Contribution of Genetic Test to Early Diagnosis of Methylenetetrahydrofolate Reductase (MTHFR) Deficiency: The Experience of a Reference Center in Southern Italy
Abstract
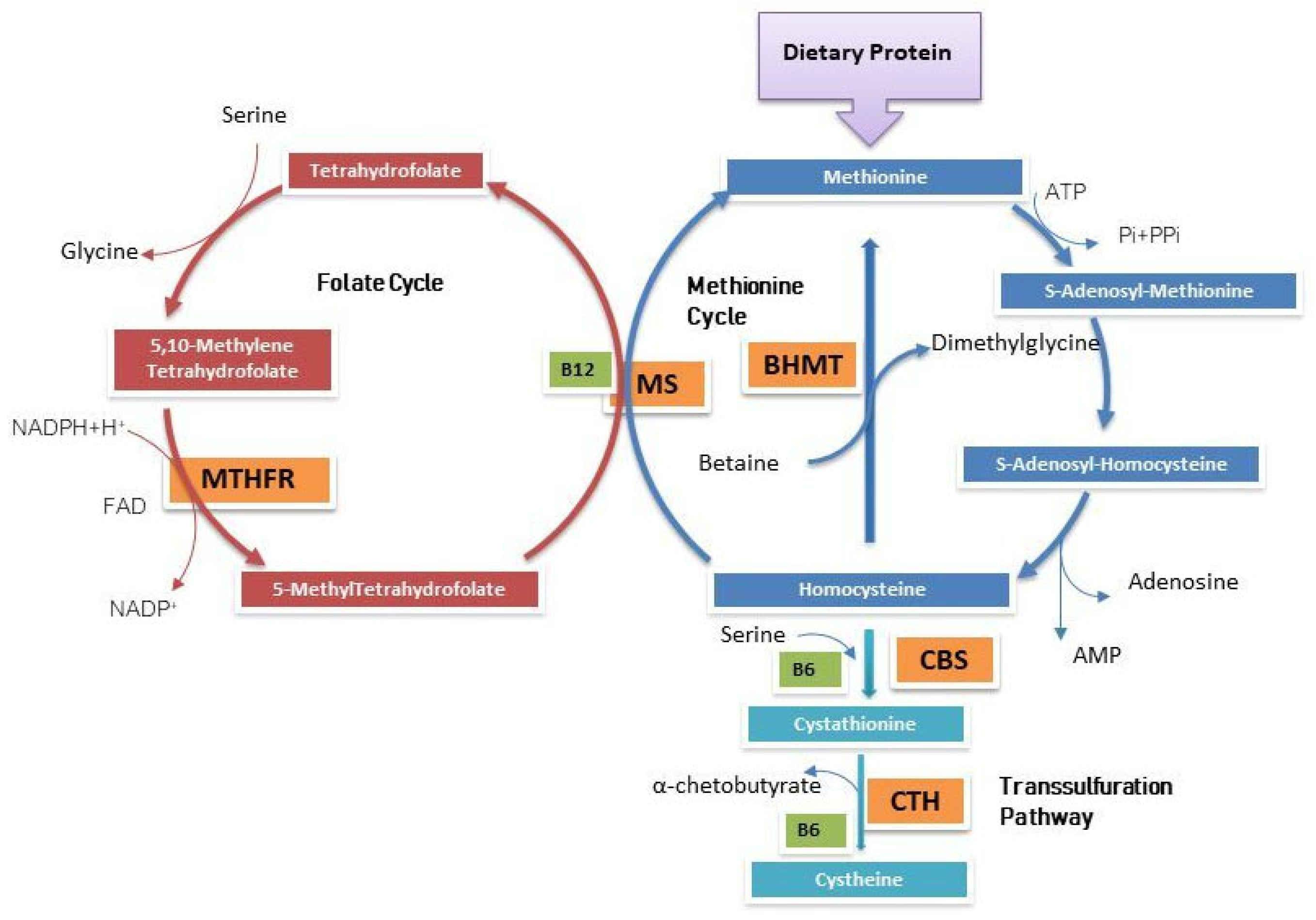
:1. Introduction
2. Materials and Methods
2.1. Newborn Screening
2.2. Genetic Analysis
2.3. Bioinformatics Analysis
2.4. Homology Modelling
3. Results
3.1. Clinical and Biochemical Findings
3.2. Molecular Analysis of the MTHFR Gene and In Silico Evaluation of the Novel Variant
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Scolamiero, E.; Villani, G.R.; Ingenito, L.; Pecce, R.; Albano, L.; Caterino, M.; di Girolamo, M.G.; Di Stefano, C.; Franzese, I.; Gallo, G.; et al. Maternal vitamin B12 deficiency detected in expanded newborn screening. Clin. Biochem. 2014, 47, 312–317. [Google Scholar] [CrossRef]
- Scolamiero, E.; Cozzolino, C.; Albano, L.; Ansalone, A.; Caterino, M.; Corbo, G.; di Girolamo, M.G.; Di Stefano, C.; Durante, A.; Franzese, G.; et al. Targeted metabolomics in the expanded newborn screening for inborn errors of metabolism. Mol. Biosyst. 2015, 11, 1525–1535. [Google Scholar] [CrossRef] [PubMed]
- Di Dato, F.; Spadarella, S.; Puoti, M.G.; Caprio, M.G.; Pagliardini, S.; Zuppaldi, C.; Vallone, G.; Fecarotta, S.; Esposito, G.; Iorio, R.; et al. Daily Fructose Traces Intake and Liver Injury in Children with Hereditary Fructose Intolerance. Nutrients 2019, 11, 2397. [Google Scholar] [CrossRef] [PubMed]
- Therrell, B.L.; Padilla, C.D.; Loeber, J.G.; Kneisser, I.; Saadallah, A.; Borrajo, G.J.C.; Adams, J. Current status of newborn screening worldwide: 2015. Semin. Perinatol 2015, 39, 171–187. [Google Scholar] [CrossRef]
- Huemer, M.; Kozich, V.; Rinaldo, P.; Baumgartner, M.R.; Merinero, B.; Pasquini, E.; Ribes, A.; Blom, H.J. Newborn screening for homocystinurias and methylation disorders: Systematic review and proposed guidelines. J. Inherit. Metab. Dis. 2015, 38, 1007–1019. [Google Scholar] [CrossRef] [PubMed]
- Villoria, J.G.; Pajares, S.; Lopez, R.M.; Marin, J.L.; Ribes, A. Neonatal Screening for Inherited Metabolic Diseases in 2016s. Semin. Pediatr. Neurol. 2016, 23, 257–272. [Google Scholar] [CrossRef] [PubMed]
