Integration of Non-Coding RNA and mRNA Profiles Reveals the Mechanisms of Rumen Development Induced by Different Types of Diet in Calves
Abstract
:1. Introduction
2. Materials and Methods
2.1. Ethics Statement
2.2. Animals and Feeding Strategy
2.3. Detection of Rumen Papillae Height, Width and Number
2.4. IgG Content and Digestive Enzyme Activity Detection
2.5. Sample Collection and Transcriptomic Sequencing
2.5.1. Total RNA Extraction from Calf Tissue
2.5.2. Transcriptomic Sequencing
2.6. Bioinformatics Analysis
2.6.1. Quality Control
2.6.2. ncRNA Identity
2.6.3. Quantification of Gene Expression Levels
2.6.4. Differential Expression Analysis
2.6.5. Function Annotations of RNA
2.7. Validation of RNA-Seq Results Analyzed by Quantitative PCR (qPCR)
2.8. The Data Statistical Analysis
3. Results
3.1. Development of Rumen Dorsal and Ventral Papillae
3.2. Effects of IgG Content and Digestive Enzyme Activity
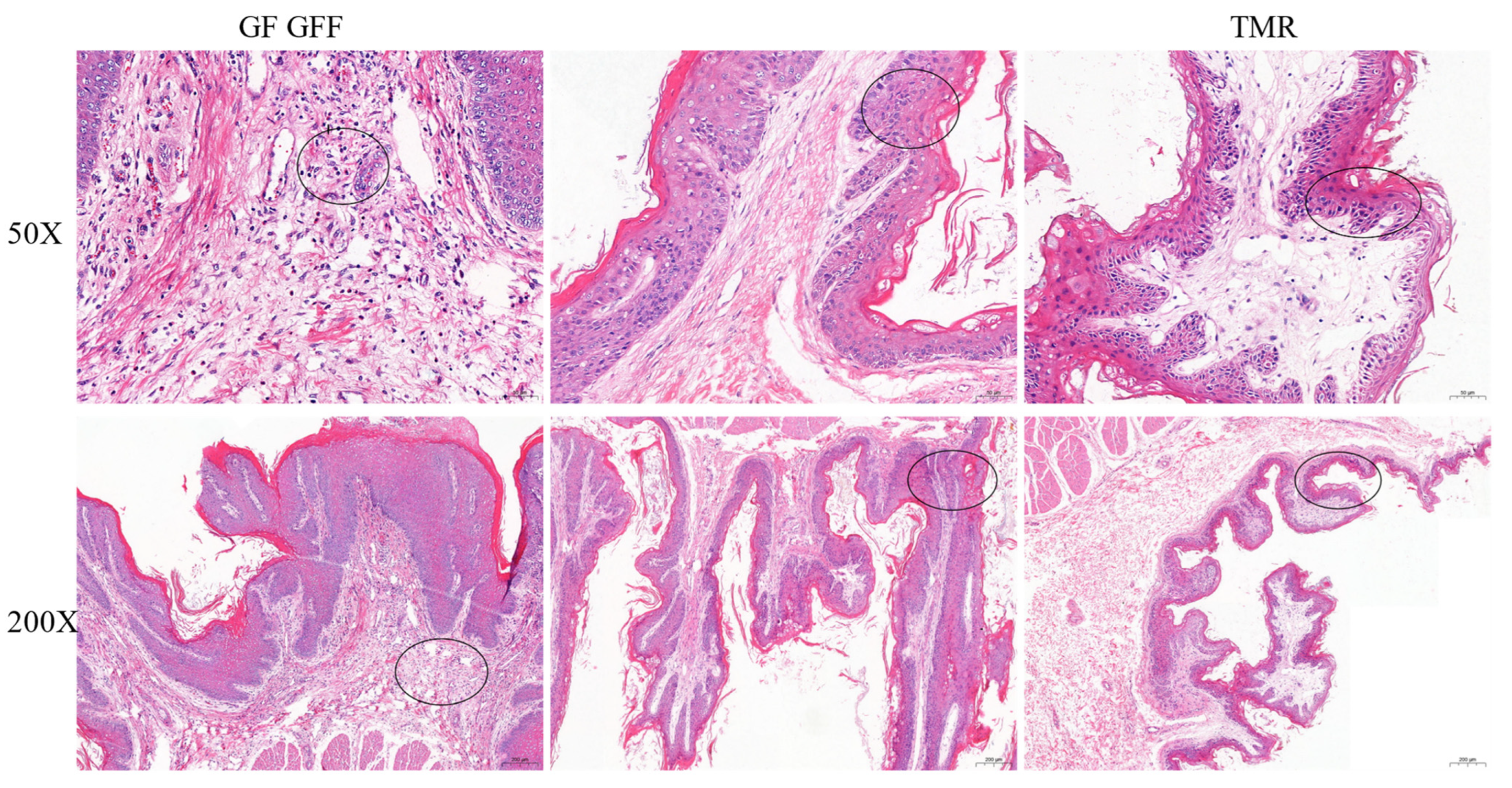
3.3. Histopathological Examination (HE) for Pathological Change
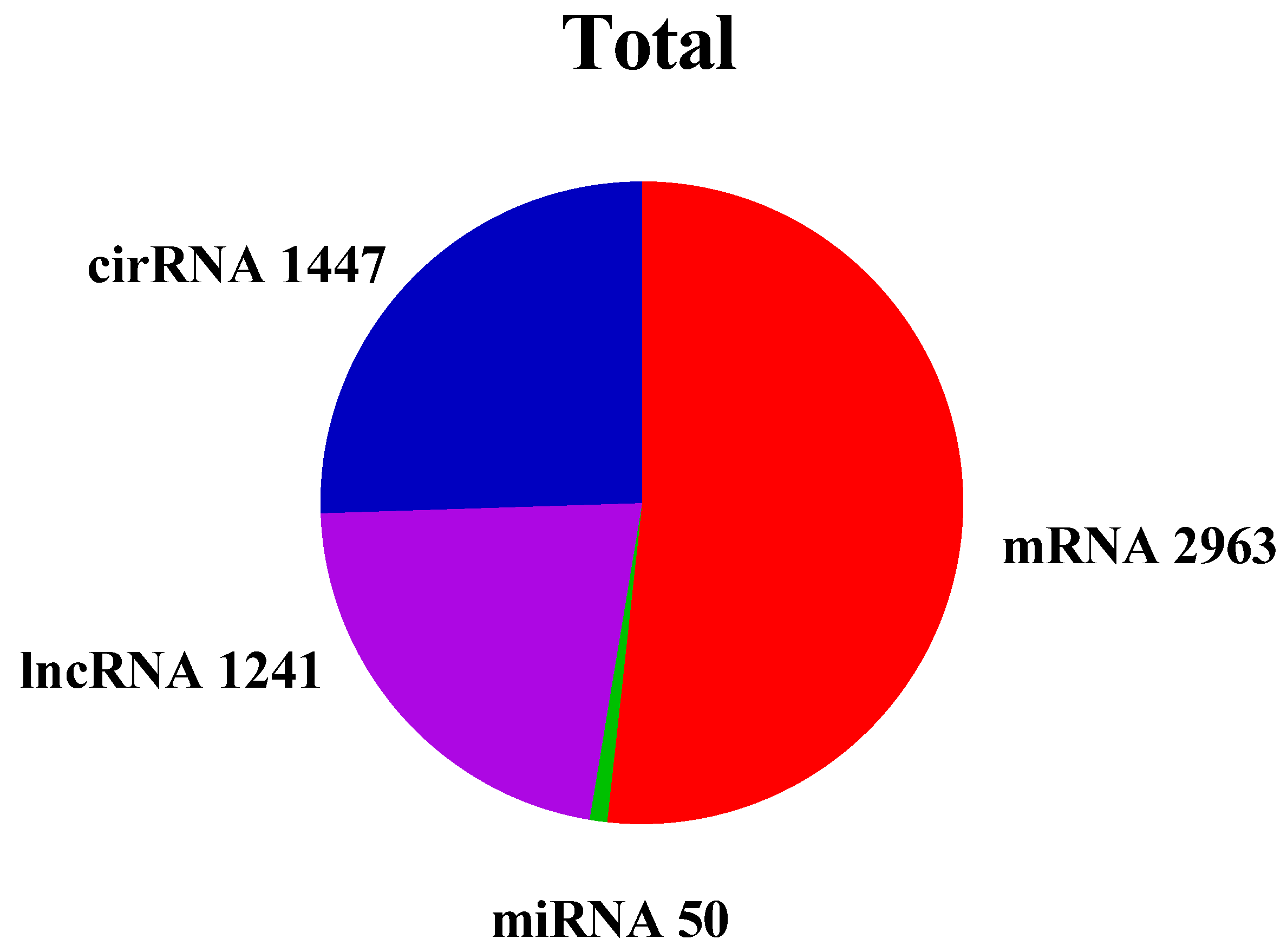
3.4. Overview of Identified ncRNA and mRNA
3.5. Expression Patterns of mRNAs and ncRNAs
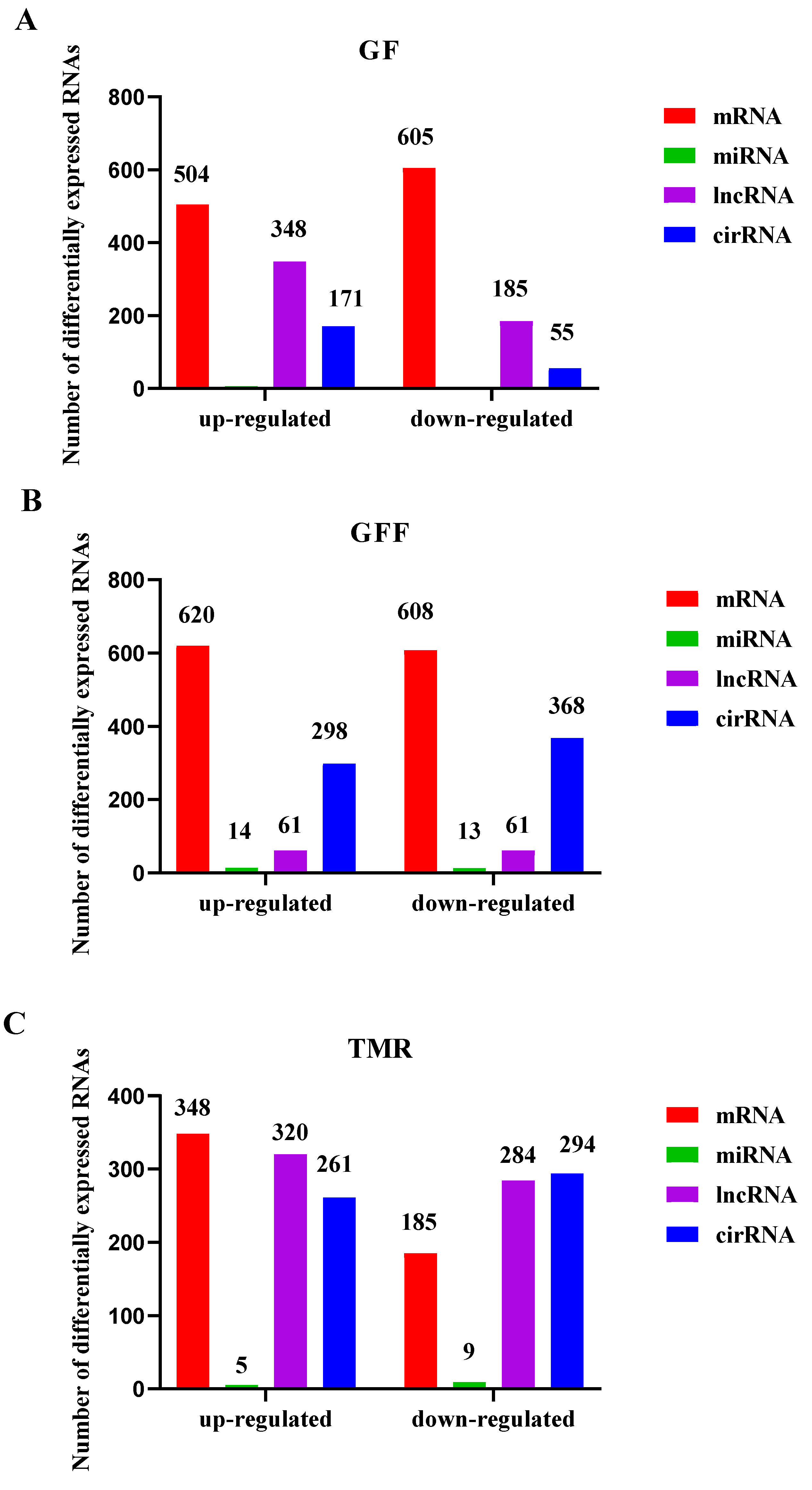
3.6. Differentially Expressed ncRNAs and mRNA
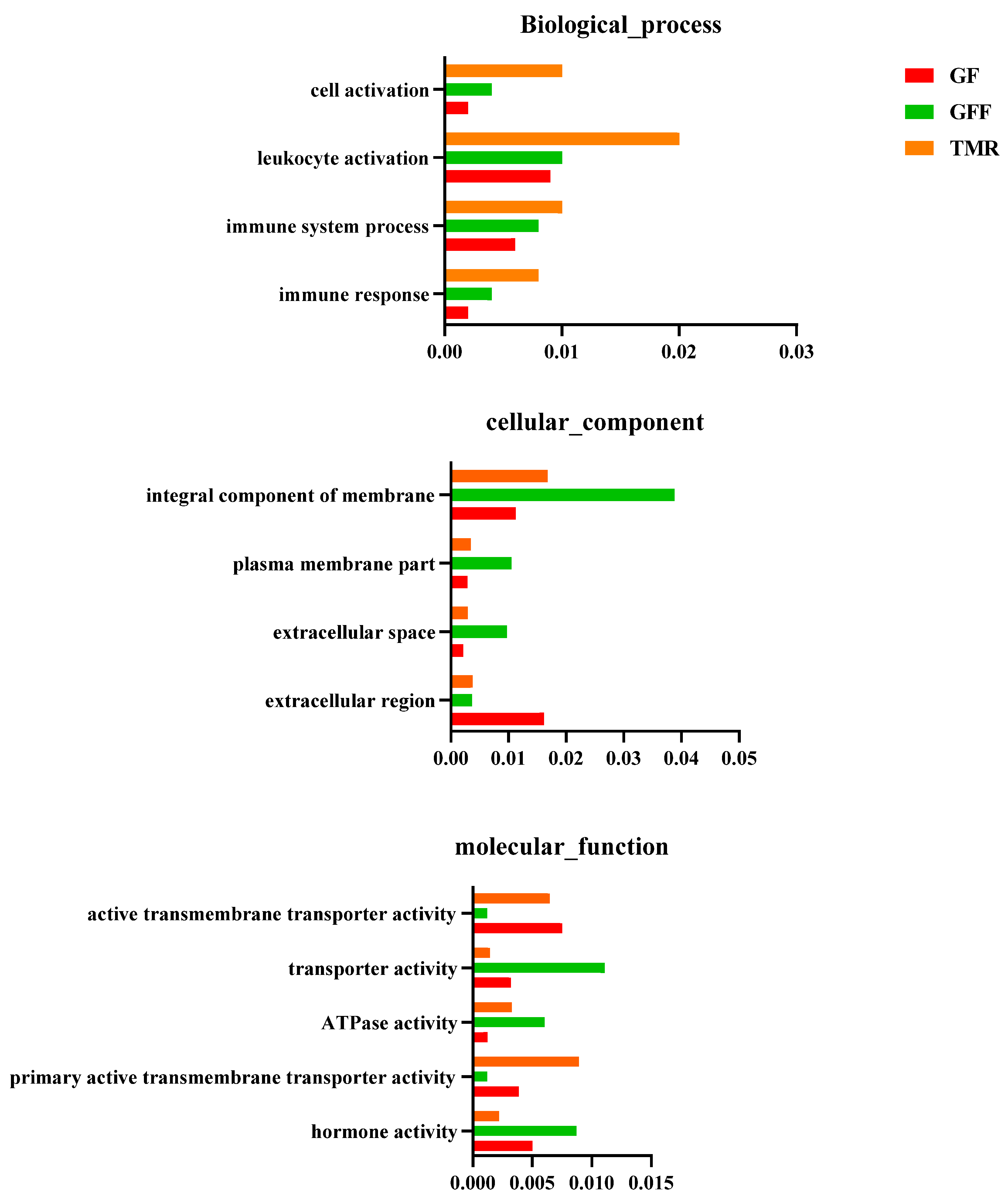
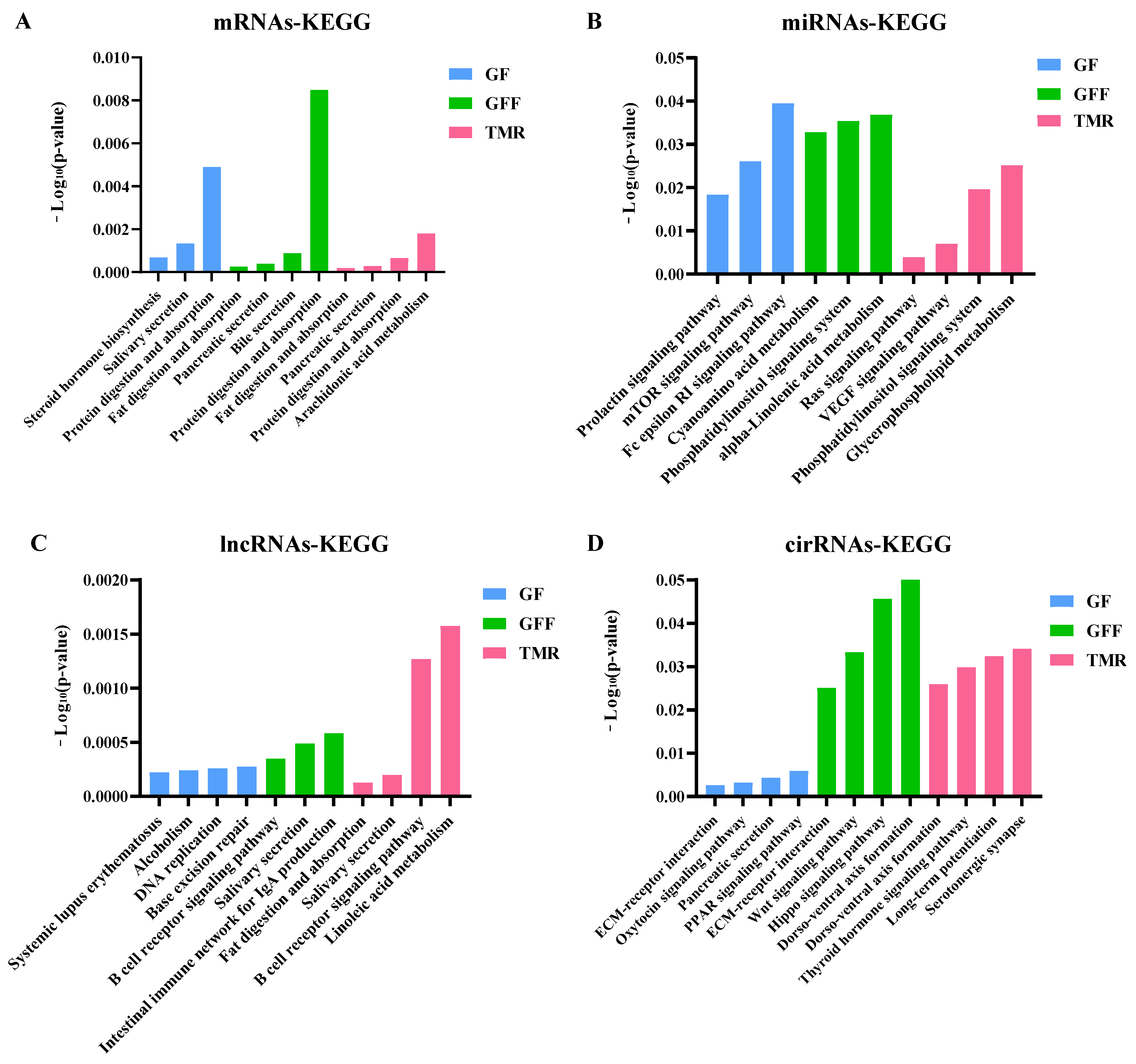
3.7. Differentially Expressed ncRNAs and mRNA Functional Enrichment Analysis
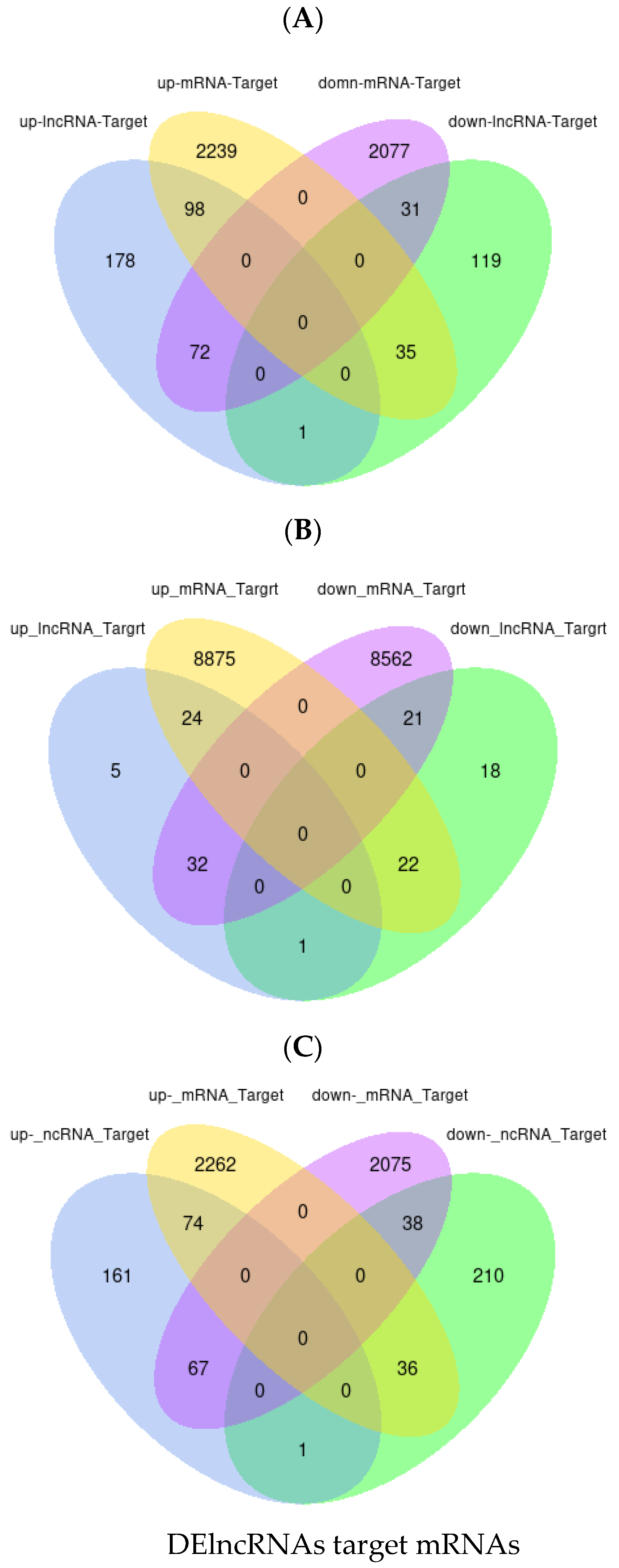
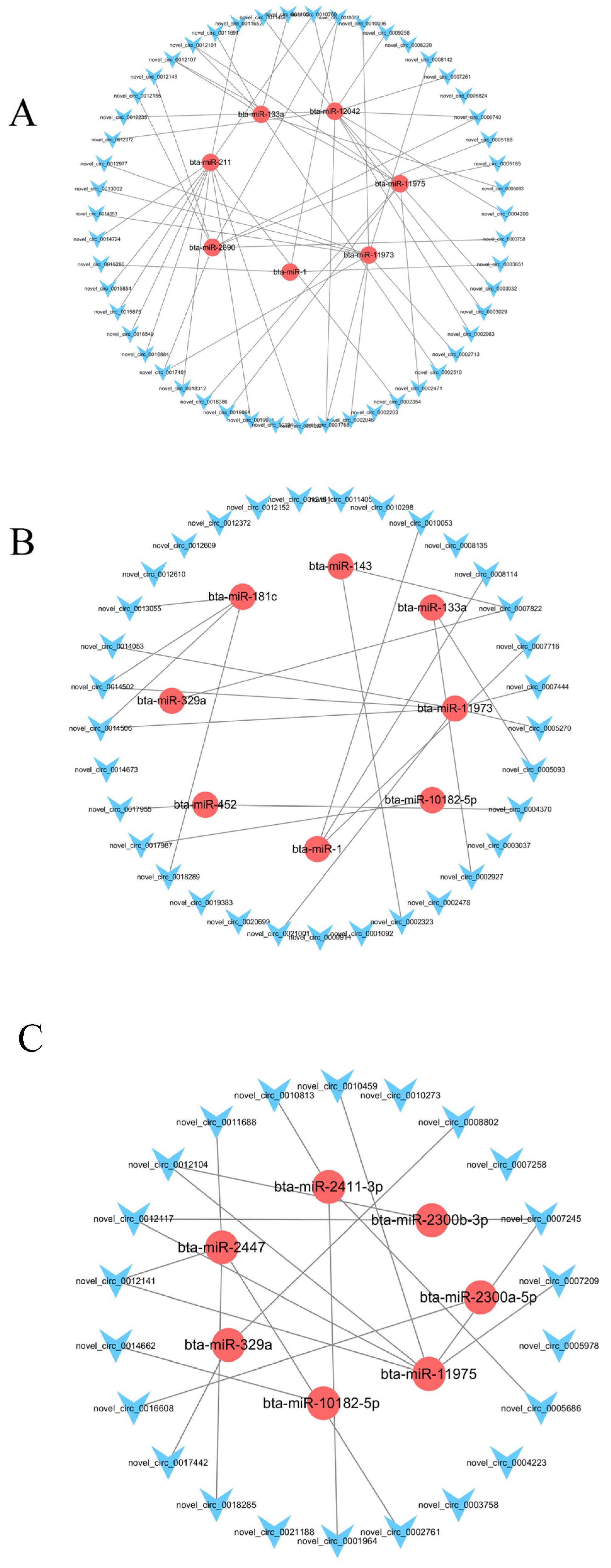
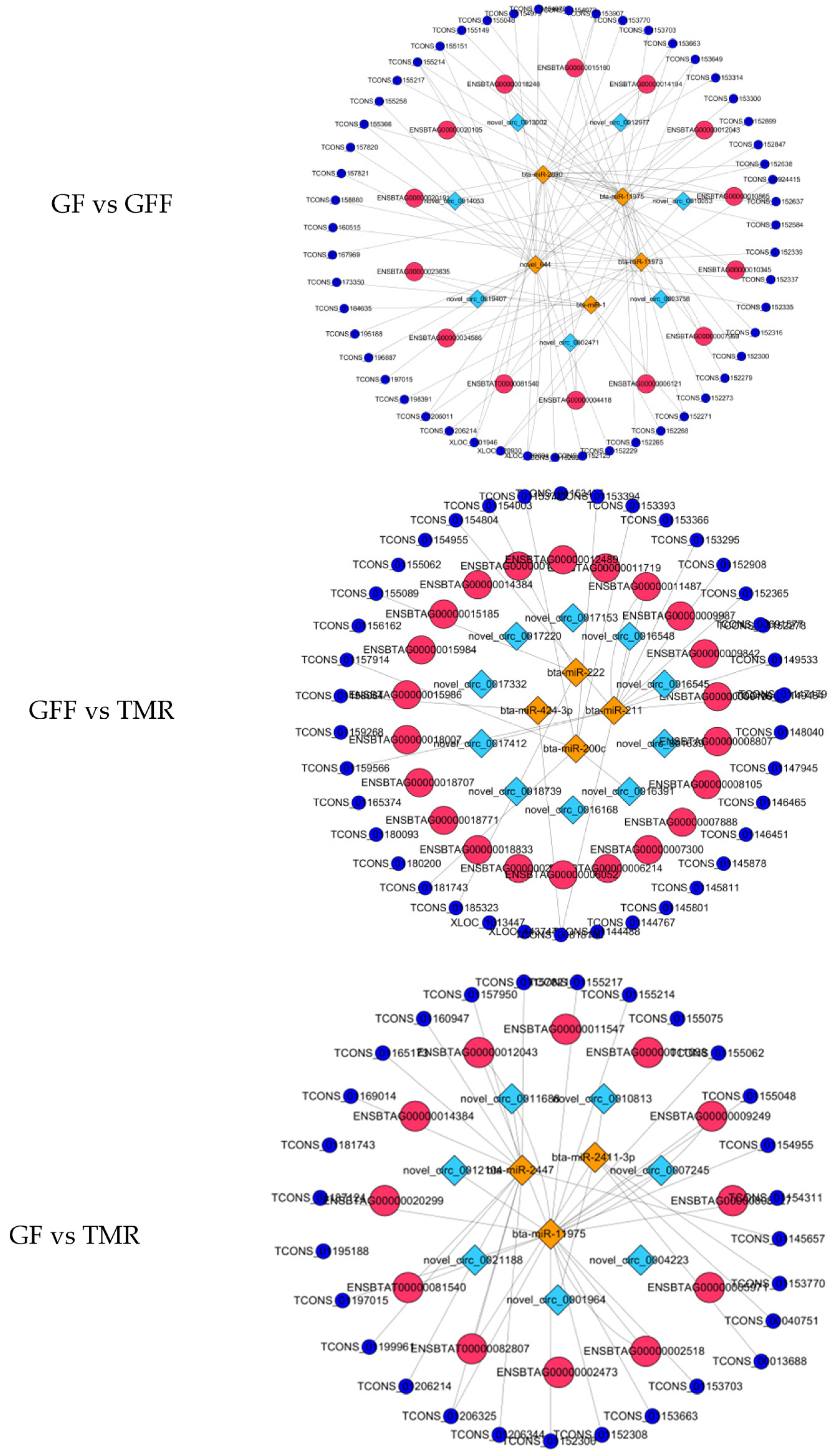
3.8. Construction of a Potential lncRNA/circRNA-miRNA-mRNA Regulatory Network
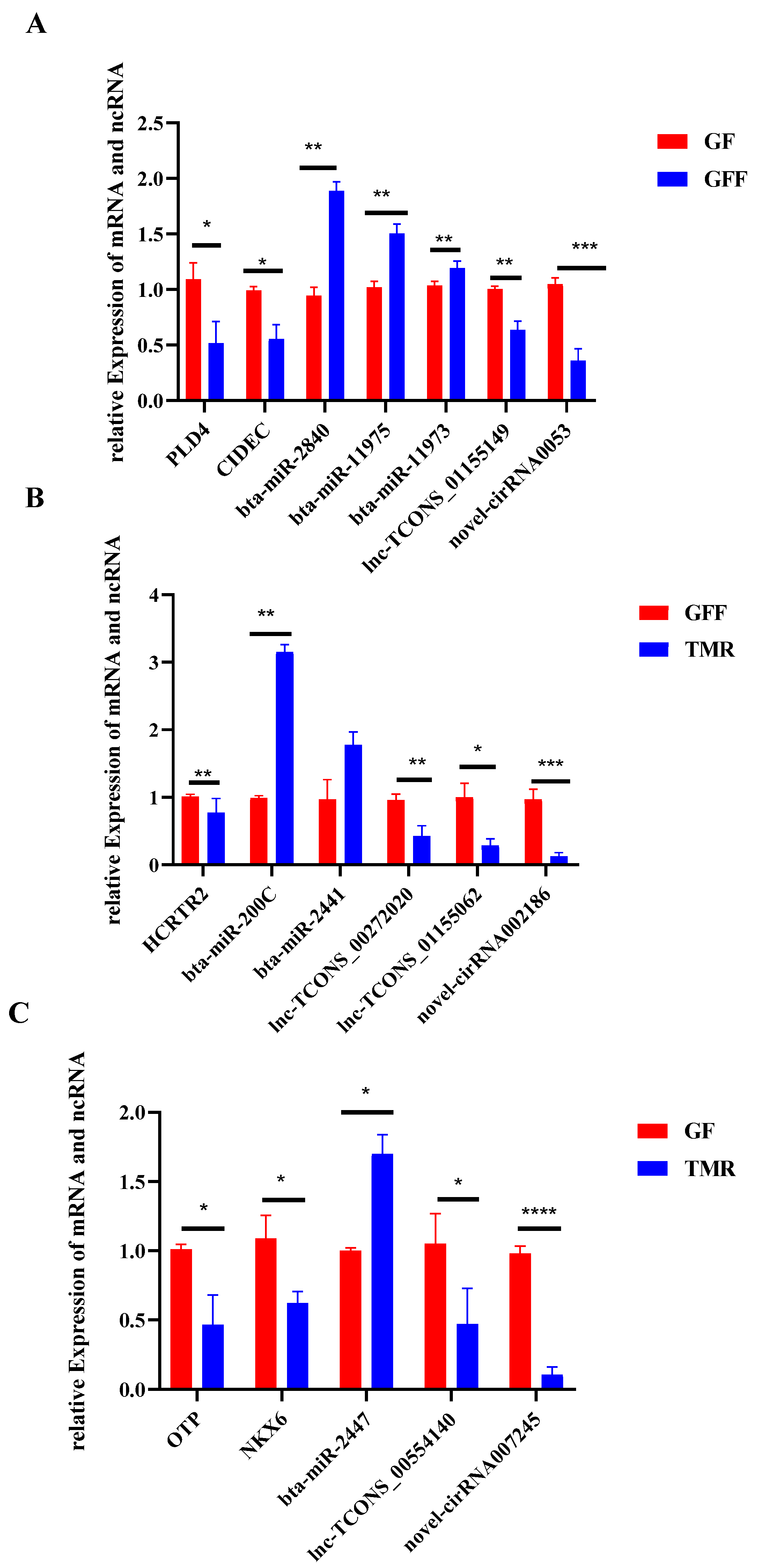
3.9. Validation of DE Genes by qRT-PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
List of Abbreviations
| Item | Unit |
| GF | Concentrate |
| GFF | Concentrate + hay |
| TMR | Concentrate + alfalfa grass + oat grass: water |
| mRNA | Messenger ribonucleic acid |
| miRNA | MicroRNA |
| lncRNA | Long non-coding RNA |
| circRNA | Circular RNA |
| ncRNA | Non-coding RNA |
| mRNAs | Differentially expressed mRNA |
| miRNAs | Differentially expressed miRNA |
| lncRNAs | Differentially expressed lncRNA |
| circRNAs | Differentially expressed circRNA |
| ncRNAs | Differentially expressed non-coding RNA |
| SNP | Single Nucleotide Polymorphisms |
| InDel | Insertion–deletion |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| CeRNAs | Competing endogenous RNAs |
| PPI | Protein–protein interaction |
| qRT-PCR | Quantitative real-time PCR |
| SD | Standard deviation |
| DMI | Dry matter intake |
| ADF | Acid detergent fiber |
| VFA | Volatile fatty acid |
| PH | Potential of hydrogen |
| RNA-seq | RNA sequencing |
| IgG | Immunoglobulin G |
| AMS | α-amylase |
| CP | Ceruloplasmin |
| LZM | Lysozyme |
| SARA | Subacute ruminal acidosis |
| MRE | miRNA response element |
| mean ± SD | mean ± standard deviation |
References
