Donkey Oil-Based Ketogenic Diet Prevents Tumor Progression by Regulating Intratumor Inflammation, Metastasis and Angiogenesis in CT26 Tumor-Bearing Mice
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Line and Culturing
2.2. Mice and Tumor Implantation
2.3. DOKD Treatments
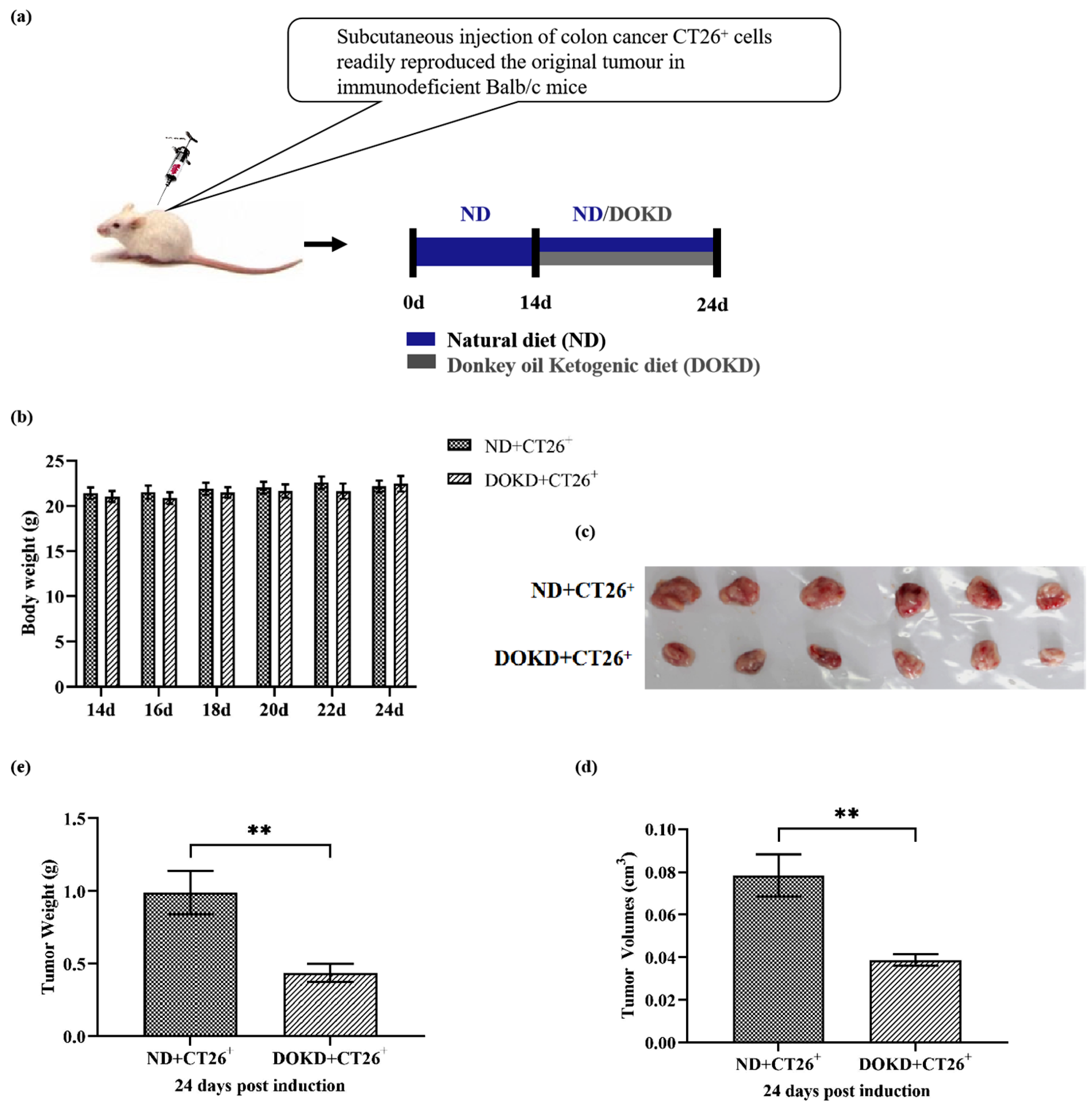
2.4. Measurement of Body Weight and Tumor Volume
2.5. Measurement of β-Hydroxybutyrate and Glucose Levels
