G-Quadruplexes in Nuclear Biomolecular Condensates
Abstract
:1. Introduction
2. RNA-Binding LLPS Drivers Are Abundant in the G4 Interactome
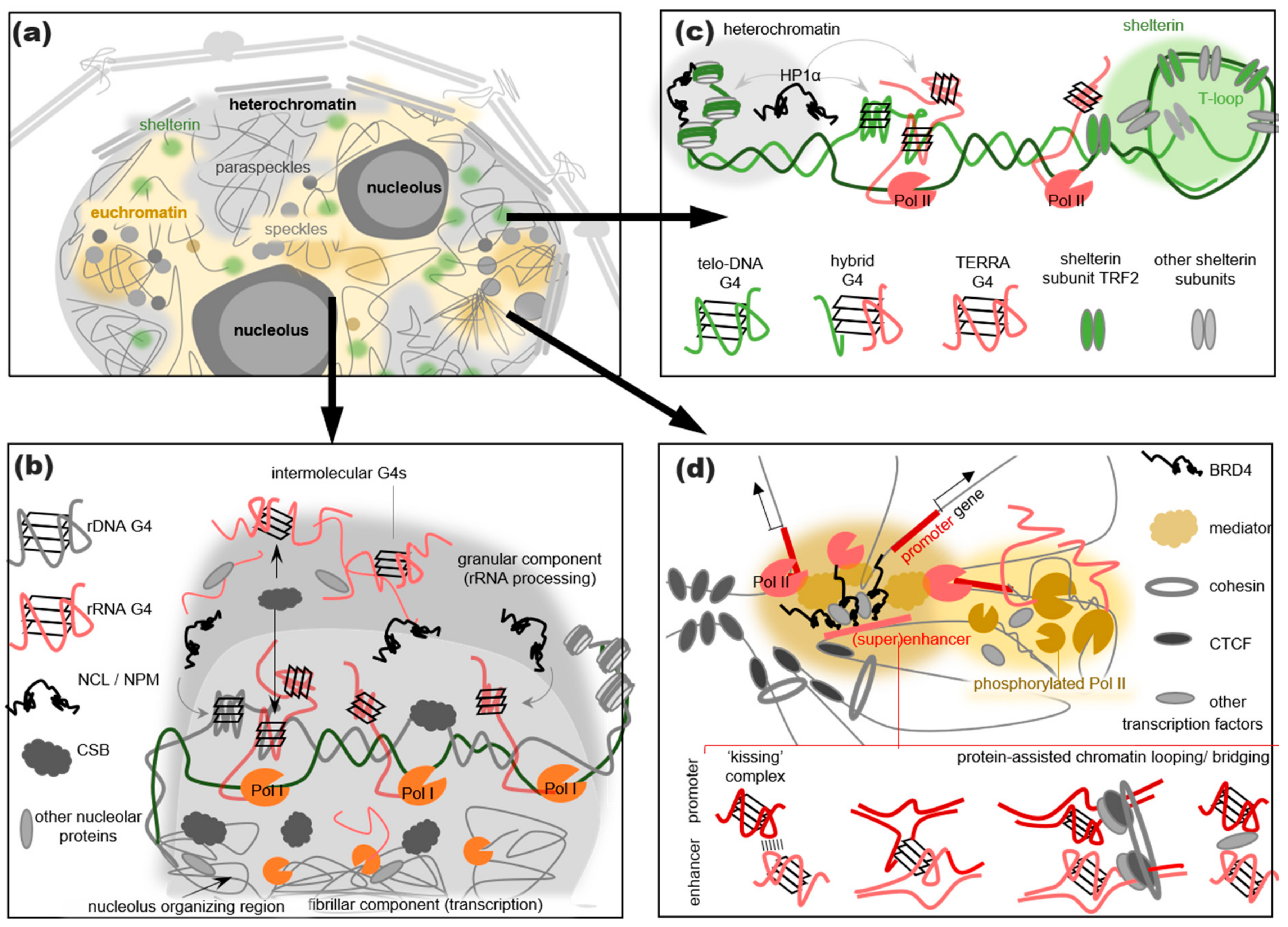
3. G4s Are Abundant in RNA Processing-Related Nuclear Condensates
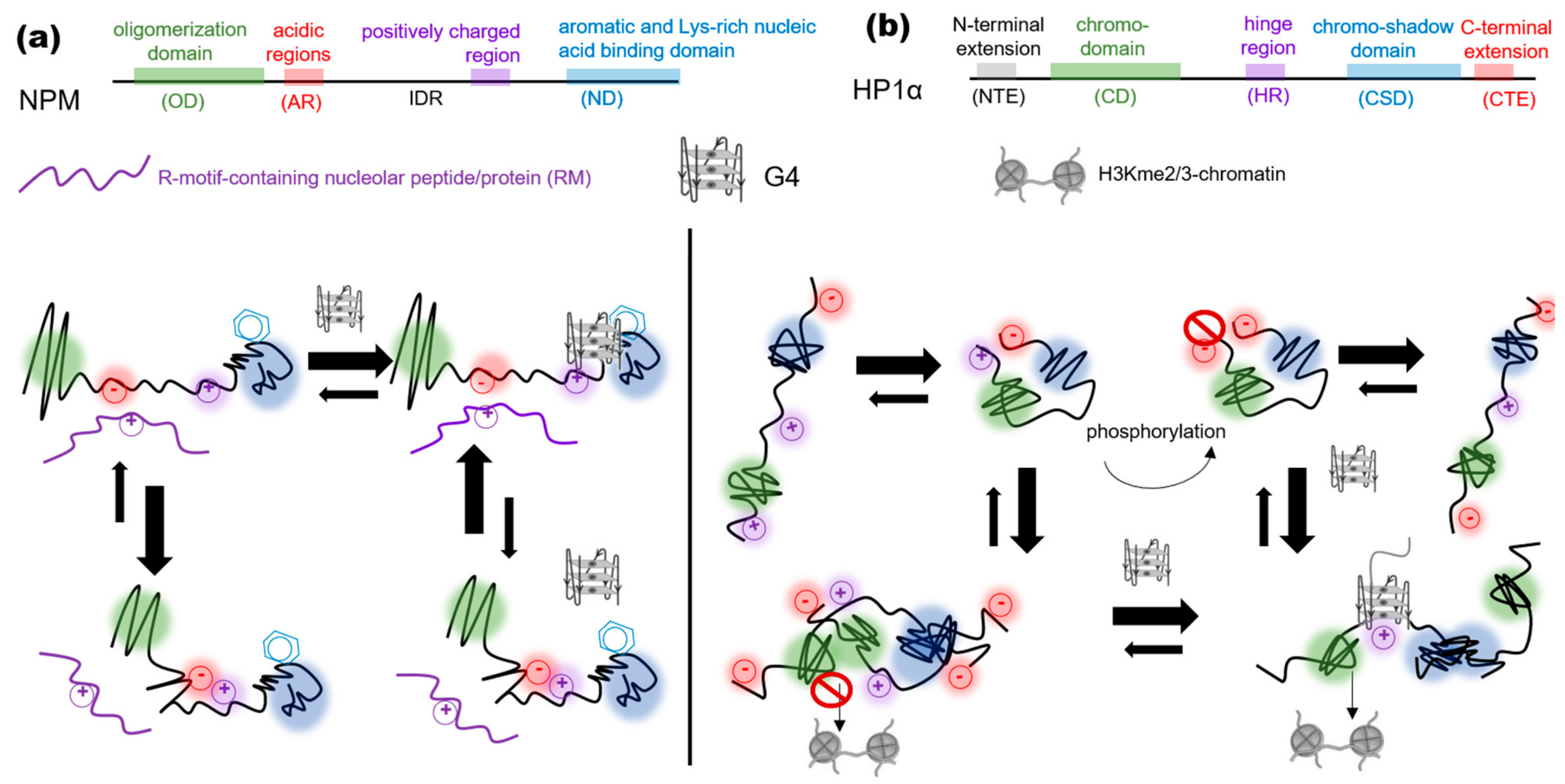
4. G4s Promote the LLPS of Heterochromatin- and Shelterin-Assembling Proteins
5. G4s May Assist in Assembling Transcription Initiation- and Reparation-Related Condensates
6. Conclusions and Open Questions
- G4s promote the LLPS of heterochromatin-associated proteins in artificial systems, but the biological relevance of these findings awaits verification.