- Zhang, Z.; deqing, Z.; Zhang, X.; Ta, N.; Gesang, J.; Luosang, C.; Pingcuo, B. Different feeding strategies can affect growth performance and rumen functions in Gangba sheep as revealed by integrated transcriptome and microbiome analyses. Front. Microbiol. 2022, 13, 908326. [Google Scholar] [CrossRef]
- Khan, M.; Bach, A.; Weary, D.; von Keyserlingk, M.A.G. Invited review: Transitioning from milk to solid feed in dairy heifers. J. Dairy Sci. 2016, 99, 885–902. [Google Scholar] [CrossRef] [PubMed]
- Terler, G.; Poier, G.; Klevenhusen, F.; Zebeli, Q.J. Replacing concentrates with a high-quality hay in the starter feed in dairy calves: I. Effects on nutrient intake, growth performance, and blood metabolic profile. J. Dairy Sci. 2022, 105, 2326–2342. [Google Scholar] [CrossRef] [PubMed]
- Sutton, J.D.; Mcgilliard, A.D.; Jacobson, N.L. Functional Development of Rumen Mucosa. I. Absorptive Ability1. J. Dairy Sci. 1963, 46, 426–436. [Google Scholar] [CrossRef]
- Tamate, H.; Mcgilliard, A.D.; Jacobson, N.L.; Getty, R. Effect of Various Dietaries on the Anatomical Development of the Stomach in the Calf1. J. Dairy Sci. 1962, 45, 408–420. [Google Scholar] [CrossRef]
- Greenwood, R.; Morrill, J.; Titgemeyer, E.; Kennedy, G. A new method of measuring diet abrasion and its effect on the development of the forestomach. J. Dairy Sci. 1997, 80, 2534–2541. [Google Scholar] [CrossRef] [PubMed]
- Kay, M.; Fell, B.F.; Boyne, R. The relationship between the acidity of the rumen contents and rumenitis, in calves fed on barley. Res. Vet. Sci. 1969, 53, 181–187. [Google Scholar] [CrossRef]
- McGavin, M.; Morrill, J. Dissection technique for examination of the bovine ruminoreticulum. J. Anim. Sci. 1976, 42, 535–538. [Google Scholar] [CrossRef]
- Khan, M.; Weary, D.; von Keyserlingk, M. Hay intake improves performance and rumen development of calves fed higher quantities of milk. J. Dairy Sci. 2011, 94, 3547–3553. [Google Scholar] [CrossRef]
- Knott, K.K.; Bowyer, P.J. Growth in Arctic Ungulates: Postnatal Development and Organ Maturation in Rangifer tarandus and Ovibos moschatus. J. Mammal. 2005, 86, 121–130. [Google Scholar] [CrossRef]
- Kosiorowska, A.; Puggaard, L.; Hedemann, M.; Sehested, J.; Jensen, S.; Kristensen, N.; Kuropka, P.; Marycz, K.; Vestergaard, M.J.A. Gastrointestinal development of dairy calves fed low- or high-starch concentrate at two milk allowances. Anim. Int. J. Anim. Biosci. 2011, 5, 211–219. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Zhang, X.; Li, F.; Li, X.; Zhao, Y.; Zhang, Y.; Zhao, L.; Xu, D.; Wang, J.; Yang, X.; et al. Identification and characterization of circular RNAs in association with the feed efficiency in Hu lambs. BMC Genom. 2022, 23, 288. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Kuang, R.; Guo, Y.; Ma, R.; Hou, Y.; Xu, Y.; Qi, X.; Wang, D.; Zhou, H.; Xiong, Y.; et al. Epigenomics analysis of miRNA cis-regulatory elements in pig muscle and fat tissues. Genomics 2022, 114, 110276. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Jiang, Z.; Han, Y.; Li, Y.; Yang, X. Regulation of Pyroptosis by ncRNA: A Novel Research Direction. Front. Cell Dev. Biol. 2022, 10, 840576. [Google Scholar] [CrossRef] [PubMed]
