2009, 3, S68. [Google Scholar] [CrossRef] [PubMed]
- Kuriakose, S.V.; Pushker, R.; Hyde, E.M. Data-driven decisions for accelerated plant breeding. In Accelerated Plant Breeding; Springer: Berlin/Heidelberg, Germany, 2020; Volume 1, pp. 89–119. [Google Scholar]
- Mishra, S.; Mishra, D.; Santra, G.H. Applications of machine learning techniques in agricultural crop production: A review paper. Indian J. Sci. Technol. 2016, 9, 1–14. [Google Scholar] [CrossRef]




| ML Method | Hyperparameter Tuning | Overfitting Risk | Explainability | Comparative Accuracy | Complexity | Samples Needed | Computation Cost | Implementation Time |
|---|---|---|---|---|---|---|---|---|
| Deep Neural Network | Very high | Low | Low | Very high | Very high | Very high | Very high | Very high |
| Artificial Neural Network | High | High | Low | High | High | Medium | High | High |
| Random Forest | High | Medium | High | High | Medium | Low | Medium | Low |
| Non-linear (Kernel) SVM | High | High | Low | High | High | High | High | High |
| Linear SVM | High | High | Low | High | Medium | High | Medium | Medium |
| Decision Tree | Medium | High | High | Medium | Medium | Medium | Medium | Low |
| Self-Organizing Maps | High | High | Medium | Medium | High | Medium | Low | Low |
| K nearest neighbor | Medium | High | Medium | Medium | High | Medium | Low | Low |
| Gradient Boosting | Very high | High | Low | High | Very high | Medium | High | Medium |
| Naive Bayes | Medium | Medium | Medium | Medium | High | Medium | High | Medium |
| Bayesian Network | High | Medium | Medium | Medium | High | Medium | High | Medium |
| Partial Linear Regression | Medium | Low | High | Medium | Medium | High | Medium | Low |
| Logistic Regression | Low | High | High | Low | Medium | Medium | Medium | Low |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoosefzadeh Najafabadi, M.; Hesami, M.; Eskandari, M. Machine Learning-Assisted Approaches in Modernized Plant Breeding Programs. Genes 2023, 14, 777. https://doi.org/10.3390/genes14040777
Yoosefzadeh Najafabadi M, Hesami M, Eskandari M. Machine Learning-Assisted Approaches in Modernized Plant Breeding Programs. Genes. 2023; 14(4):777. https://doi.org/10.3390/genes14040777
Chicago/Turabian StyleYoosefzadeh Najafabadi, Mohsen, Mohsen Hesami, and Milad Eskandari. 2023. "Machine Learning-Assisted Approaches in Modernized Plant Breeding Programs" Genes 14, no. 4: 777. https://doi.org/10.3390/genes14040777
APA StyleYoosefzadeh Najafabadi, M., Hesami, M., & Eskandari, M. (2023). Machine Learning-Assisted Approaches in Modernized Plant Breeding Programs. Genes, 14(4), 777. https://doi.org/10.3390/genes14040777