2.6. Transcriptome Sequencing (RNA-Seq)
2.7. Western Blot (WB)
2.8. Cell Counting Kit-8 (CCK8) Assay
2.9. Statistical Analysis
3. Results
3.1. KD Inhibits CT26+ Colon Tumor Growth
3.2. Effects of DOKD on Blood β-Hydroxybutyrate and Blood Glucose in CT26+ Bearing Mice
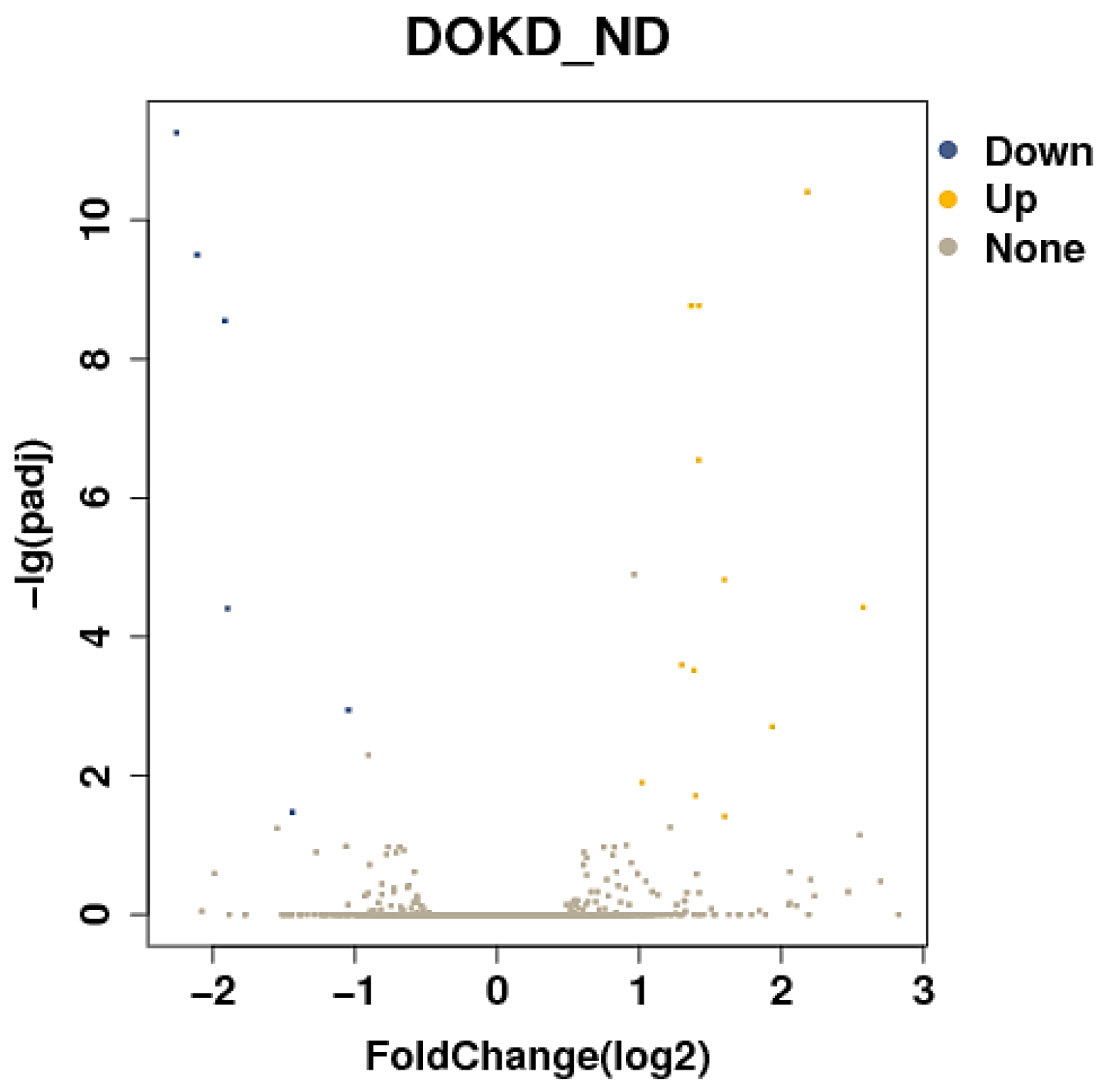
3.3. Differentially Expressed Genes (DEGs) of the DOKD/ND Group
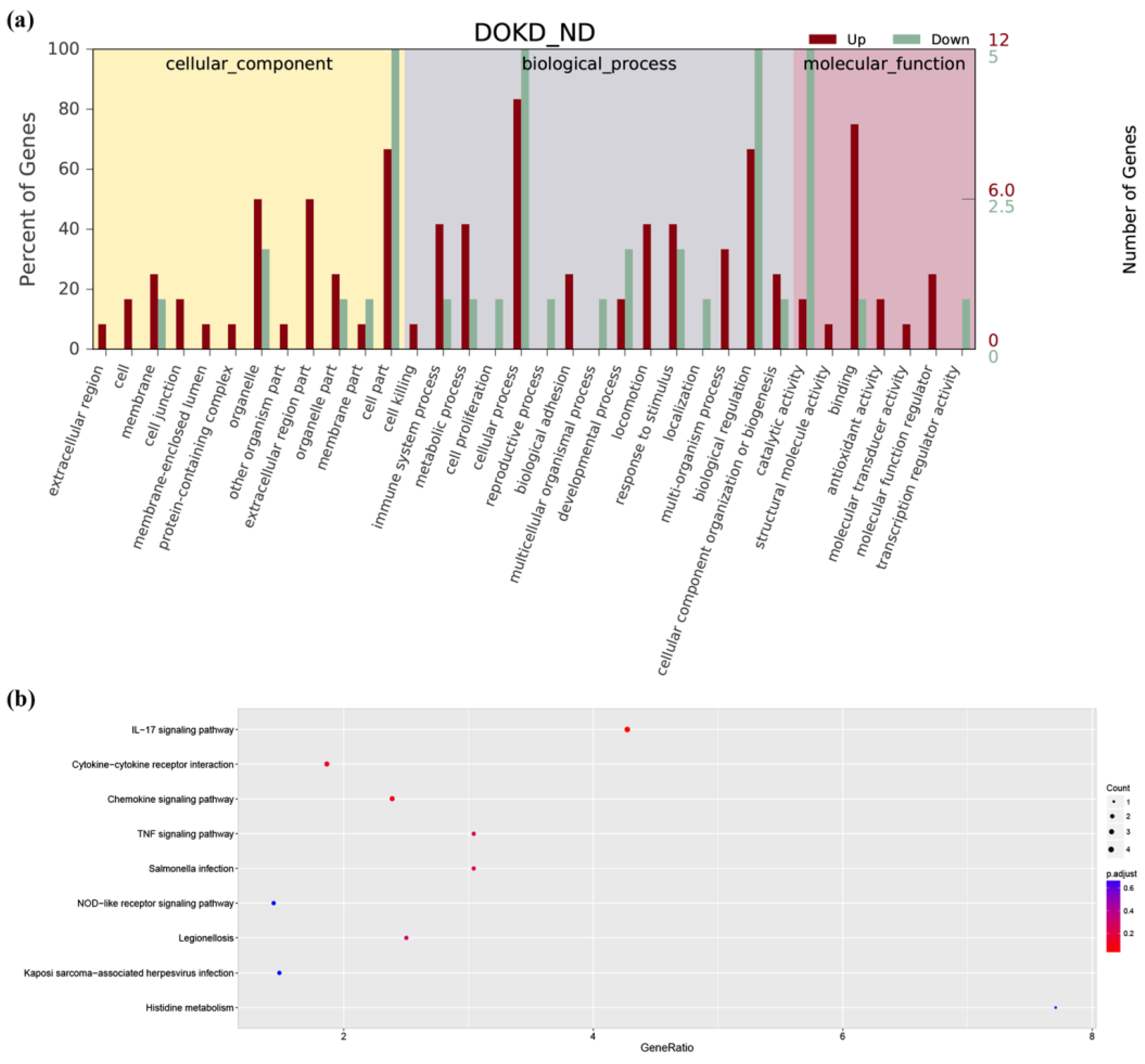