- Chen, Q.; Wu, C.; Yao, Z.; Cai, L.; Ni, Y.; Mao, S.; Zhao, R. Whole transcriptome analysis of RNA expression profiles reveals the potential regulating action of long noncoding RNA in lactating cows fed a high concentrate diet. Anim. Nutr. 2021, 7, 1315–1328. [Google Scholar] [CrossRef]
- Minuti, A.; Palladino, A.; Khan, M.J.; Alqarni, S.; Agrawal, A.; Piccioli-Capelli, F.; Hidalgo, F.; Cardoso, F.C.; Trevisi, E.; Loor, J.J. Abundance of ruminal bacteria, epithelial gene expression, and systemic biomarkers of metabolism and inflammation are altered during the peripartal period in dairy cows. J. Dairy Sci. 2015, 98, 8940–8951. [Google Scholar] [CrossRef]
- Morris, N.L.; Choudhry, M.A. Maintenance of gut barrier integrity after injury: Trust your gut microRNAs. J. Leukoc. Biol. 2021, 110, 979–986. [Google Scholar] [CrossRef]
- Wei, J.W.; Huang, K.; Yang, C.; Kang, C.S. Non-coding RNAs as regulators in epigenetics (Review). Oncol. Rep. 2017, 37, 3–9. [Google Scholar] [CrossRef]
- Liu, Z.; Zhang, Z.; Song, G.; Wang, X.; Xing, H.; Wang, C. Resveratrol Alleviates Skeletal Muscle Insulin Resistance by Downregulating Long Noncoding RNA. Int. J. Endocrinol. 2022, 2022, 2539519. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, Y.; Zhang, S.; Huang, D.; Li, B.; Liang, G.; Wu, Y.; Jiang, Q.; Li, L.; Lin, C.; et al. circRNA circARNT2 Suppressed the Sensitivity of Hepatocellular Carcinoma Cells to Cisplatin by Targeting the miR-155-5p/PDK1 Axis. Mol. Ther. Nucleic Acids 2021, 23, 244–254. [Google Scholar] [CrossRef]
- Tay, Y.; Rinn, J.; Pandolfi, P.P. The multilayered complexity of ceRNA crosstalk and competition. Nature 2014, 505, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Parkhomchuk, D.; Borodina, T.; Amstislavskiy, V.; Banaru, M.; Hallen, L.; Krobitsch, S.; Lehrach, H.; Soldatov, A. Transcriptome analysis by strand-specific sequencing of complementary DNA. Nucleic Acids Res. 2009, 37, e123. [Google Scholar] [CrossRef] [PubMed]
- Rebenku, I.; Bartha, F.A.; Katona, T.; Zsebik, B.; Antalffy, G.; Takács, L.; Molnár, B.; Vereb, G. Taking molecular pathology to the next level: Whole slide multicolor confocal imaging with the Pannoramic Confocal digital pathology scanner. Cytometry. Part A 2022, 103, 198–207. [Google Scholar] [CrossRef]
- Tang, P.; Cui, E.H.; Chang, W.C.; Yu, C.; Wang, H.; Du, E.Q.; Wang, J.Y. Nanoparticle-Based Bivalent Swine Influenza Virus Vaccine Induces Enhanced Immunity and Effective Protection against Drifted H1N1 and H3N2 Viruses in Mice. Viruses 2022, 14, 2443. [Google Scholar] [CrossRef] [PubMed]