- G4s promote the LLPS of RNA-binding proteins in the pseudo-cellular environment. These findings are in line with the studies of cytoplasmic condensates and may be relevant to the assembly of nuclear RNA processing factor-rich condensates, namely nucleoli, speckles, and paraspeckles.
- The integrity and/or functions of speckles/paraspeckles are disrupted by G4 mutations and G4-stabilizing ligands. The shelterin integrity and function are also disrupted by G4 ligands. The effects of these ligands are attributed to their interference with G4 protein interactions.
- The colocalization with Pol II clusters, TFs, and chromatin loop boundaries supports the idea that G4s assist in the transcription initiation. However, conclusive evidence is lacking. A comparison of transcription burst rates at G4-rich and non-G4 SEs could probably clarify this matter.
- The nucleobase exposure in the G4 outer tetrads and the adjoining ssDNA regions for transient π–π interactions with aromatic amino acid-rich proteins and cation–π interactions with Arg-rich ones (Figure 1).
- The exposure of a protein IDR/LCD for transient interactions with other macromolecules following a G4-binding-induced conformational transition (Figure 2).
- The accumulation of multiple IDR/LCD-containing proteins at the G4 repeats through the G4 recognition by the structured domains of these proteins or their partners (Figure 3b–d).
- The assembly of a transient nucleic acid “net” through the formation of G4–G4 kissing complexes, intermolecular G4 folding, or chromatin looping mediated by G4-binding proteins (Figure 3b–d).
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Burge, S.; Parkinson, G.N.; Hazel, P.; Todd, A.K.; Neidle, S. Quadruplex DNA: Sequence, topology and structure. Nucleic Acids Res. 2006, 34, 5402–5415. [Google Scholar] [CrossRef] [PubMed]
- Spiegel, J.; Adhikari, S.; Balasubramanian, S. The Structure and Function of DNA G-Quadruplexes. Trends Chem. 2020, 2, 123–136. [Google Scholar] [CrossRef]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef] [PubMed]
- Biffi, G.; Tannahill, D.; McCafferty, J.; Balasubramanian, S. Quantitative visualization of DNA G-quadruplex structures in human cells. Nat. Chem. 2013, 5, 182–186. [Google Scholar] [CrossRef]
- Henderson, A.; Wu, Y.; Huang, Y.C.; Chavez, E.A.; Platt, J.; Johnson, F.B.; Brosh, R.M., Jr.; Sen, D.; Lansdorp, P.M. Detection of G-quadruplex DNA in mammalian cells. Nucleic Acids Res. 2014, 42, 860–869. [Google Scholar] [CrossRef] [PubMed]
- Zheng, K.-W.; Zhang, J.-Y.; He, Y.-D.; Gong, J.-Y.; Wen, C.-J.; Chen, J.-N.; Hao, Y.-H.; Zhao, Y.; Tan, Z. Detection of genomic G-quadruplexes in living cells using a small artificial protein. Nucleic Acids Res. 2020, 48, 11706–11720. [Google Scholar] [CrossRef]
- Rodriguez, R.; Miller, K.M.; Forment, J.; Bradshaw, C.; Nikan, M.; Britton, S.; Oelschlaegel, T.; Xhemalce, B.; Balasubramanian, S.; Jackson, S.P. Small-molecule–induced DNA damage identifies alternative DNA structures in human genes. Nat. Chem. Biol. 2012, 8, 301–310. [Google Scholar] [CrossRef]
- Haensel-Hertsch, R.; Beraldi, D.; Lensing, S.V.; Marsico, G.; Zyner, K.; Parry, A.; Di Antonio, M.; Pike, J.; Kimura, H.; Narita, M.; et al. G-quadruplex structures mark human regulatory chromatin. Nat. Genet. 2016, 48, 1267–1272. [Google Scholar] [CrossRef]
- Lago, S.; Nadai, M.; Cernilogar, F.M.; Kazerani, M.; Moreno, H.D.; Schotta, G.; Richter, S.N. Promoter G-quadruplexes and transcription factors cooperate to shape the cell type-specific transcriptome. Nat. Commun. 2021, 12, 3885. [Google Scholar] [CrossRef]
- Liu, H.-Y.; Zhao, Q.; Zhang, T.-P.; Wu, Y.; Xiong, Y.-X.; Wang, S.-K.; Ge, Y.-L.; He, J.-H.; Lv, P.; Ou, T.-M.; et al. Conformation Selective Antibody Enables Genome Profiling and Leads to Discovery of Parallel G-Quadruplex in Human Telomeres. Cell Chem. Biol. 2016, 23, 1261–1270. [Google Scholar] [CrossRef]
- Hou, Y.; Li, F.; Zhang, R.; Li, S.; Liu, H.; Qin, Z.S.; Sun, X. Integrative characterization of G-Quadruplexes in the three-dimensional chromatin structure. Epigenetics 2019, 14, 894–911. [Google Scholar] [CrossRef]
- Robinson, J.; Raguseo, F.; Nuccio, S.P.; Liano, D.; Di Antonio, M. DNA G-quadruplex structures: More than simple roadblocks to transcription? Nucleic Acids Res. 2021, 49, 8419–8431. [Google Scholar] [CrossRef]
- Kim, N. The Interplay between G-quadruplex and Transcription. Curr. Med. Chem. 2019, 26, 2898–2917. [Google Scholar] [CrossRef]
- Valton, A.-L.; Prioleau, M.-N. G-Quadruplexes in DNA Replication: A Problem or a Necessity? Trends Genet. 2016, 32, 697–706. [Google Scholar] [CrossRef] [PubMed]
- Linke, R.; Limmer, M.; Juranek, S.A.; Heine, A.; Paeschke, K. The Relevance of G-Quadruplexes for DNA Repair. Int. J. Mol. Sci. 2021, 22, 12599. [Google Scholar] [CrossRef] [PubMed]
- Pavlova, A.V.; Kubareva, E.A.; Monakhova, M.V.; Zvereva, M.I.; Dolinnaya, N.G. Impact of G-Quadruplexes on the Regulation of Genome Integrity, DNA Damage and Repair. Biomolecules 2021, 11, 1284. [Google Scholar] [CrossRef]
- Varizhuk, A.; Isaakova, E.; Pozmogova, G. DNA G-Quadruplexes (G4s) Modulate Epigenetic (Re)Programming and Chromatin Remodeling. Bioessays 2019, 41, e1900091. [Google Scholar] [CrossRef] [PubMed]
- Mao, Y.S.; Zhang, B.; Spector, D.L. Biogenesis and function of nuclear bodies. Trends Genet. 2011, 27, 295–306. [Google Scholar] [CrossRef] [PubMed]