3.4. GO and KEGG Analysis
3.5. Therapeutic Pathways and Potential Targets of DOKD on Mouse CT26+ Colon Cancer
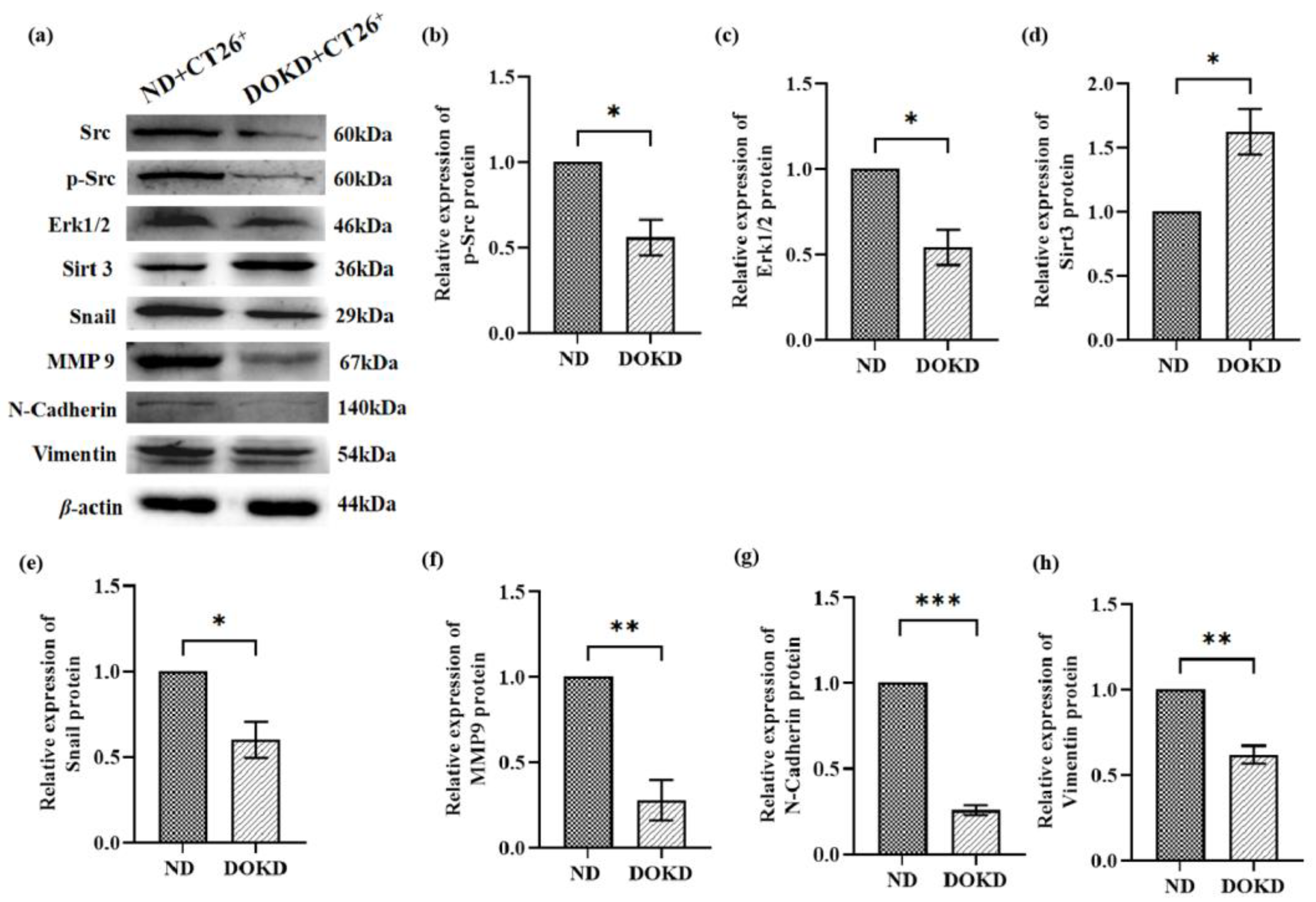
3.6. DOKD Up-Regulate the Expressions of IL-17, TLR4, NF-κB p65 and TNF-α and Down-Regulate the Expressions of HIF-1α in CT26+ Bearing Mice
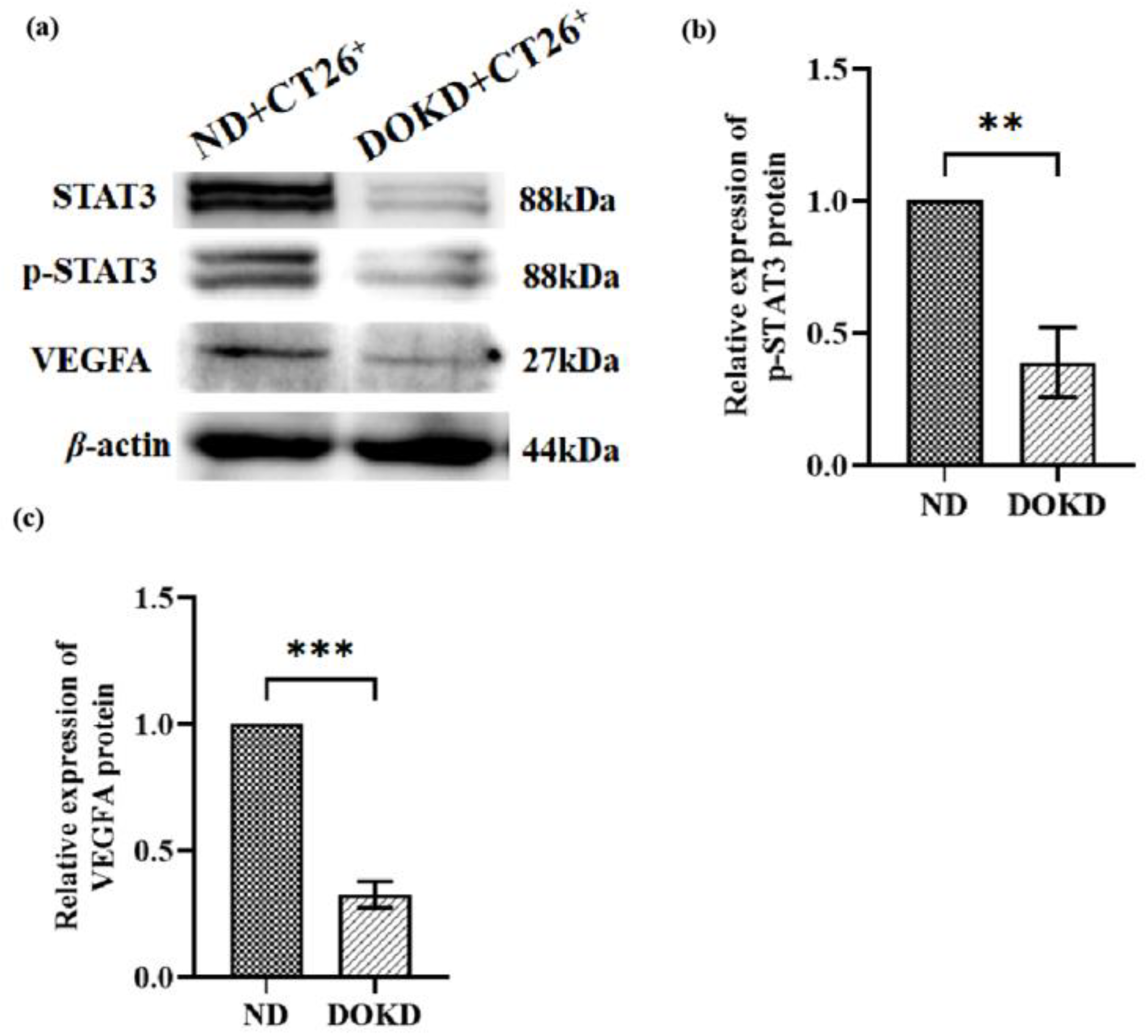
3.7. DOKD Inhibits the Expression of Metastasis-Related Proteins in CT26+ Bearing Mice