- Sun, R.; Xu, K.; Yu, L.; Pu, Y.; Xiong, F.; He, Y.; Huang, Q.; Tang, M.; Chen, M.; Yin, L.; et al. Preliminary study on impacts of polystyrene microplastics on the hematological system and gene expression in bone marrow cells of mice. Ecotoxicol. Environ. Saf. 2021, 218, 112296. [Google Scholar] [CrossRef] [PubMed]
- Zhan, L.; Zhang, J.; Zhao, B.; Li, X.; Zhang, X.; Hu, R.; Elken, E.M.; Kong, L.; Gao, Y. Genomic and Transcriptomic Analysis of Bovine Pasteurella multocida Serogroup A Strain Reveals Insights Into Virulence Attenuation. Front. Vet. Sci. 2021, 8, 765495. [Google Scholar] [CrossRef] [PubMed]
- Sun, S.; Cao, C.; Li, J.; Meng, Q.; Cheng, B.; Shi, B.; Shan, A. Lycopene Modulates Placental Health and Fetal Development Under High-Fat Diet During Pregnancy of Rats. Mol. Nutr. Food Res. 2021, 65, e2001148. [Google Scholar] [CrossRef]
- Singla, D.; Yadav, I. GAAP: A GUI-based Genome Assembly and Annotation Package. Curr. Genom. 2022, 23, 77–82. [Google Scholar] [CrossRef]
- Tang, R.; Liu, F.; Lan, Y.; Wang, J.; Wang, L.; Li, J.; Liu, X.; Fan, Z.; Guo, T.; Yue, B. Transcriptomics and metagenomics of common cutworm (Spodoptera litura) and fall armyworm (Spodoptera frugiperda) demonstrate differences in detoxification and development. BMC Genom. 2022, 23, 388. [Google Scholar] [CrossRef]
- Garousi, S.; Jahanbakhsh Godehkahriz, S.; Esfahani, K.; Lohrasebi, T.; Mousavi, A.; Hatef Salmanian, A.; Rezvani, M.; Moein, M. Meta-Analysis of EGF-Stimulated Normal and Cancer Cell Lines to Discover EGF-Associated Oncogenic Signaling Pathways and Prognostic Biomarkers. Iran. J. Biotechnol. 2022, 20, e3245. [Google Scholar] [CrossRef]
- Long, W.; Liu, S.; Li, X.X.; Shen, X.; Zeng, J.; Luo, J.S.; Li, K.R.; Wu, A.G.; Yu, L.; Qin, D.L.; et al. Whole transcriptome sequencing and integrated network analysis elucidates the effects of 3,8-Di-O-methylellagic acid 2-O-glucoside derived from Sanguisorba offcinalis L., a novel differentiation inducer on erythroleukemia cells. Pharmacol. Res. 2021, 166, 105491. [Google Scholar] [CrossRef] [PubMed]
- Ai, Y.; Luo, S.; Wang, B.; Xiao, S.; Wang, Y. MiR-126-5p Promotes Tumor Cell Proliferation, Metastasis and Invasion by Targeting TDO2 in Hepatocellular Carcinoma. Molecules 2022, 27, 443. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.; Lin, Y.; Jiang, L.; Xi, J.; Wang, X.; Guan, J.; Chen, J.; Pan, Y.; Luo, J.; Ye, C.; et al. Comparison analysis of microRNAs in response to dengue virus type 2 infection between the Vero cell-adapted strain and its source, the clinical C6/36 isolated strain. Virus Res. 2018, 250, 65–74. [Google Scholar] [CrossRef]
- Friedländer, M.R.; Mackowiak, S.D.; Li, N.; Chen, W.; Rajewsky, N. miRDeep2 accurately identifies known and hundreds of novel microRNA genes in seven animal clades. Nucleic Acids Res. 2012, 40, 37–52. [Google Scholar] [CrossRef] [PubMed]