- Rippe, K. Liquid–Liquid Phase Separation in Chromatin. Cold Spring Harb. Perspect. Biol. 2022, 14, a040683. [Google Scholar] [CrossRef]
- Banani, S.F.; Lee, H.O.; Hyman, A.A.; Rosen, M.K. Biomolecular condensates: Organizers of cellular biochemistry. Nat. Rev. Mol. Cell Biol. 2017, 18, 285–298. [Google Scholar] [CrossRef]
- Shin, Y.; Brangwynne, C.P. Liquid phase condensation in cell physiology and disease. Science 2017, 357, eaaf4382. [Google Scholar] [CrossRef]
- Bergeron-Sandoval, L.-P.; Safaee, N.; Michnick, S.W. Mechanisms and Consequences of Macromolecular Phase Separation. Cell 2016, 165, 1067–1079. [Google Scholar] [CrossRef]
- Lee, J.; Cho, H.; Kwon, I. Phase separation of low-complexity domains in cellular function and disease. Exp. Mol. Med. 2022, 54, 1412–1422. [Google Scholar] [CrossRef] [PubMed]
- Dignon, G.L.; Best, R.B.; Mittal, J. Biomolecular Phase Separation: From Molecular Driving Forces to Macroscopic Properties. Annu. Rev. Phys. Chem. 2020, 71, 53–75. [Google Scholar] [CrossRef] [PubMed]
- Borcherds, W.; Bremer, A.; Borgia, M.B.; Mittag, T. How do intrinsically disordered protein regions encode a driving force for liquid–liquid phase separation? Curr. Opin. Struct. Biol. 2021, 67, 41–50. [Google Scholar] [CrossRef] [PubMed]
- O’Flynn, B.G.; Mittag, T. The role of liquid–liquid phase separation in regulating enzyme activity. Curr. Opin. Cell Biol. 2021, 69, 70–79. [Google Scholar] [CrossRef]
- Frottin, F.; Schueder, F.; Tiwary, S.; Gupta, R.; Körner, R.; Schlichthaerle, T.; Cox, J.; Jungmann, R.; Hartl, F.U.; Hipp, M.S. The nucleolus functions as a phase-separated protein quality control compartment. Science 2019, 365, 342–347. [Google Scholar] [CrossRef]
- Zhang, Y.; Yang, M.; Duncan, S.; Yang, X.; Abdelhamid, M.; Huang, L.; Zhang, H.; Benfey, P.N.; Waller, Z.A.E.; Ding, Y. G-quadruplex structures trigger RNA phase separation. Nucleic Acids Res. 2019, 47, 11746–11754. [Google Scholar] [CrossRef]
- Kharel, P.; Becker, G.; Tsvetkov, V.; Ivanov, P. Properties and biological impact of RNA G-quadruplexes: From order to turmoil and back. Nucleic Acids Res. 2020, 48, 12534–12555. [Google Scholar] [CrossRef]
- Ishiguro, A.; Ishihama, A. Essential Roles and Risks of G-Quadruplex Regulation: Recognition Targets of ALS-Linked TDP-43 and FUS. Front. Mol. Biosci. 2022, 9, 957502. [Google Scholar] [CrossRef]
- Wang, E.; Thombre, R.; Shah, Y.; Latanich, R.; Wang, J. G-Quadruplexes as pathogenic drivers in neurodegenerative disorders. Nucleic Acids Res. 2021, 49, 4816–4830. [Google Scholar] [CrossRef]
- Teng, Y.; Zhu, M.; Qiu, Z. G-Quadruplexes in Repeat Expansion Disorders. Int. J. Mol. Sci. 2023, 24, 2375. [Google Scholar] [CrossRef]
- Brázda, V.; Hároníková, L.; Liao, J.C.C.; Fojta, M. DNA and RNA Quadruplex-Binding Proteins. Int. J. Mol. Sci. 2014, 15, 17493–17517. [Google Scholar] [CrossRef] [PubMed]
- Vlasenok, M.; Levchenko, O.; Basmanov, D.; Klinov, D.; Varizhuk, A.; Pozmogova, G. Data set on G4 DNA interactions with human proteins. Data Brief 2018, 18, 348–359. [Google Scholar] [CrossRef] [PubMed]
- Makowski, M.M.; Gräwe, C.; Foster, B.M.; Nguyen, N.V.; Bartke, T.; Vermeulen, M. Global profiling of protein–DNA and protein–nucleosome binding affinities using quantitative mass spectrometry. Nat. Commun. 2018, 9, 1653. [Google Scholar] [CrossRef] [PubMed]
- Meier-Stephenson, V. G4-quadruplex-binding proteins: Review and insights into selectivity. Biophys. Rev. 2022, 14, 635–654. [Google Scholar] [CrossRef]
- Sanchez-Martin, V. DNA G-Quadruplex-Binding Proteins: An Updated Overview. DNA 2023, 3, 1–12. [Google Scholar] [CrossRef]
- Mishra, S.K.; Tawani, A.; Mishra, A.; Kumar, A. G4IPDB: A database for G-quadruplex structure forming nucleic acid interacting proteins. Sci. Rep. 2016, 6, 38144. [Google Scholar] [CrossRef]
- Zhang, X.; Spiegel, J.; Martínez Cuesta, S.; Adhikari, S.; Balasubramanian, S. Chemical profiling of DNA G-quadruplex-interacting proteins in live cells. Nat. Chem. 2021, 13, 626–633. [Google Scholar] [CrossRef]
- Su, H.; Xu, J.; Chen, Y.; Wang, Q.; Lu, Z.; Chen, Y.; Chen, K.; Han, S.; Fang, Z.; Wang, P.; et al. Photoactive G-Quadruplex Ligand Identifies Multiple G-Quadruplex-Related Proteins with Extensive Sequence Tolerance in the Cellular Environment. J. Am. Chem. Soc. 2021, 143, 1917–1923. [Google Scholar] [CrossRef]
- Wu, G.; Xing, Z.; Tran, E.J.; Yang, D. DDX5 helicase resolves G-quadruplex and is involved in MYC gene transcriptional activation. Proc. Natl. Acad. Sci. USA 2019, 116, 20453–20461. [Google Scholar] [CrossRef]
- Gao, J.; Gao, Z.; Putnam, A.A.; Byrd, A.K.; Venus, S.L.; Marecki, J.C.; Edwards, A.D.; Lowe, H.M.; Jankowsky, E.; Raney, K.D. G-quadruplex DNA inhibits unwinding activity but promotes liquid–liquid phase separation by the DEAD-box helicase Ded1p. Chem. Commun. 2021, 57, 7445–7448. [Google Scholar] [CrossRef] [PubMed]
- Song, Q.-X.; Lai, C.-W.; Liu, N.-N.; Hou, X.-M.; Xi, X.-G. DEAD-box RNA helicase Dbp2 binds to G-quadruplex nucleic acids and regulates different conformation of G-quadruplex DNA. Biochem. Biophys. Res. Commun. 2022, 634, 182–188. [Google Scholar] [CrossRef] [PubMed]
- Ali, M.A. DEAD-box RNA helicases: The driving forces behind RNA metabolism at the crossroad of viral replication and antiviral innate immunity. Virus Res. 2021, 296, 198352. [Google Scholar] [CrossRef] [PubMed]
- Taschuk, F.; Cherry, S. DEAD-Box Helicases: Sensors, Regulators, and Effectors for Antiviral Defense. Viruses 2020, 12, 181. [Google Scholar] [CrossRef] [PubMed]