3.8. DOKD Inhibits Tumor Angiogenesis in a Colon Cancer CT26 Tumor Model
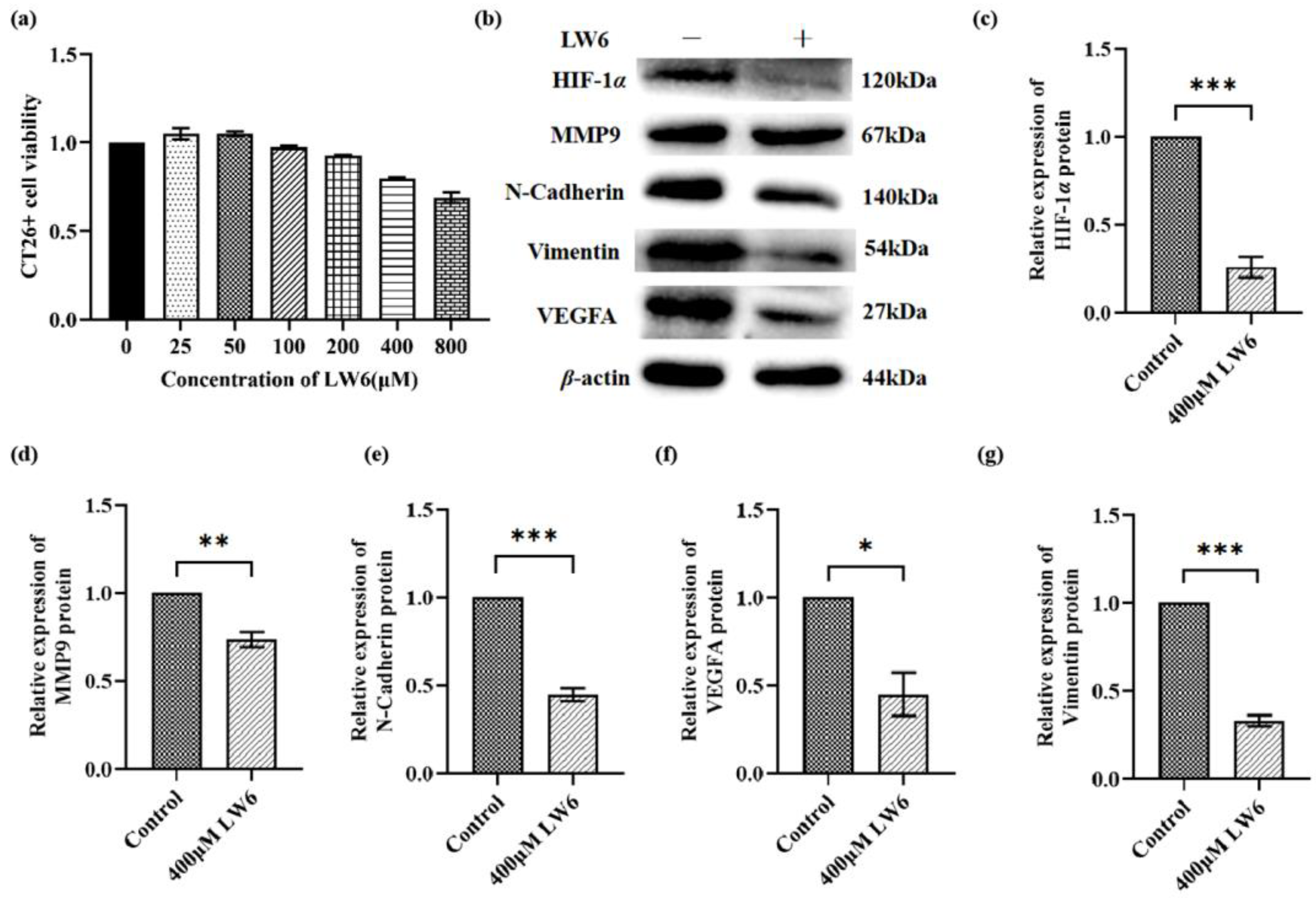
3.9. HIF-1α Regulated Tumor Angiogenesis and Metastasis by Vimentin, MMP9 and VEGFA Pathway in CT26+ Colon Cancer Cells
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Colon Cancer. Am. Fam. Physician 2018, 10. Available online: https://pubmed.ncbi.nlm.nih.gov/29763282/ (accessed on 3 March 2023).