- Wen, M.; Shen, Y.; Shi, S.; Tang, T. miREvo: An integrative microRNA evolutionary analysis platform for next-generation sequencing experiments. BMC Bioinform. 2012, 13, 140. [Google Scholar] [CrossRef]
- Zhou, J.; Liu, L.; Wu, P.; Zhao, L.; Wu, Y. Identification and characterization of non-coding RNA networks in infected macrophages revealing the pathogenesis of F. nucleatum-associated diseases. BMC Genom. 2022, 23, 826. [Google Scholar] [CrossRef]
- Gao, Y.; Wang, J.; Zhao, F. CIRI: An efficient and unbiased algorithm for de novo circular RNA identification. Genome Biol. 2015, 16, 4. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Bao, Y.; Wang, L.; Shi, L.; Yun, F.; Liu, X.; Chen, Y.; Chen, C.; Ren, Y.; Jia, Y. Transcriptome profiling revealed multiple genes and ECM-receptor interaction pathways that may be associated with breast cancer. Cell. Mol. Biol. Lett. 2019, 24, 38. [Google Scholar] [CrossRef]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef]
- Young, M.D.; Wakefield, M.J.; Smyth, G.K.; Oshlack, A. Gene ontology analysis for RNA-seq: Accounting for selection bias. Genome Biol. 2010, 11, R14. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Shen, H.; Wang, P.; Min, J.; Yu, Y.; Wang, Q.; Wang, S.; Xi, W.; Nguyen, Q.M.; Xiao, J.; et al. Identification and characterization of circular RNAs in atrial appendage of patients with atrial fibrillation. Exp. Cell Res. 2020, 389, 111821. [Google Scholar] [CrossRef] [PubMed]
- Kanehisa, M.; Furumichi, M.; Tanabe, M.; Sato, Y.; Morishima, K. KEGG: New perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res. 2017, 45, D353–D361. [Google Scholar] [CrossRef]
- Zhou, G.; Kang, D.; Ma, S.; Wang, X.; Gao, Y.; Yang, Y.; Wang, X.; Chen, Y. Integrative analysis reveals ncRNA-mediated molecular regulatory network driving secondary hair follicle regression in cashmere goats. BMC Genom. 2018, 19, 222. [Google Scholar] [CrossRef]
- Wang, J.; Fan, H.; Xia, S.; Shao, J.; Tang, T.; Chen, L.; Bai, X.; Sun, W.; Jia, X.; Chen, S.; et al. Microbiome, Transcriptome, and Metabolomic Analyses Revealed the Mechanism of Immune Response to Diarrhea in Rabbits Fed Antibiotic-Free Diets. Front. Microbiol. 2022, 13, 888984. [Google Scholar] [CrossRef]
- Garcia-Martin, R.; Wang, G.; Brandão, B.B.; Zanotto, T.M.; Shah, S.; Kumar Patel, S.; Schilling, B.; Kahn, C.R. MicroRNA sequence codes for small extracellular vesicle release and cellular retention. Nature 2022, 601, 446–451. [Google Scholar] [CrossRef]
- Singh, A.; Agarwal, A.; Mohan, R.; Singh, S.; Tewari, P.; Srivastava, S. The Effect of Timing of Debridement and Surgical Intervention in Open Fractures on the Rate of Infection and Surgical Outcomes: A Prospective Study in a Tertiary Care Setup. Cureus 2023, 15, e37204. [Google Scholar] [CrossRef]
- Li, B.; Yang, J.; He, J.; Peng, X.; Zeng, Q.; Song, Y.; Xu, K.; Ma, H. Characterization of the whole transcriptome of spleens from Chinese indigenous breed Ningxiang pig reveals diverse coding and non-coding RNAs for immunity regulation. Genomics 2021, 113, 2468–2482. [Google Scholar] [CrossRef] [PubMed]