- Overwijn, D.; Hondele, M. DEAD-box ATPases as regulators of biomolecular condensates and membrane-less organelles. Trends Biochem. Sci. 2023, 48, 244–258. [Google Scholar] [CrossRef] [PubMed]
- Hondele, M.; Sachdev, R.; Heinrich, S.; Wang, J.; Vallotton, P.; Fontoura, B.M.A.; Weis, K. DEAD-box ATPases are global regulators of phase-separated organelles. Nature 2019, 573, 144–148. [Google Scholar] [CrossRef]
- Liu, X.; Xiong, Y.; Zhang, C.; Lai, R.; Liu, H.; Peng, R.; Fu, T.; Liu, Q.; Fang, X.; Mann, S.; et al. G-Quadruplex-Induced Liquid–Liquid Phase Separation in Biomimetic Protocells. J. Am. Chem. Soc. 2021, 143, 11036–11043. [Google Scholar] [CrossRef]
- Gallo, A.; Sterzo, C.L.; Mori, M.; Di Matteo, A.; Bertini, I.; Banci, L.; Brunori, M.; Federici, L. Structure of Nucleophosmin DNA-binding Domain and Analysis of Its Complex with a G-quadruplex Sequence from the c-MYC Promoter. J. Biol. Chem. 2012, 287, 26539–26548. [Google Scholar] [CrossRef]
- Dempsey, L.A.; Sun, H.; Hanakahi, L.A.; Maizels, N. G4 DNA Binding by LR1 and Its Subunits, Nucleolin and hnRNP D, A Role for G-G pairing in Immunoglobulin Switch Recombination. J. Biol. Chem. 1999, 274, 1066–1071. [Google Scholar] [CrossRef]
- Ghosh, M.; Singh, M. RGG-box in hnRNPA1 specifically recognizes the telomere G-quadruplex DNA and enhances the G-quadruplex unfolding ability of UP1 domain. Nucleic Acids Res. 2018, 46, 10246–10261. [Google Scholar] [CrossRef] [PubMed]
- Paramasivam, M.; Membrino, A.; Cogoi, S.; Fukuda, H.; Nakagama, H.; Xodo, L.E. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: Implications for transcription. Nucleic Acids Res. 2009, 37, 2841–2853. [Google Scholar] [CrossRef] [PubMed]
- Yagi, R.; Miyazaki, T.; Oyoshi, T. G-quadruplex binding ability of TLS/FUS depends on the β-spiral structure of the RGG domain. Nucleic Acids Res. 2018, 46, 5894–5901. [Google Scholar] [CrossRef] [PubMed]
- Takahama, K.; Miyawaki, A.; Shitara, T.; Mitsuya, K.; Morikawa, M.; Hagihara, M.; Kino, K.; Yamamoto, A.; Oyoshi, T. G-Quadruplex DNA- and RNA-Specific-Binding Proteins Engineered from the RGG Domain of TLS/FUS. ACS Chem. Biol. 2015, 10, 2564–2569. [Google Scholar] [CrossRef] [PubMed]
- Martini, S.; Davis, K.; Faraway, R.; Elze, L.; Lockwood, N.; Jones, A.; Xie, X.; McDonald, N.Q.; Mann, D.J.; Armstrong, A.; et al. A genetically-encoded crosslinker screen identifies SERBP1 as a PKCε substrate influencing translation and cell division. Nat. Commun. 2021, 12, 6934. [Google Scholar] [CrossRef] [PubMed]
- Baudin, A.; Moreno-Romero, A.K.; Xu, X.; Selig, E.E.; Penalva, L.O.F.; Libich, D.S. Structural Characterization of the RNA-Binding Protein SERBP1 Reveals Intrinsic Disorder and Atypical RNA Binding Modes. Front. Mol. Biosci. 2021, 8, 744707. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Cika, J.A.; Stanley, C.B.; Nourse, A.; Onuchic, P.L.; Banerjee, P.R.; Phillips, A.H.; Park, C.-G.; Deniz, A.A.; Kriwacki, R.W. Self-interaction of NPM1 modulates multiple mechanisms of liquid–liquid phase separation. Nat. Commun. 2018, 9, 842. [Google Scholar] [CrossRef]
- Arcovito, A.; Chiarella, S.; Della Longa, S.; Di Matteo, A.; Sterzo, C.L.; Scaglione, G.L.; Federici, L. Synergic Role of Nucleophosmin Three-helix Bundle and a Flanking Unstructured Tail in the Interaction with G-quadruplex DNA. J. Biol. Chem. 2014, 289, 21230–21241. [Google Scholar] [CrossRef]
- Mitrea, D.M.; Cika, J.A.; Guy, C.S.; Ban, D.; Banerjee, P.R.; Stanley, C.B.; Nourse, A.; Deniz, A.A.; Kriwacki, R.W. Nucleophosmin integrates within the nucleolus via multi-modal interactions with proteins displaying R-rich linear motifs and rRNA. eLife 2016, 5, 13571. [Google Scholar] [CrossRef]
- Nishikawa, T.; Kuwano, Y.; Takahara, Y.; Nishida, K.; Rokutan, K. HnRNPA1 interacts with G-quadruplex in the TRA2B promoter and stimulates its transcription in human colon cancer cells. Sci. Rep. 2019, 9, 10276. [Google Scholar] [CrossRef]
- Molliex, A.; Temirov, J.; Lee, J.; Coughlin, M.; Kanagaraj, A.P.; Kim, H.J.; Mittag, T.; Taylor, J.P. Phase Separation by Low Complexity Domains Promotes Stress Granule Assembly and Drives Pathological Fibrillization. Cell 2015, 163, 123–133. [Google Scholar] [CrossRef] [PubMed]
- Gui, X.; Luo, F.; Li, Y.; Zhou, H.; Qin, Z.; Liu, Z.; Gu, J.; Xie, M.; Zhao, K.; Dai, B.; et al. Structural basis for reversible amyloids of hnRNPA1 elucidates their role in stress granule assembly. Nat. Commun. 2019, 10, 2006. [Google Scholar] [CrossRef] [PubMed]
- Tonello, F.; Massimino, M.L.; Peggion, C. Nucleolin: A cell portal for viruses, bacteria, and toxins. Cell. Mol. Life Sci. 2022, 79, 271. [Google Scholar] [CrossRef] [PubMed]
- Serin, G.; Joseph, G.; Ghisolfi, L.; Bauzan, M.; Erard, M.; Amalric, F.; Bouvet, P. Two RNA-binding Domains Determine the RNA-binding Specificity of Nucleolin. J. Biol. Chem. 1997, 272, 13109–13116. [Google Scholar] [CrossRef]
- Masuzawa, T.; Oyoshi, T. Roles of the RGG Domain and RNA Recognition Motif of Nucleolin in G-Quadruplex Stabilization. ACS Omega Am. Chem. Soc. 2020, 5, 5202–5208. [Google Scholar] [CrossRef]
- Spiegel, J.; Cuesta, S.M.; Adhikari, S.; Hänsel-Hertsch, R.; Tannahill, D.; Balasubramanian, S. G-quadruplexes are transcription factor binding hubs in human chromatin. Genome Biol. 2021, 22, 117. [Google Scholar] [CrossRef]
- Ji, Y.; Li, F.; Qiao, Y. Modulating liquid–liquid phase separation of FUS: Mechanisms and strategies. J. Mater. Chem. B 2022, 10, 8616–8628. [Google Scholar] [CrossRef]