- Gupta, R.; Bhatt, L.K.; Johnston, T.P.; Prabhavalkar, K.S. Colon cancer stem cells: Potential target for the treatment of colorectal cancer. Cancer Biol. Ther. 2019, 20, 1068–1082. [Google Scholar] [CrossRef] [PubMed]
- Pallag, A.; Roşca, E.; Ţiţ, D.M.; Muţiu, G.; Bungău, S.G.; Pop, O.L. Monitoring the effects of treatment in colon cancer cells using immunohistochemical and histoenzymatic techniques. Rom. J. Morphol. Embryol. 2015, 56, 1103–1109. [Google Scholar]
- Weber, D.D.; Aminzadeh-Gohari, S.; Tulipan, J.; Catalano, L.; Feichtinger, R.G.; Kofler, B. Ketogenic diet in the treatment of cancer-Where do we stand? Mol. Metab. 2020, 33, 102–121. [Google Scholar] [CrossRef]
- Feng, S.; Wang, H.; Liu, J.; Aa, J.; Zhou, F.; Wang, G. Multi-dimensional roles of ketone bodies in cancer biology: Opportunities for cancer therapy. Pharmacol. Res. 2019, 150, 104500. [Google Scholar] [CrossRef] [PubMed]
- Sperry, J.; Condro, M.C.; Guo, L.; Braas, D.; Vanderveer-Harris, N.; Kim, K.K.O.; Pope, W.B.; Divakaruni, A.S.; Lai, A.; Christofk, H.; et al. Glioblastoma Utilizes Fatty Acids and Ketone Bodies for Growth Allowing Progression during Ketogenic Diet Therapy. iScience 2020, 23, 101453. [Google Scholar] [CrossRef]
- Mann, S.D.O.; Sidhu, M.D.O.; Gowin, K.D.O. Understanding the Mechanisms of Diet and Outcomes in Colon, Prostate, and Breast Cancer; Malignant Gliomas; and Cancer Patients on Immunotherapy. Nutrients 2020, 12, 2226. [Google Scholar] [CrossRef]
- Okechukwu, C.E. Cross Talk between the Ketogenic Diet and Metastatic Prostate Cancer Cells. World J. Men’s Health 2022, 40, 162–163. [Google Scholar] [CrossRef]
- Cohen, C.W.; Fontaine, K.R.; Arend, R.C.; Gower, B.A. A Ketogenic Diet Is Acceptable in Women with Ovarian and Endometrial Cancer and Has No Adverse Effects on Blood Lipids: A Randomized, Controlled Trial. Nutr. Cancer 2020, 72, 584–594. [Google Scholar] [CrossRef]
- Nakamura, K.; Tonouchi, H.; Sasayama, A.; Ashida, K. A Ketogenic Formula Prevents Tumor Progression and Cancer Cachexia by Attenuating Systemic Inflammati on in Colon 26 Tumor-Bearing Mice. Nutrients 2018, 10, 206. [Google Scholar] [CrossRef]
- Batch, J.T.; Lamsal, S.P.; Adkins, M.; Sultan, S.; Ramirez, M.N. Advantages and Disadvantages of the Ketogenic Diet: A Review Article. Cureus 2020, 12, e9639. [Google Scholar] [CrossRef] [PubMed]
- Paoli, A. Ketogenic diet for obesity: Friend or foe? Int. J. Environ. Res. Public Health 2014, 11, 2092–2107. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.; Zhang, L.; Cao, Y.; Yang, L.; Zhang, H.; Li, Y.; Zhao, H.; Li, Y. Chemical and physical characterization of donkey abdo minal fat in comparisonwith cow, pig and sheep fats. J. Am. Oil Chem. Soc. 2013, 90, 1371–1376. [Google Scholar] [CrossRef]
- Li, L.; Wei, Z.; Zhang, J.; Long, Y.; Shi, J.; Jiao, Z.; Hun, J.; Liu, G. Comparison of Fatty Acid Component between Donkey Bones, Donkey Skin and E-jiao. Mod. Food Sci. Technol. 2020, 36, 82–87. [Google Scholar] [CrossRef]
- Lee, G.Y.; Han, S.N. The Role of Vitamin E in Immunity. Nutrients 2018, 10, 1614. [Google Scholar] [CrossRef]
- Shin, M.K.; Jeon, Y.D.; Hong, S.H.; Kang, S.H.; Kee, J.Y.; Jin, J.S.; Kuzu, O.F.; Noory, M.A.; Robertson, G.P. The Role of Cholesterol in Cancer. Cancer Res. 2016, 76, 2063–2070. [Google Scholar] [CrossRef]
- Kumar, S.; Behl, T.; Sachdeva, M.; Sehgal, A.; Kumari, S.; Kumar, A.; Kaur, G.; Yadav, H.N.; Bungau, S. Implicating the effect of ketogenic diet as a preventive measure to obesity and diabetes mellitus. Life Sci. 2021, 264, 118661. [Google Scholar] [CrossRef]
- Mitchell, J.P.; Carmody, R.J. NF-κB and the Transcriptional Control of Inflammation. Int. Rev. Cell Mol. Biol. 2018, 335, 41–84. [Google Scholar] [CrossRef]
- Singh, N.; Baby, D.; Rajguru, J.P.; Patil, P.B.; Thakkannavar, S.S.; Pujari, V.B. Inflammation and cancer. Ann. Afr. Med. 2019, 18, 121–126. [Google Scholar] [CrossRef]
- Wang, X.; Yang, L.; Huang, F.; Zhang, Q.; Liu, S.; Ma, L.; You, Z. Inflammatory cytokines IL-17 and TNF-α up-regulate PD-L1 expression in human prostate and colon cancer cells. Immunol. Lett. 2017, 184, 7–14. [Google Scholar] [CrossRef]
- Lussier, D.M.; Woolf, E.C.; Johnson, J.L.; Brooks, K.S.; Blattman, J.N.; Scheck, A.C. Enhanced immunity in a mouse model of malignant glioma is mediated by a therapeutic ketogenic diet. BMC Cancer 2016, 16, 310. [Google Scholar] [CrossRef]
- Korbecki, J.; Simińska, D.; Gąssowska-Dobrowolska, M.; Listos, J.; Gutowska, I.; Chlubek, D.; Baranowska-Bosiacka, I. Chronic and Cycling Hypoxia: Drivers of Cancer Chronic Inflammation through HIF-1 and NF-κB Activation: A Review of the Molecular Mechanisms. Int. J. Mol. Sci. 2021, 22, 10701. [Google Scholar] [CrossRef] [PubMed]
- Ioannou, M.; Paraskeva, E.; Baxevanidou, K.; Simos, G.; Papamichali, R.; Papacharalambous, C.; Samara, M.; Koukoulis, G. HIF-1α in colorectal carcinoma: Review of the literature. J. BUON 2015, 20, 680–689. [Google Scholar] [PubMed]