- Hollmann, M.; Knowlton, K.F.; Hanigan, M.D. Evaluation of solids, nitrogen, and phosphorus excretion models for lactating dairy cows. J. Dairy Sci. 2008, 91, 1245–1257. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Hidaka, K.; Mishima, T.; Nibe, K.; Kitahara, G.; Hidaka, Y.; Katamoto, H.; Kamimura, S. Effect of sugar supplementation on rumen protozoa profile and papillae development in retarded growth calves. J. Vet. Med. Sci. 2010, 72, 1471–1474. [Google Scholar] [CrossRef]
- Kristensen, N.B.; Sehested, J.; Jensen, S.K.; Vestergaard, M. Effect of milk allowance on concentrate intake, ruminal environment, and ruminal development in milk-fed Holstein calves. J. Dairy Sci. 2007, 90, 4346–4355. [Google Scholar] [CrossRef]
- Cao, Y.; Liu, S.; Yang, X.; Guo, L.; Cai, C.; Yao, J. Effects of dietary leucine and phenylalanine on gastrointestinal development and small intestinal enzyme activities in milk-fed holstein dairy calves. Biosci. Rep. 2019, 39, BSR20181733. [Google Scholar] [CrossRef]
- Dai, D.; Pang, K.; Liu, S.; Wang, X.; Yang, Y.; Chai, S.; Wang, S. Effects of Concentrate Supplementation on Growth Performance, Rumen Fermentation, and Bacterial Community Composition in Grazing Yaks during the Warm Season. Animals 2022, 12, 1398. [Google Scholar] [CrossRef]
- Zhao, L.; Ding, Y.; Yang, C.; Wang, P.; Zhao, Z.; Ma, Y.; Shi, Y.; Kang, X. Identification and characterization of hypothalamic circular RNAs associated with bovine residual feed intake. Gene 2023, 851, 147017. [Google Scholar] [CrossRef]
- Patience, J.F.; Rossoni-Serão, M.C.; Gutiérrez, N.A. A review of feed efficiency in swine: Biology and application. J. Anim. Sci. Biotechnol. 2015, 6, 33. [Google Scholar] [CrossRef]
- Horodyska, J.; Hamill, R.M.; Reyer, H.; Trakooljul, N.; Lawlor, P.G.; McCormack, U.M.; Wimmers, K. RNA-Seq of Liver From Pigs Divergent in Feed Efficiency Highlights Shifts in Macronutrient Metabolism, Hepatic Growth and Immune Response. Front. Genet. 2019, 10, 117. [Google Scholar] [CrossRef]
- Bernatik, O.; Paclikova, P.; Sri Ganji, R.; Bryja, V. Activity of Smurf2 Ubiquitin Ligase Is Regulated by the Wnt Pathway Protein Dishevelled. Cells 2020, 9, 1147. [Google Scholar] [CrossRef]
- Boyer, A.; Goff, A.K.; Boerboom, D. WNT signaling in ovarian follicle biology and tumorigenesis. Trends Endocrinol. Metab. TEM 2010, 21, 25–32. [Google Scholar] [CrossRef]
- Li, J.; Xue, M.; Zhang, L.; Li, L.; Lian, H.; Li, M.; Gao, T.; Fu, T.; Tu, Y. Integration of Long Non-Coding RNA and mRNA Profiling Reveals the Mechanisms of Different Dietary NFC/NDF Ratios Induced Rumen Development in Calves. Animals 2022, 12, 650. [Google Scholar] [CrossRef]
- Wu, H.; Zhu, N.; Liu, J.; Ma, J.; Jiao, R. Shaggy regulates tissue growth through Hippo pathway in Drosophila. Sci. China Life Sci. 2022, 65, 2131–2144. [Google Scholar] [CrossRef]