- Murthy, A.C.; Tang, W.S.; Jovic, N.; Janke, A.M.; Seo, D.H.; Perdikari, T.M.; Mittal, J.; Fawzi, N.L. Molecular interactions contributing to FUS SYGQ LC-RGG phase separation and co-partitioning with RNA polymerase II heptads. Nat. Struct. Mol. Biol. 2021, 28, 923–935. [Google Scholar] [CrossRef]
- Murray, D.T.; Kato, M.; Lin, Y.; Thurber, K.R.; Hung, I.; McKnight, S.L.; Tycko, R. Structure of FUS Protein Fibrils and Its Relevance to Self-Assembly and Phase Separation of Low-Complexity Domains. Cell 2017, 171, 615–627.e16. [Google Scholar] [CrossRef]
- Qamar, S.; Wang, G.; Randle, S.J.; Ruggeri, F.; Varela, J.; Lin, Q.; Phillips, E.C.; Miyashita, A.; Williams, D.; Ströhl, F.; et al. FUS Phase Separation Is Modulated by a Molecular Chaperone and Methylation of Arginine Cation-π Interactions. Cell 2018, 173, 720–734.e15. [Google Scholar] [CrossRef]
- Monahan, Z.; Ryan, V.H.; Janke, A.M.; Burke, K.A.; Rhoads, S.N.; Zerze, G.H.; O’Meally, R.; Dignon, G.L.; Conicella, A.E.; Zheng, W.; et al. Phosphorylation of the FUS low-complexity domain disrupts phase separation, aggregation, and toxicity. EMBO J. 2017, 36, 2951–2967. [Google Scholar] [CrossRef] [PubMed]
- Hofweber, M.; Hutten, S.; Bourgeois, B.; Spreitzer, E.; Niedner-Boblenz, A.; Schifferer, M.; Ruepp, M.D.; Simons, M.; Niessing, D.; Madl, T.; et al. Phase Separation of FUS Is Suppressed by Its Nuclear Import Receptor and Arginine Methylation. Cell 2018, 173, 706–719.e13. [Google Scholar] [CrossRef] [PubMed]
- Singatulina, A.S.; Hamon, L.; Sukhanova, M.V.; Desforges, B.; Joshi, V.; Bouhss, A.; Lavrik, O.I.; Pastré, D. PARP-1 Activation Directs FUS to DNA Damage Sites to Form PARG-Reversible Compartments Enriched in Damaged DNA. Cell Rep. 2019, 27, 1809–1821.e5. [Google Scholar] [CrossRef] [PubMed]
- Youn, J.-Y.; Dyakov, B.J.; Zhang, J.; Knight, J.D.; Vernon, R.M.; Forman-Kay, J.D.; Gingras, A.-C. Properties of Stress Granule and P-Body Proteomes. Mol. Cell 2019, 76, 286–294. [Google Scholar] [CrossRef]
- Paiz, E.A.; Allen, J.H.; Correia, J.J.; Fitzkee, N.C.; Hough, L.E.; Whitten, S.T. Beta turn propensity and a model polymer scaling exponent identify intrinsically disordered phase-separating proteins. J. Biol. Chem. 2021, 297, 101343. [Google Scholar] [CrossRef]
- Vernon, R.M.; Forman-Kay, J.D. First-generation predictors of biological protein phase separation. Curr. Opin. Struct. Biol. 2019, 58, 88–96. [Google Scholar] [CrossRef]
- Li, Q.; Wang, X.; Dou, Z.; Yang, W.; Huang, B.; Lou, J.; Zhang, Z. Protein Databases Related to Liquid–Liquid Phase Separation. Int. J. Mol. Sci. 2020, 21, 6796. [Google Scholar] [CrossRef]
- Raiber, E.-A.; Kranaster, R.; Ni Lam, E.Y.; Nikan, M.; Balasubramanian, S. A non-canonical DNA structure is a binding motif for the transcription factor SP1 in vitro. Nucleic Acids Res. 2012, 40, 1499–1508. [Google Scholar] [CrossRef]
- Pavlova, I.I.; Tsvetkov, V.B.; Isaakova, E.A.; Severov, V.V.; Khomyakova, E.A.; Lacis, I.A.; Lazarev, V.N.; Lagarkova, M.A.; Pozmogova, G.E.; Varizhuk, A.M. Transcription-facilitating histone chaperons interact with genomic and synthetic G4 structures. Int. J. Biol. Macromol. 2020, 160, 1144–1157. [Google Scholar] [CrossRef]
- Cogoi, S.; Paramasivam, M.; Membrino, A.; Yokoyama, K.K.; Xodo, L.E. The KRAS Promoter Responds to Myc-associated Zinc Finger and Poly(ADP-ribose) Polymerase 1 Proteins, Which Recognize a Critical Quadruplex-forming GA-element. J. Biol. Chem. 2010, 285, 22003–22016. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y. HnRNPA1 Specifically Recognizes the Base of Nucleotide at the Loop of RNA G-Quadruplex. Molecules 2018, 23, 237. [Google Scholar] [CrossRef] [PubMed]
- Mimura, M.; Tomita, S.; Shinkai, Y.; Hosokai, T.; Kumeta, H.; Saio, T.; Shiraki, K.; Kurita, R. Quadruplex Folding Promotes the Condensation of Linker Histones and DNAs via Liquid–Liquid Phase Separation. J. Am. Chem. Soc. 2021, 143, 9849–9857. [Google Scholar] [CrossRef] [PubMed]
- Simko, E.A.J.; Liu, H.; Zhang, T.; Velasquez, A.; Teli, S.; Haeusler, A.R.; Wang, J. G-quadruplexes offer a conserved structural motif for NONO recruitment to NEAT1 architectural lncRNA. Nucleic Acids Res. 2020, 48, 7421–7438. [Google Scholar] [CrossRef] [PubMed]
- Mou, X.; Liew, S.W.; Kwok, C.K. Identification and targeting of G-quadruplex structures in MALAT1 long non-coding RNA. Nucleic Acids Res. 2022, 50, 397–410. [Google Scholar] [CrossRef]
- Cogoi, S.; Paramasivam, M.; Spolaore, B.; Xodo, L.E. Structural polymorphism within a regulatory element of the human KRAS promoter: Formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008, 36, 3765–3780. [Google Scholar] [CrossRef]
- Edwards, A.D.; Marecki, J.C.; Byrd, A.K.; Gao, J.; Raney, K.D. G-Quadruplex loops regulate PARP-1 enzymatic activation. Nucleic Acids Res. 2021, 49, 416–431. [Google Scholar] [CrossRef]
- De Almeida, C.R.; Dhir, S.; Dhir, A.; Moghaddam, A.E.; Sattentau, Q.; Meinhart, A.; Proudfoot, N.J. RNA Helicase DDX1 Converts RNA G-Quadruplex Structures into R-Loops to Promote IgH Class Switch Recombination. Mol. Cell 2018, 70, 650–662.e8. [Google Scholar] [CrossRef]
- Figueiredo, J.; Miranda, A.; Lopes-Nunes, J.; Carvalho, J.; Alexandre, D.; Valente, S.; Mergny, J.-L.; Cruz, C. Targeting nucleolin by RNA G-quadruplex-forming motif. Biochem. Pharmacol. 2021, 189, 114418. [Google Scholar] [CrossRef]