- Anderson, R.L.; Balasas, T.; Callaghan, J.; Coombes, R.C.; Evans, J.; Hall, J.A.; Kinrade, S.; Jones, D.; Jones, P.S.; Jones, R.; et al. Cancer Research UK and Cancer Therapeutics CRC Australia Metastasis Working Group. A framework for the development of effective anti-metastatic agents. Nat. Rev. Clin. Oncol. 2019, 16, 185–204. [Google Scholar] [CrossRef] [PubMed]
- Wheelock, M.J.; Shintani, Y.; Maeda, M.; Fukumoto, Y.; Johnson, K.R. Cadherin switching. J. Cell Sci. 2008, 121, 727–735. [Google Scholar] [CrossRef] [PubMed]
- Gheldof, A.; Berx, G. Cadherins and epithelial-to-mesenchymal transition. Prog. Mol. Biol. Transl. Sci. 2013, 116, 317–336. [Google Scholar] [CrossRef] [PubMed]
- Satelli, A.; Li, S. Vimentin in cancer and its potential as a molecular target for cancer therapy. Cell. Mol. Life Sci. 2011, 68, 3033–3046. [Google Scholar] [CrossRef]
- Mrozik, K.M.; Blaschuk, O.W.; Cheong, C.M.; Zannettino, A.C.W.; Vandyke, K. N-cadherin in cancer metastasis, its emerging role in haematological malignancies and potential as a therapeutic target in cancer. BMC Cancer 2018, 18, 939. [Google Scholar] [CrossRef]
- Lambert, A.W.; Pattabiraman, D.R.; Weinberg, R.A. Emerging Biological Principles of Metastasis. Cell 2017, 168, 670–691. [Google Scholar] [CrossRef] [PubMed]
- Roche, J. The Epithelial-to-Mesenchymal Transition in Cancer. Cancers 2018, 10, 52, Erratum in Cancers 2018, 10, 79. [Google Scholar] [CrossRef]
- Gialeli, C.; Theocharis, A.D.; Karamanos, N.K. Roles of matrix metalloproteinases in cancer progression and their pharmacological targeting. FEBS J. 2011, 278, 16–27. [Google Scholar] [CrossRef] [PubMed]
- Mehner, C.; Hockla, A.; Miller, E.; Ran, S.; Radisky, D.C.; Radisky, E.S. Tumor cell-produced matrix metalloproteinase 9 (MMP-9) drives malignant progression and metastasis of basal-like triple negative breast cancer. Oncotarget 2014, 5, 2736–2749. [Google Scholar] [CrossRef] [PubMed]
- Joseph, C.; Alsaleem, M.; Orah, N.; Narasimha, P.L.; Miligy, I.M.; Kurozumi, S.; Ellis, I.O.; Mongan, N.P.; Green, A.R.; Rakha, E.A. Elevated MMP9 expression in breast cancer is a predictor of shorter patient survival. Breast Cancer Res. Treat. 2020, 182, 267–282. [Google Scholar] [CrossRef]
- Nishida, N.; Yano, H.; Nishida, T.; Kamura, T.; Kojiro, M. Angiogenesis in cancer. Vasc. Health Risk Manag. 2006, 2, 213–219. [Google Scholar] [CrossRef] [PubMed]
- Backer, M.V.; Backer, J.M. Imaging key biomarkers of tumor angiogenesis. Theranostics 2012, 2, 502–515. [Google Scholar] [CrossRef]
- Zhang, Q.; Lu, S.; Li, T.; Yu, L.; Zhang, Y.; Zeng, H.; Qian, X.; Bi, J.; Lin, Y. ACE2 inhibits breast cancer angiogenesis via suppressing the VEGFa/VEGFR2/ERK pathway. J. Exp. Clin. Cancer Res. 2019, 38, 173. [Google Scholar] [CrossRef] [PubMed]
- Liang, L.; Hui, K.; Hu, C.; Wen, Y.; Yang, S.; Zhu, P.; Wang, L.; Xia, Y.; Qiao, Y.; Sun, W.; et al. Autophagy inhibition potentiates the anti-angiogenic property of multikinase inhibitor anlotinib through JAK2/STAT3/VEGFA signaling in non-small cell lung cancer cells. J. Exp. Clin. Cancer Res. 2019, 38, 71. [Google Scholar] [CrossRef]
- Lai, S.W.; Chen, M.Y.; Bamodu, O.A.; Hsieh, M.S.; Huang, T.Y.; Yeh, C.T.; Lee, W.H.; Cherng, Y.G. Exosomal lncRNA PVT1/VEGFA Axis Promotes Colon Cancer Metastasis and Stemness by Downregulation of Tumor Suppressor miR-152-3p. Oxid. Med. Cell Longev. 2021, 2021, 9959807. [Google Scholar] [CrossRef]
- Wang, H.; Jia, R.; Lv, T.; Wang, M.; He, S.; Zhang, X. Resveratrol Suppresses Tumor Progression via Inhibiting STAT3/HIF-1α/VEGF Pathway in an Orthotopic Rat Model of Non-Small-Cell Lung Cancer (NSCLC). Onco. Targets Ther. 2020, 13, 7057–7063. [Google Scholar] [CrossRef]
- Chen, J.; Liu, X.; Jiao, H.; Peng, L.; Huo, Z.; Yang, W.; Shen, Q.; Li, T.; Liu, Q. Prognostic and clinical significance of STAT3 and MMP9 in patients with gastric cancer: A meta-analysis of a Chinese cohort. Int. J. Clin. Exp. Med. 2015, 8, 546–557. [Google Scholar]
- Wang, C.; Liu, H.; Yang, M.; Bai, Y.; Ren, H.; Zou, Y.; Yao, Z.; Zhang, B.; Li, Y. RNA-Seq Based Transcriptome Analysis of Endothelial Differentiation of Bone Marrow Mesenchymal Stem Cells. Eur. J. Vasc. Endovasc. Surg. 2020, 59, 834–842. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef]
- Zhang, N.; Liu, C.; Jin, L.; Zhang, R.; Wang, T.; Wang, Q.; Chen, J.; Yang, F.; Siebert, H.C.; Zheng, X. Ketogenic Diet Elicits Antitumor Properties through Inducing Oxidative Stress, Inhibiting MMP-9 Expression, and Rebalancing M1/M2 Tumor-Associated Macrophage Phenotype in a Mouse Model of Colon Cancer. J. Agric. Food Chem. 2020, 68, 11182–11196. [Google Scholar] [CrossRef]
- Zhou, J.; Jiang, Y.Y.; Chen, H.; Wu, Y.C.; Zhang, L. Tanshinone I attenuates the malignant biological properties of ovarian cancer by inducing apoptosis and autophagy via the inactivation of PI3K/AKT/mTOR pathway. Cell Prolif. 2020, 53, e12739. [Google Scholar] [CrossRef]
- Hu, L.; Liu, Y.; Kong, X.; Wu, R.; Peng, Q.; Zhang, Y.; Zhou, L.; Duan, L. Fusobacterium nucleatum Facilitates M2 Macrophage Polarization and Colorectal Carcinoma Progression by Activating TLR4/NF-κB/S100A9 Cascade. Front. Immunol. 2021, 12, 658681. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Fan, H.; Zhao, Y.; Chen, X.; Zhu, Z.; Zha, X.; Zhao, Y.; Chai, X.; Li, J.; Tu, P.; et al. An immune-stimulating proteoglycan from the medicinal mushroom Huaier up-regulates NF-κB and MAPK signaling via Toll-like receptor 4. J. Biol. Chem. 2019, 294, 2628–2641. [Google Scholar] [CrossRef] [PubMed]