- Ruoppolo, M.; Malvagia, S.; Boenzi, S.; Carducci, C.; Dionisi-Vici, C.; Teofoli, F.; Burlina, A.; Angeloni, A.; Aronica, T.; Bordugo, A.; et al. Expanded Newborn Screening in Italy Using Tandem Mass Spectrometry: Two Years of National Experience. Int. J. Neonatal Screen. 2022, 8, 47. [Google Scholar] [CrossRef]
- Huemer, M.; Mulder-Bleile, R.; Burda, P.; Froese, D.S.; Suormala, T.; Ben Zeev, B.; Chinnery, P.F.; Dionisi-Vici, C.; Dobbelaere, D.; Gokcay, G.; et al. Clinical pattern, mutations and in vitro residual activity in 33 patients with severe 5, 10 methylenetetrahydrofolate reductase (MTHFR) deficiency. J. Inherit. Metab. Dis. 2016, 39, 115–124. [Google Scholar] [CrossRef]
- Lossos, A.; Teltsh, O.; Milman, T.; Meiner, V.; Rozen, R.; Leclerc, D.; Schwahn, B.C.; Karp, N.; Rosenblatt, D.S.; Watkins, D.; et al. Severe Methylenetetrahydrofolate Reductase Deficiency Clinical Clues to a Potentially Treatable Cause of Adult-Onset Hereditary Spastic Paraplegia. JAMA Neurol. 2014, 71, 901–904. [Google Scholar] [CrossRef]
- Cappuccio, G.; Cozzolino, C.; Frisso, G.; Romanelli, R.; Parenti, G.; D’Amico, A.; Carotenuto, B.; Salvatore, F.; Del Giudice, E. Pearls & Oy-sters: Familial epileptic encephalopathy due to methylenetetrahydrofolate reductase deficiency. Neurology 2014, 83, E41–E44. [Google Scholar] [CrossRef]
- Gales, A.; Masingue, M.; Millecamps, S.; Giraudier, S.; Grosliere, L.; Adam, C.; Salim, C.; Navarro, V.; Nadjar, Y. Adolescence/adult onset MTHFR deficiency may manifest as isolated and treatable distinct neuro-psychiatric syndromes. Orphanet J. Rare Dis. 2018, 13, 29. [Google Scholar] [CrossRef] [PubMed]
- Hiraoka, M.; Kagawa, Y. Genetic polymorphisms and folate status. Congenit. Anom. 2017, 57, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Zaric, B.L.; Obradovic, M.; Bajic, V.; Haidar, M.A.; Jovanovic, M.; Isenovic, E.R. Homocysteine and Hyperhomocysteinaemia. Curr. Med. Chem. 2019, 26, 2948–2961. [Google Scholar] [CrossRef] [PubMed]
- Froese, D.S.; Kopec, J.; Rembeza, E.; Bezerra, G.A.; Oberholzer, A.E.; Suormala, T.; Lutz, S.; Chalk, R.; Borkowska, O.; Baumgartner, M.R.; et al. Structural basis for the regulation of human 5,10-methylenetetrahydrofolate reductase by phosphorylation and S-adenosylmethionine inhibition. Nat. Commun. 2018, 9, 2261. [Google Scholar] [CrossRef]