- Mukherjee, A.K.; Sharma, S.; Bagri, S.; Kutum, R.; Kumar, P.; Hussain, A.; Singh, P.; Saha, D.; Kar, A.; Dash, D.; et al. Telomere repeat–binding factor 2 binds extensively to extra-telomeric G-quadruplexes and regulates the epigenetic status of several gene promoters. J. Biol. Chem. 2019, 294, 17709–17722. [Google Scholar] [CrossRef]
- Mei, Y.; Deng, Z.; Vladimirova, O.; Gulve, N.; Johnson, F.B.; Drosopoulos, W.C.; Schildkraut, C.L.; Lieberman, P.M. TERRA G-quadruplex RNA interaction with TRF2 GAR domain is required for telomere integrity. Sci. Rep. 2021, 11, 3509. [Google Scholar] [CrossRef]
- Roach, R.J.; Garavís, M.; González, C.; Jameson, G.B.; Filichev, V.V.; Hale, T.K. Heterochromatin protein 1α interacts with parallel RNA and DNA G-quadruplexes. Nucleic Acids Res. 2020, 48, 682–693. [Google Scholar] [CrossRef]
- Vernon, R.M.; Chong, P.A.; Tsang, B.; Kim, T.H.; Bah, A.; Farber, P.; Lin, H.; Forman-Kay, J.D. Pi-Pi contacts are an overlooked protein feature relevant to phase separation. eLife 2018, 7, e31486. [Google Scholar] [CrossRef] [PubMed]
- Hong, Y.; Najafi, S.; Casey, T.; Shea, J.-E.; Han, S.-I.; Hwang, D.S. Hydrophobicity of arginine leads to reentrant liquid-liquid phase separation behaviors of arginine-rich proteins. Nat. Commun. 2022, 13, 7326. [Google Scholar] [CrossRef] [PubMed]
- Liano, D.; Chowdhury, S.; Di Antonio, M. Cockayne Syndrome B Protein Selectively Resolves and Interact with Intermolecular DNA G-Quadruplex Structures. J. Am. Chem. Soc. 2021, 143, 20988–21002. [Google Scholar] [CrossRef] [PubMed]
- Mestre-Fos, S.; Penev, P.I.; Suttapitugsakul, S.; Hu, M.; Ito, C.; Petrov, A.S.; Wartell, R.M.; Wu, R.; Williams, L.D. G-Quadruplexes in Human Ribosomal RNA. J. Mol. Biol. 2019, 431, 1940–1955. [Google Scholar] [CrossRef]
- Gałgański, Ł.; Urbanek-Trzeciak, M.O.; Krzyzosiak, W.J. Nuclear speckles: Molecular organization, biological function and role in disease. Nucleic Acids Res. 2017, 45, 10350–10368. [Google Scholar] [CrossRef]
- Bond, C.S.; Fox, A.H. Paraspeckles: Nuclear bodies built on long noncoding RNA. J. Cell Biol. 2009, 186, 637–644. [Google Scholar] [CrossRef]
- Bhatt, U.; Kretzmann, A.L.; Guédin, A.; Ou, A.; Kobelke, S.; Bond, C.S.; Evans, C.W.; Hurley, L.H.; Mergny, J.-L.; Iyer, K.S.; et al. The role of G-Quadruplex DNA in Paraspeckle formation in cancer. Biochimie 2021, 190, 124–131. [Google Scholar] [CrossRef]
- Singh, Y.K.; Ekka, M.K.; Chakraborty, D.; Maiti, S.; Sale, J.E.; Stoddard, B.L. Editorial Expression of Concern on article ‘Alternative splicing modulation mediated by G-quadruplex structures in MALAT1 lncRNA’. Nucleic Acids Res. 2022, 50, 4199. [Google Scholar]
- Lindström, M.S.; Jurada, D.; Bursac, S.; Oršolić, I.; Bartek, J.; Volarevic, S. Nucleolus as an emerging hub in maintenance of genome stability and cancer pathogenesis. Oncogene 2018, 37, 2351–2366. [Google Scholar] [CrossRef]
- Cong, R.; Das, S.; Ugrinova, I.; Kumar, S.; Mongelard, F.; Wong, J.; Bouvet, P. Interaction of nucleolin with ribosomal RNA genes and its role in RNA polymerase I transcription. Nucleic Acids Res. 2012, 40, 9441–9454. [Google Scholar] [CrossRef]
- Chiarella, S.; De Cola, A.; Scaglione, G.L.; Carletti, E.; Graziano, V.; Barcaroli, D.; Sterzo, C.L.; Di Matteo, A.; Di Ilio, C.; Falini, B.; et al. Nucleophosmin mutations alter its nucleolar localization by impairing G-quadruplex binding at ribosomal DNA. Nucleic Acids Res. 2013, 41, 3228–3239. [Google Scholar] [CrossRef]
- Guillen-Chable, F.; Bayona, A.; Rodríguez-Zapata, L.C.; Castano, E. Phase Separation of Intrinsically Disordered Nucleolar Proteins Relate to Localization and Function. Int. J. Mol. Sci. 2021, 22, 13095. [Google Scholar] [CrossRef]
- Liano, D.; Monti, L.; Chowdhury, S.; Raguseo, F.; Di Antonio, M. Long-range DNA interactions: Inter-molecular G-quadruplexes and their potential biological relevance. Chem. Commun. 2022, 58, 12753–12762. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Chen, H.; Li, Q.; Guan, A.; Wang, L.; Shi, Y.; Xu, S.; Liu, M.; Tang, Y. Direct visualization of nucleolar G-quadruplexes in live cells by using a fluorescent light-up probe. Biochim. Biophys. Acta (BBA)—Gen. Subj. 2018, 1862, 1101–1106. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, H.; Wang, L.; Liu, Y.; Chen, H.; Li, Q.; Guan, A.; Liu, M.; Tang, Y. Real-time monitoring of DNA G-quadruplexes in living cells with a small-molecule fluorescent probe. Nucleic Acids Res. 2018, 46, 7522–7532. [Google Scholar] [CrossRef]
- Hoffmann, R.F.; Moshkin, Y.M.; Mouton, S.; Grzeschik, N.A.; Kalicharan, R.D.; Kuipers, J.; Wolters, A.H.; Nishida, K.; Romashchenko, A.V.; Postberg, J.; et al. Guanine quadruplex structures localize to heterochromatin. Nucleic Acids Res. 2016, 44, 152–163. [Google Scholar] [CrossRef]
- Hanna, R.; Flamier, A.; Barabino, A.; Bernier, G. G-quadruplexes originating from evolutionary conserved L1 elements interfere with neuronal gene expression in Alzheimer’s disease. Nat. Commun. 2021, 12, 1828. [Google Scholar] [CrossRef]
- Kaya-Okur, H.S.; Wu, S.J.; Codomo, C.A.; Pledger, E.S.; Bryson, T.D.; Henikoff, J.G.; Ahmad, K.; Henikoff, S. CUT&Tag for efficient epigenomic profiling of small samples and single cells. Nat. Commun. 2019, 10, 1930. [Google Scholar] [CrossRef]
- Lyu, J.; Shao, R.; Yung, P.Y.K.; Elsässer, S.J. Genome-wide mapping of G-quadruplex structures with CUT&Tag. Nucleic Acids Res. 2022, 50, e13. [Google Scholar] [CrossRef]