- McGeachy, M.J.; Cua, D.J.; Gaffen, S.L. The IL-17 Family of Cytokines in Health and Disease. Immunity 2019, 50, 892–906. [Google Scholar] [CrossRef]
- Shao, J.; Ding, Z.; Peng, J.; Zhou, R.; Li, L.; Qian, Q.; Chen, Y. MiR-146a-5p promotes IL-1β-induced chondrocyte apoptosis through the TRAF6-mediated NF-kB pathway. Inflamm. Res. 2020, 69, 619–630. [Google Scholar] [CrossRef]
- Warburg, O.; Wind, F.; Negelein, E. The Metabolism of Tumors in the Body. J. Gen. Physiol. 1927, 8, 519–530. [Google Scholar] [CrossRef] [PubMed]
- Bhattacharya, B.; Mohd Omar, M.F.; Soong, R. The Warburg effect and drug resistance. Br. J. Pharmacol. 2016, 173, 970–979. [Google Scholar] [CrossRef]
- Uddin, M.S.; Kabir, M.T.; Tewari, D.; Al Mamun, A.; Barreto, G.E.; Bungau, S.G.; Bin-Jumah, M.N.; Abdel-Daim, M.M.; Ashraf, G.M. Emerging Therapeutic Promise of Ketogenic Diet to Attenuate Neuropathological Alterations in Alzheimer’s Disease. Mol. Neurobiol. 2020, 57, 4961–4977. [Google Scholar] [CrossRef] [PubMed]
- Diakos, C.I.; Charles, K.A.; McMillan, D.C.; Clarke, S.J. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014, 15, e493–e503. [Google Scholar] [CrossRef] [PubMed]
- Narumi, K.; Miyakawa, R.; Ueda, R.; Hashimoto, H.; Yamamoto, Y.; Yoshida, T.; Aoki, K. Proinflammatory Proteins S100A8/S100A9 Activate NK Cells via Interaction with RAGE. J. Immunol. 2015, 194, 5539–5548. [Google Scholar] [CrossRef] [PubMed]
- Litak, J.; Grochowski, C.; Litak, J.; Osuchowska, I.; Gosik, K.; Radzikowska, E.; Kamieniak, P.; Rolinski, J. TLR-4 Signaling vs. Immune Checkpoints, miRNAs Molecules, Cancer Stem Cells, and Wingless-Signaling Interplay in Glioblastoma Multiforme-Future Perspectives. Int. J. Mol. Sci. 2020, 21, 3114. [Google Scholar] [CrossRef] [PubMed]
- Eyerich, K.; Dimartino, V.; Cavani, A. IL-17 and IL-22 in immunity: Driving protection and pathology. Eur. J. Immunol. 2017, 47, 607–614. [Google Scholar] [CrossRef]
- Housseau, F.; Wu, S.; Wick, E.C.; Fan, H.; Wu, X.; Llosa, N.J.; Smith, K.N.; Tam, A.; Ganguly, S.; Wanyiri, J.W.; et al. Redundant Innate and Adaptive Sources of IL17 Production Drive Colon Tumorigenesis. Cancer Res. 2016, 76, 2115–2124. [Google Scholar] [CrossRef]
- Abusleme, L.; Moutsopoulos, N.M. IL-17: Overview and role in oral immunity and microbiome. Oral. Dis. 2017, 23, 854–865. [Google Scholar] [CrossRef]
- Lei, R.; Li, J.; Liu, F.; Li, W.; Zhang, S.; Wang, Y.; Chu, X.; Xu, J. HIF-1α promotes the keloid development through the activation of TGF-β/Smad and TLR4/MyD88/NF-κB pathways. Cell Cycle 2019, 18, 3239–3250. [Google Scholar] [CrossRef]
- Wang, B.; Li, K.; Wang, H.; Shen, X.; Zheng, J. Systemic chemotherapy promotes HIF-1α-mediated glycolysis and IL-17F pathways in cutaneous T-cell lymphoma. Exp. Dermatol. 2020, 29, 987–992. [Google Scholar] [CrossRef]
- Li, H.; Jia, Y.; Wang, Y. Targeting HIF-1α signaling pathway for gastric cancer treatment. Pharmazie 2019, 74, 3–7. [Google Scholar] [CrossRef]
- Li, Y.Y.; Zheng, Y.L. Hypoxia promotes invasion of retinoblastoma cells in vitro by upregulating HIF-1α/MMP9 signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2017, 21, 5361–5369. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.N.; Xu, Y.Y.; Ma, Q.; Li, M.Q.; Guo, J.X.; Wang, X.; Jin, X.; Shang, J.; Jiao, L.X. Dextran Sulfate Effects EMT of Human Gastric Cancer Cells by Reducing HIF-1α/ TGF-β. J. Cancer 2021, 12, 3367–3377. [Google Scholar] [CrossRef]
- Suh, Y.; Yoon, C.H.; Kim, R.K.; Lim, E.J.; Oh, Y.S.; Hwang, S.G.; An, S.; Yoon, G.; Gye, M.C.; Yi, J.M.; et al. Claudin-1 induces epithelial-mesenchymal transition through activation of the c-Abl-ERK signaling pathway in human liver cells. Oncogene 2013, 32, 4873–4882. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.B.; Sharma, A.; Dhawan, P. Claudin-1 expression confers resistance to anoikis in colon cancer cells in a Src-dependent manner. Carcinogenesis 2012, 33, 2538–2547. [Google Scholar] [CrossRef]
- Smith, B.N.; Burton, L.J.; Henderson, V.; Randle, D.D.; Morton, D.J.; Smith, B.A.; Taliaferro-Smith, L.; Nagappan, P.; Yates, C.; Zayzafoon, M.; et al. Snail promotes epithelial mesenchymal transition in breast cancer cells in part via activation of nuclear ERK2. PLoS ONE 2014, 9, e104987. [Google Scholar] [CrossRef]
- Derycke, L.D.; Bracke, M.E. N-cadherin in the spotlight of cell-cell adhesion, differentiation, embryogenesis, invasion and signalling. Int. J. Dev. Biol. 2004, 48, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Usman, S.; Waseem, N.H.; Nguyen, T.K.N.; Mohsin, S.; Jamal, A.; Teh, M.T.; Waseem, A. Vimentin Is at the Heart of Epithelial Mesenchymal Transition (EMT) Mediated Metastasis. Cancers 2021, 13, 4985. [Google Scholar] [CrossRef]
- Riva, M.; Källberg, E.; Björk, P.; Hancz, D.; Vogl, T.; Roth, J.; Ivars, F.; Leanderson, T. Induction of nuclear factor-κB responses by the S100A9 protein is Toll-like receptor-4-dependent. Immunology 2012, 137, 172–182. [Google Scholar] [CrossRef]