- Li, X.; Tian, R.; Gao, H.; Yang, Y.; Williams, B.R.G.; Gantier, M.P.; McMillan, N.A.J.; Xu, D.; Hu, Y.; Gao, Y. Identification of a histone family gene signature for predicting the prognosis of cervical cancer patients. Sci. Rep. 2017, 7, 16495. [Google Scholar] [CrossRef]
- Vigors, S.; JV, O.D.; Ryan, M.; Sweeney, T. Analysis of the basal colonic innate immune response of pigs divergent in feed efficiency and following an ex vivo lipopolysaccharide challenge. Physiol. Genom. 2019, 51, 443–448. [Google Scholar] [CrossRef]
- Yang, L.; He, T.; Xiong, F.; Chen, X.; Fan, X.; Jin, S.; Geng, Z. Identification of key genes and pathways associated with feed efficiency of native chickens based on transcriptome data via bioinformatics analysis. BMC Genom. 2020, 21, 292. [Google Scholar] [CrossRef]

| Terms | GF | GFF | TMR | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Papillae Height (μm) | Papillae Width (μm) | Papillae Number | Papillae Height (μm) | Papillae Width (μm) | Papillae Number | Papillae Height (μm) | Papillae Width (μm) | Nipple Number | |
| anterior dorsum | 1906.43 ± 1257.35 a | 400.66 ± 149.18 a | 6.89 ± 1.24 | 1501.97 ± 1060.40 b | 274.47 ± 96.73 b | 8.61 ± 3.95 | 1540.82 ± 933.12 b | 269.94 ± 102.15 b | 6.83 ± 2.22 |
| middle dorsum | 2128.66 ± 1451.40 a | 419.27 ± 174.35 a | 4.50 ± 1.62 a | 1922.92 ± 1423.43 a | 333.18 ± 120.577 b | 7.72 ± 3.68 a | 1171.69 ± 587.37 b | 317.49 ± 135.73 b | 9.28 ± 1.18 b |
| back dorsum | 1483.10 ± 1174.22 a | 316.11 ± 125.19 | 6.39 ± 1.80 | 2217.58 ± 1398.27 b | 341.29 ± 255.09 | 7.11 ± 2.87 | 1725.35 ± 870.44 b | 319.22 ± 87.99 | 8.50 ± 1.30 |
| anterior abdomen | 1326.79 ± 1116.87 a | 330.39 ± 153.94 | 5.11 ± 1.92 a | 2286.27 ± 1329.40 a | 306.41 ± 153.94 | 7.67 ± 1.44 a | 1907.41 ± 1485.78 b | 262.73 ± 92.01 | 7.17 ± 1.89 b |
| middle abdomen | 1668.26 ± 1162.20 a | 315.91 ± 125.38 a | 4.50 ± 1.12 a | 2198.94 ± 1309.87 b | 44.66 ± 87.85 b | 6.56 ± 2.60 ab | 1631.82 ± 899.75 b | 244.66 ± 87.85 b | 8.38 ± 2.43 b |
| posterior abdomen | 2037.77 ± 1566.81 a | 336.90 ± 146.27 a | 4.28 ± 0.83 a | 1726.60 ± 1211.84 a | 275.17 ± 88.79 b | 6.56 ± 2.13 b | 1025.13 ± 649.82 b | 224.38 ± 71.08 | 9.45 ± 2.51 |
| Fluency Index | GF | GFF | TMR |
|---|---|---|---|
| Mean ± SD (%) | Mean ± SD (%) | Mean ± SD (%) | |
| α-amylase content (nmol/mL) | 35.65 ± 3.40 b | 57.96 ± 9.68 b | 38.55 ± 10.68 a |
| Ceruloplasmin activity (U/L) | 9.19 ± 0.06 b | 8.81 ± 1.65 b | 12.73 ± 0.13 a |
| Trypsin activity (U/mL) | 213.56 ± 187.52 | 89.56 ± 50.97 | 265.22 ± 173.63 |
| Lysozyme content (μg/mL) | 1.50 ± 0.09 | 1.72 ± 0.17 | 1.62 ± 0.43 |
| IgG (mg/mL) | 0.56 ± 0.11 | 0.58 ± 0.08 | 0.60 ± 0.32 |
| Sample Name | Raw Reads | Clean Reads | Clean Bases | Error Rate (%) | Q20 (%) | Q30 (%) | GC Content (%) |
|---|---|---|---|---|---|---|---|