- Larson, A.G.; Elnatan, D.; Keenen, M.M.; Trnka, M.J.; Johnston, J.B.; Burlingame, A.L.; Agard, D.A.; Redding, S.; Narlikar, G.J. Liquid droplet formation by HP1α suggests a role for phase separation in heterochromatin. Nature 2017, 547, 236–240. [Google Scholar] [CrossRef]
- Shakya, A.; Park, S.; Rana, N.; King, J.T. Liquid-Liquid Phase Separation of Histone Proteins in Cells: Role in Chromatin Organization. Biophys. J. 2020, 118, 753–764. [Google Scholar] [CrossRef]
- Erdel, F.; Rippe, K. Formation of Chromatin Subcompartments by Phase Separation. Biophys. J. 2018, 114, 2262–2270. [Google Scholar] [CrossRef]
- Sridhar, A.; Farr, S.E.; Portella, G.; Schlick, T.; Orozco, M.; Collepardo-Guevara, R. Emergence of chromatin hierarchical loops from protein disorder and nucleosome asymmetry. Proc. Natl. Acad. Sci. USA 2020, 117, 7216–7224. [Google Scholar] [CrossRef]
- Farr, S.E.; Woods, E.J.; Joseph, J.A.; Garaizar, A.; Collepardo-Guevara, R. Nucleosome plasticity is a critical element of chromatin liquid–liquid phase separation and multivalent nucleosome interactions. Nat. Commun. 2021, 12, 2883. [Google Scholar] [CrossRef]
- Pavlova, I.; Barinov, N.; Novikov, R.; Severov, V.; Iudin, M.; Vedekhina, T.; Larin, A.; Babenko, V.; Aralov, A.; Gnuchikh, E.; et al. Modeling G4s in chromatin context confirms partial nucleosome exclusion and reveals nucleosome-disrupting effects of the least selective G4 ligands. Biochimie 2023, 204, 8–21. [Google Scholar] [CrossRef]
- Her, C.; Phan, T.M.; Jovic, N.; Kapoor, U.; Ackermann, B.E.; Rizuan, A.; Kim, Y.C.; Mittal, J.; Debelouchina, G.T. Molecular interactions underlying the phase separation of HP1α: Role of phosphorylation, ligand and nucleic acid binding. Nucleic Acids Res. 2022, 50, 12702–12722. [Google Scholar] [CrossRef]
- Wang, J.; Cohen, A.L.; Letian, A.; Tadeo, X.; Moresco, J.J.; Liu, J.; Yates, J.R.; Qiao, F.; Jia, S. The proper connection between shelterin components is required for telomeric heterochromatin assembly. Genes Dev. 2016, 30, 827–839. [Google Scholar] [CrossRef]
- Jack, A.; Kim, Y.; Strom, A.R.; Lee, D.S.; Williams, B.; Schaub, J.M.; Kellogg, E.H.; Finkelstein, I.J.; Ferro, L.S.; Yildiz, A.; et al. Compartmentalization of telomeres through DNA-scaffolded phase separation. Dev. Cell 2022, 57, 277–290.e9. [Google Scholar] [CrossRef]
- Rodriguez, R.; Müller, S.; Yeoman, J.A.; Trentesaux, C.; Riou, J.-F.; Balasubramanian, S. A Novel Small Molecule That Alters Shelterin Integrity and Triggers a DNA-Damage Response at Telomeres. J. Am. Chem. Soc. 2008, 130, 15758–15759. [Google Scholar] [CrossRef]
- Biffi, G.; Tannahill, D.; Balasubramanian, S. An Intramolecular G-Quadruplex Structure Is Required for Binding of Telomeric Repeat-Containing RNA to the Telomeric Protein TRF2. J. Am. Chem. Soc. 2012, 134, 11974–11976. [Google Scholar] [CrossRef]
- Komůrková, D.; Kovaříková, A.S.; Bártová, E. G-Quadruplex Structures Colocalize with Transcription Factories and Nuclear Speckles Surrounded by Acetylated and Dimethylated Histones H3. Int. J. Mol. Sci. 2021, 22, 1995. [Google Scholar] [CrossRef] [PubMed]
- Cook, P.R.; Marenduzzo, D. Transcription-driven genome organization: A model for chromosome structure and the regulation of gene expression tested through simulations. Nucleic Acids Res. 2018, 46, 9895–9906. [Google Scholar] [CrossRef] [PubMed]
- Ishov, A.M.; Gurumurthy, A.; Bungert, J. Coordination of transcription, processing, and export of highly expressed RNAs by distinct biomolecular condensates. Emerg. Top. Life Sci. 2020, 4, 281–291. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Belmont, A.S. Genome organization around nuclear speckles. Curr. Opin. Genet. Dev. 2019, 55, 91–99. [Google Scholar] [CrossRef] [PubMed]
- Cheung, K.L.; Kim, C.; Zhou, M.-M. The Functions of BET Proteins in Gene Transcription of Biology and Diseases. Front. Mol. Biosci. 2021, 8, 728777. [Google Scholar] [CrossRef]
- Cheung, K.L.; Zhang, F.; Jaganathan, A.; Sharma, R.; Zhang, Q.; Konuma, T.; Shen, T.; Lee, J.Y.; Ren, C.; Chen, C.H.; et al. Distinct Roles of Brd2 and Brd4 in Potentiating the Transcriptional Program for Th17 Cell Differentiation. Mol. Cell 2017, 65, 1068–1080.e5. [Google Scholar] [CrossRef]
- Donati, B.; Lorenzini, E.; Ciarrocchi, A. BRD4 and Cancer: Going beyond transcriptional regulation. Mol. Cancer 2018, 17, 164. [Google Scholar] [CrossRef]
- Monfils, K.; Barakat, T.S. Models behind the mystery of establishing enhancer-promoter interactions. Eur. J. Cell Biol. 2021, 100, 151170. [Google Scholar] [CrossRef] [PubMed]
- Williams, J.D.; Houserova, D.; Johnson, B.R.; Dyniewski, B.; Berroyer, A.; French, H.; Barchie, A.A.; Bilbrey, D.D.; Demeis, J.D.; Ghee, K.R.; et al. Characterization of long G4-rich enhancer-associated genomic regions engaging in a novel loop:loop ‘G4 Kissing’ interaction. Nucleic Acids Res. 2020, 48, 5907–5925. [Google Scholar] [CrossRef]
- Li, L.; Williams, P.; Ren, W.; Wang, M.Y.; Gao, Z.; Miao, W.; Huang, M.; Song, J.; Wang, Y. YY1 interacts with guanine quadruplexes to regulate DNA looping and gene expression. Nat. Chem. Biol. 2021, 17, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Weintraub, A.S.; Li, C.H.; Zamudio, A.V.; Sigova, A.A.; Hannett, N.M.; Day, D.S.; Abraham, B.J.; Cohen, M.A.; Nabet, B.; Buckley, D.L.; et al. YY1 Is a Structural Regulator of Enhancer-Promoter Loops. Cell 2017, 171, 1573–1588.e28. [Google Scholar] [CrossRef]