- Jia, Y.; Yan, X.; Cao, Y.; Song, W.; Zhang, G.; Hu, X. Anticancer. Activity of Modified Tongyou Decoction on Eca109 Esophageal Cancer Cell Invasion and Metastasis through Regulation of the Epithelial-Mesenchymal Transition Mediated by the HIF-1α-Snail Axis. Evid. Based Complement. Altern. Med. 2020, 2020, 3053506. [Google Scholar] [CrossRef]
- Zou, G.; Zhang, X.; Wang, L.; Li, X.; Xie, T.; Zhao, J.; Yan, J.; Wang, L.; Ye, H.; Jiao, S.; et al. Herb-sourced emodin inhibits angiogenesis of breast cancer by targeting VEGFA transcription. Theranostics 2020, 10, 6839–6853. [Google Scholar] [CrossRef]
- Mu, G.; Zhu, Y.; Dong, Z.; Shi, L.; Deng, Y.; Li, H. Calmodulin 2 Facilitates Angiogenesis and Metastasis of Gastric Cancer via STAT3/HIF-1A/VEGF-A Mediated Macrophage Polarization. Front. Oncol. 2021, 11, 727306. [Google Scholar] [CrossRef]
- Kitajima, Y.; Miyazaki, K. The Critical Impact of HIF-1a on Gastric Cancer Biology. Cancers 2013, 5, 15–26. [Google Scholar] [CrossRef] [PubMed]
- Schneider, M.; Korzeniewski, N.; Merkle, K.; Schüler, J.; Grüllich, C.; Hadaschik, B.; Hohenfellner, M.; Duensing, S. The tyrosine kinase inhibitor nilotinib has antineoplastic activity in prostate cancer cells but up-regulates the ERK survival signal-Implications for targeted therapies. Urol. Oncol. 2015, 33, 72.e1–72.e7. [Google Scholar] [CrossRef] [PubMed]
- Mo, C.; Liu, T.; Zhang, S.; Guo, K.; Li, M.; Qin, X.; Liu, Y. Reduced N-acetylglucosaminyltransferase III expression via Smad3 and Erk signaling in TGF-β1-induced HCC EMT model. Discov. Med. 2017, 23, 7–17. [Google Scholar] [PubMed]
- Lin, J.; Cao, S.; Wang, Y.; Hu, Y.; Liu, H.; Li, J.; Chen, J.; Li, P.; Liu, J.; Wang, Q.; et al. Long non-coding RNA UBE2CP3 enhances HCC cell secretion of VEGFA and promotes angiogenesis by activating ERK1/2/HIF-1α/VEGFA signalling in hepatocellular carcinoma. J. Exp. Clin. Cancer Res. 2018, 37, 113. [Google Scholar] [CrossRef] [PubMed]
- Liu, P.; Atkinson, S.J.; Akbareian, S.E.; Zhou, Z.; Munsterberg, A.; Robinson, S.D.; Bao, Y. Sulforaphane exerts anti-angiogenesis effects against hepatocellular carcinoma through inhibition of STAT3/HIF-1α/VEGF signalling. Sci. Rep. 2017, 7, 12651. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, W.; Ding, J.; Wang, J.; Zhang, J. Downregulation of Rab17 promotes cell proliferation and invasion in non-small cell lung cancer through STAT3/HIF-1α/VEGF signaling. Thorac. Cancer 2020, 11, 379–388. [Google Scholar] [CrossRef]
- Morscher, R.J.; Aminzadeh-Gohari, S.; Hauser-Kronberger, C.; Feichtinger, R.G.; Sperl, W.; Kofler, B. Combination of metronomic cyclophosphamide and dietary intervention inhibits neuroblastoma growth in a CD1-nu mouse model. Oncotarget 2016, 7, 17060–17073. [Google Scholar] [CrossRef]



| Composition | ND | DOKD | ||
|---|---|---|---|---|
| Weight (grams/kg) | Energy Density (kcal/g) | Weight (grams/kg) | Energy Density (kcal/g) | |
| Casein | 200.000 | 0.800 | 162.500 | 0.650 |
| Carbohydrate | 626.000 | 2.504 | - | - |
| -Corn Starch | 394.000 | 1.576 | - | - |
| -Maltodextrin | 132.000 | 0.528 | - | - |
| -Sucrose | 100.000 | 0.400 | - | - |
| Fat | 70.000 | 0.630 | 690.000 | 6.210 |
| -Soybean oil | 70.000 | 0.630 | - | - |
| -Donkey oil | - | - | 690.000 | 6.210 |
| Cellulose | 53.500 | 0.000 | 97.000 | 0.000 |
| L-cysteine | 3.000 | 0.012 | 3.000 | 0.012 |
| Mineral mixture | 35.000 | 0.000 | 35.000 | 0.000 |
| a Fiber mixture | 10.000 | 0.040 | 10.000 | 0.040 |
| Choline tartrate | 2.500 | 0.000 | 2.500 | 0.000 |
| Antioxidants (TBGQ) | 0.014 | 0.000 | 0.014 | 0.000 |
| Total (g) | 1000.014 | - | 1000.014 | - |
| The energy density (kcal/g) | 3.9 | 6.912 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, H.; Xie, L.; Zhang, N.; Qi, X.; Lu, T.; Xing, J.; Akhtar, M.F.; Li, L.; Liu, G. Donkey Oil-Based Ketogenic Diet Prevents Tumor Progression by Regulating Intratumor Inflammation, Metastasis and Angiogenesis in CT26 Tumor-Bearing Mice. Genes 2023, 14, 1024. https://doi.org/10.3390/genes14051024
Zhang H, Xie L, Zhang N, Qi X, Lu T, Xing J, Akhtar MF, Li L, Liu G. Donkey Oil-Based Ketogenic Diet Prevents Tumor Progression by Regulating Intratumor Inflammation, Metastasis and Angiogenesis in CT26 Tumor-Bearing Mice. Genes. 2023; 14(5):1024. https://doi.org/10.3390/genes14051024
Chicago/Turabian StyleZhang, Huachen, Lan Xie, Ning Zhang, Xingzhen Qi, Ting Lu, Jingya Xing, Muhammad Faheem Akhtar, Lanjie Li, and Guiqin Liu. 2023. "Donkey Oil-Based Ketogenic Diet Prevents Tumor Progression by Regulating Intratumor Inflammation, Metastasis and Angiogenesis in CT26 Tumor-Bearing Mice" Genes 14, no. 5: 1024. https://doi.org/10.3390/genes14051024
APA StyleZhang, H., Xie, L., Zhang, N., Qi, X., Lu, T., Xing, J., Akhtar, M. F., Li, L., & Liu, G. (2023). Donkey Oil-Based Ketogenic Diet Prevents Tumor Progression by Regulating Intratumor Inflammation, Metastasis and Angiogenesis in CT26 Tumor-Bearing Mice. Genes, 14(5), 1024. https://doi.org/10.3390/genes14051024