| GF_R_1 | 95151460 | 94466002 | 12.79G | 0.03 | 97.58 | 93.48 | 52.04 |
| GF_R_2 | 83295944 | 82575946 | 14.17G | 0.03 | 97.62 | 93.66 | 55.92 |
| GF_R_3 | 99995438 | 99117250 | 12.39G | 0.03 | 97.62 | 93.60 | 52.20 |
| GFF_R_1 | 81517354 | 80087068 | 12.01G | 0.03 | 97.48 | 93.33 | 51.38 |
| GFF_R_2 | 79648920 | 78647860 | 11.80G | 0.03 | 97.59 | 93.55 | 52.30 |
| GFF_R_3 | 91568644 | 90891982 | 13.63G | 0.03 | 97.72 | 93.81 | 49.97 |
| TMR_R_1 | 85349140 | 84721348 | 12.71G | 0.03 | 97.60 | 93.48 | 48.89 |
| TMR_R_2 | 85825104 | 85003696 | 12.75G | 0.03 | 97.70 | 93.81 | 52.33 |
| TMR_R_3 | 86443542 | 85679884 | 12.85G | 0.03 | 97.71 | 93.78 | 50.36 |
| Group | miRNAs | circRNAs | lncRNAs | mRNAs |
|---|---|---|---|---|
| GF vs. GFF | bta-miR-11973 | novel_circ_0014053, novel_circ_0012977, novel_circ_0013002 | XLOC_432876, XLOC_480762, XLOC_1012137, XLOC_098896 | MAL, PLD4, CIDEC, |
| bta-miR-1 | novel_circ_0010053 | XLOC_511980, XLOC_1005675, XLOC_1005790, XLOC_062658, XLOC_122211 | CLEC6A | |
| bta-miR-2890 | novel_circ_0003758, novel_circ_0010053 | XLOC_1001781, XLOC_999381, XLOC_654245, XLOC_998773 | FEV, MGLL, CACNG2, PLA2G2E | |
| GFF vs. TMR | bta-miR-200c | novel_circ_0021078, novel_circ_0021086, novel_circ_0016391, novel_circ_0008818, | XLOC_514666, XLOC_223927, XLOC_546291, XLOC_005658 | XLOC_398026, XLOC_1006227, XLOC_629685, XLOC_1001314XLOC_1013447 |
| bta-miR-211 | novel_circ_0015818, novel_circ_0015854, novel_circ_0011652, novel_circ_0016548, novel_circ_0011061 | GTPBP4, XLOC_1006044, XLOC_998134.SPDYA | XLOC_253297, XLOC_443744, PLEKHS1, XLOC_830139 | |
| GF vs. TMR | bta-miR-11975 | novel_circ_0012104, novel_circ_0007245 | CDCP1, EFHB, XLOC_996619, XLOC_996619, XLOC_1003325, LYRM1, XLOC_1001987, FBXL19 | NKX6-1, OTP, PADI3 |
| bta-miR-2411-3p | novel_circ_0010813, novel_circ_0001964 | XLOC_999378, SLX4IP, SLX4IP, XLOC_59297 | XLOC_1005215, XLOC_830139 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, J.; Fan, H.; Li, M.; Zhao, K.; Xia, S.; Chen, Y.; Shao, J.; Tang, T.; Bai, X.; Liu, Z.; et al. Integration of Non-Coding RNA and mRNA Profiles Reveals the Mechanisms of Rumen Development Induced by Different Types of Diet in Calves. Genes 2023, 14, 1093. https://doi.org/10.3390/genes14051093
Wang J, Fan H, Li M, Zhao K, Xia S, Chen Y, Shao J, Tang T, Bai X, Liu Z, et al. Integration of Non-Coding RNA and mRNA Profiles Reveals the Mechanisms of Rumen Development Induced by Different Types of Diet in Calves. Genes. 2023; 14(5):1093. https://doi.org/10.3390/genes14051093
Chicago/Turabian StyleWang, Jie, Huimei Fan, Mianying Li, Kaisen Zhao, Siqi Xia, Yang Chen, Jiahao Shao, Tao Tang, Xue Bai, Zheliang Liu, and et al. 2023. "Integration of Non-Coding RNA and mRNA Profiles Reveals the Mechanisms of Rumen Development Induced by Different Types of Diet in Calves" Genes 14, no. 5: 1093. https://doi.org/10.3390/genes14051093