- Tikhonova, P.; Pavlova, I.; Isaakova, E.; Tsvetkov, V.; Bogomazova, A.; Vedekhina, T.; Luzhin, A.V.; Sultanov, R.; Severov, V.; Klimina, K.; et al. DNA G-Quadruplexes Contribute to CTCF Recruitment. Int. J. Mol. Sci. 2021, 22, 7090. [Google Scholar] [CrossRef] [PubMed]
- Ong, C.-T.; Corces, V.G. CTCF: An architectural protein bridging genome topology and function. Nat. Rev. Genet. 2014, 15, 234–246. [Google Scholar] [CrossRef] [PubMed]
- Xiao, T.; Li, X.; Felsenfeld, G. The Myc-associated zinc finger protein (MAZ) works together with CTCF to control cohesin positioning and genome organization. Proc. Natl. Acad. Sci. USA 2021, 118, 2023127118. [Google Scholar] [CrossRef] [PubMed]
- Prorok, P.; Artufel, M.; Aze, A.; Coulombe, P.; Peiffer, I.; Lacroix, L.; Guédin, A.; Mergny, J.-L.; Damaschke, J.; Schepers, A.; et al. Involvement of G-quadruplex regions in mammalian replication origin activity. Nat. Commun. 2019, 10, 3274. [Google Scholar] [CrossRef]
- Nathanailidou, P.; Taraviras, S.; Lygerou, Z. DNA Replication Control: Liquid-liquid Phase Separation Comes into Play. J. Mol. Biochem. 2020, 9, 54–56. [Google Scholar]
- Parker, M.W.; Bell, M.; Mir, M.; Kao, J.A.; Darzacq, X.; Botchan, M.R.; Berger, J.M. A new class of disordered elements controls DNA replication through initiator self-assembly. eLife 2019, 8, 48562. [Google Scholar] [CrossRef]
- Halder, K.; Halder, R.; Chowdhury, S. Genome-wide analysis predicts DNA structural motifs as nucleosome exclusion signals. Mol. Biosyst. 2009, 5, 1703–1712. [Google Scholar] [CrossRef]
- Lerner, L.K.; Sale, J.E. Replication of G quadruplex DNA. Genes 2019, 10, 95. [Google Scholar] [CrossRef]
- Schiavone, D.; Guilbaud, G.; Murat, P.; Papadopoulou, C.; Sarkies, P.; Prioleau, M. Determinants of G quadruplex-induced epigenetic instability in REV 1-deficient cells. EMBO J. 2014, 33, 2507–2520. [Google Scholar] [CrossRef]
- Saito, I.; Takayama, M.; Sugiyama, H.; Nakatani, K.; Tsuchida, A.; Yamamoto, M. Photoinduced DNA Cleavage via Electron Transfer: Demonstration That Guanine Residues Located 5′ to Guanine Are the Most Electron-Donating Sites. J. Am. Chem. Soc. 1995, 117, 6406–6407. [Google Scholar] [CrossRef]
- Fleming, A.M.; Burrows, C.J. Interplay of Guanine Oxidation and G-Quadruplex Folding in Gene Promoters. J. Am. Chem. Soc. 2020, 142, 1115–1136. [Google Scholar] [CrossRef]
- Alemasova, E.E.; Lavrik, O.I. A sePARate phase? Poly(ADP-ribose) versus RNA in the organization of biomolecular condensates. Nucleic Acids Res. 2022, 50, 10817–10838. [Google Scholar] [CrossRef] [PubMed]
- Sugimura, K.; Takebayashi, S.; Taguchi, H.; Takeda, S.; Okumura, K. PARP-1 ensures regulation of replication fork progression by homologous recombination on damaged DNA. J. Cell Biol. 2008, 183, 1203–1212. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Liu, R.; Jia, Z.; Li, R.; Zhu, F.; Zhu, W.; Shao, Y.; Jin, Y.; Xue, Y.; Huang, J.; et al. Poly(ADP-ribosylation) of P-TEFb by PARP1 disrupts phase separation to inhibit global transcription after DNA damage. Nat. Cell Biol. 2022, 24, 513–525. [Google Scholar] [CrossRef] [PubMed]
- Rhine, K.; Dasovich, M.; Yoniles, J.; Badiee, M.; Skanchy, S.; Ganser, L.R.; Ge, Y.; Fare, C.M.; Shorter, J.; Leung, A.K.; et al. Poly(ADP-ribose) drives condensation of FUS via a transient interaction. Mol. Cell 2022, 82, 969–985.e11. [Google Scholar] [CrossRef]

| Protein | G4 Binding (G4 Type) | LLPS Capability | ||
|---|---|---|---|---|
| Code | Name | RNA Granule DB | ParSe * | |
| SP1 | Specificity protein 1 | [78] (DNA) | 1.1 | ++ |
| FUS | Fused in sarcoma | [53,54] (DNA, RNA) | 22 | ++ |
| BRD3 | Bromodomain-containing protein 3 | [34,79] (DNA) | 0.3 | +/− |
| MAZ | MYC-associated zinc finger protein | [80] (DNA) | 0.3 | + |
| TAF15 | TATA box-binding protein-associated factor 15 | [6] (DNA) | 15.7 | ++ |
| hnRNPA1 | Heterogeneous nuclear ribonucleoprotein A1 | [51,81] (DNA, RNA) | 18.8 | + |
| H1 | Histone H1 | [82] (DNA) | 6.4 | − |
| NONO | Non-POU domain-containing octamer-binding protein | [83,84] (RNA) | 10.4 | +/− |
| PARP1 | Poly [ADP-ribose] polymerase 1 | [85,86] (DNA) | 2.5 | +/− |
| DDX1 | DEAD-box helicase 1 | [39,87] (DNA, RNA) | 29.3 | +/− |
| DDX5 | DEAD-box helicase 5 | [41] (DNA) | 4.7 | +/− |
| DDX24 | DEAD-box helicase 24 | [39] (DNA, RNA) | 0.6 | − |
| DBP1/2 | D-box-binding PAR BZIP transcription factor | [42,44] (DNA, RNA) | 1 | +/− |
| NCL | Nucleolin | [50,88] (DNA, RNA) | 3.8 | + |
| NPM1 | Nucleophosmin | [49] (DNA) | 8.7 | − |
| TRF2 | Telomeric repeat-binding factor 2 | [89,90] (DNA, RNA) | 0.1 | +/− |
| SERBP1 | SERPINE1 MRNA binding protein 1 | [48,56] (DNA, RNA) | 15.4 | + |
| HP1α | Heterochromatin protein 1, isoform α | [91] (DNA, RNA) | 0.2 | − |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pavlova, I.; Iudin, M.; Surdina, A.; Severov, V.; Varizhuk, A. G-Quadruplexes in Nuclear Biomolecular Condensates. Genes 2023, 14, 1076. https://doi.org/10.3390/genes14051076
Pavlova I, Iudin M, Surdina A, Severov V, Varizhuk A. G-Quadruplexes in Nuclear Biomolecular Condensates. Genes. 2023; 14(5):1076. https://doi.org/10.3390/genes14051076
Chicago/Turabian StylePavlova, Iuliia, Mikhail Iudin, Anastasiya Surdina, Vjacheslav Severov, and Anna Varizhuk. 2023. "G-Quadruplexes in Nuclear Biomolecular Condensates" Genes 14, no. 5: 1076. https://doi.org/10.3390/genes14051076
APA StylePavlova, I., Iudin, M., Surdina, A., Severov, V., & Varizhuk, A. (2023). G-Quadruplexes in Nuclear Biomolecular Condensates. Genes, 14(5), 1076. https://doi.org/10.3390/genes14051076