- Burda, P.; Schafer, A.; Suormala, T.; Rummel, T.; Burer, C.; Heuberger, D.; Frapolli, M.; Giunta, C.; Sokolova, J.; Vlaskova, H.; et al. Insights into Severe 5,10-Methylenetetrahydrofolate Reductase Deficiency: Molecular Genetic and Enzymatic Characterization of 76 Patients. Hum. Mutat. 2015, 36, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Burda, P.; Suormala, T.; Heuberger, D.; Schafer, A.; Fowler, B.; Froese, D.S.; Baumgartner, M.R. Functional characterization of missense mutations in severe methylenetetrahydrofolate reductase deficiency using a human expression system. J. Inherit. Metab. Dis. 2017, 40, 297–306. [Google Scholar] [CrossRef]
- Froese, D.S.; Huemer, M.; Suormala, T.; Burda, P.; Coelho, D.; Gueant, J.L.; Landolt, M.A.; Kozich, V.; Fowler, B.; Baumgartner, M.R. Mutation Update and Review of Severe Methylenetetrahydrofolate Reductase Deficiency. Hum. Mutat. 2016, 37, 427–438. [Google Scholar] [CrossRef]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Evans, K.; Hayden, M.; Heywood, S.; Hussain, M.; Phillips, A.D.; Cooper, D.N. The Human Gene Mutation Database: Towards a comprehensive repository of inherited mutation data for medical research, genetic diagnosis and next-generation sequencing studies. Hum. Genet. 2017, 136, 665–677. [Google Scholar] [CrossRef]
- Miranda-Vilela, A.L. Role of Polymorphisms in Factor V (FV Leiden), Prothrombin, Plasminogen Activator Inhibitor Type-1 (PAI-1), Methylenetetrahydrofolate Reductase (MTHFR) and Cystathionine β-Synthase (CBS) Genes as Risk Factors for Thrombophilias. Mini-Rev. Med. Chem. 2012, 12, 997–1006. [Google Scholar] [CrossRef]
- Levin, B.L.; Varga, E. MTHFR: Addressing Genetic Counseling Dilemmas Using Evidence-Based Literature. J. Genet. Couns. 2016, 25, 901–911. [Google Scholar] [CrossRef]
- Gramer, G.; Fang-Hoffmann, J.; Feyh, P.; Klinke, G.; Monostori, P.; Okun, J.G.; Hoffmann, G.F. High incidence of maternal vitamin B(12) deficiency detected by newborn screening: First results from a study for the evaluation of 26 additional target disorders for the German newborn screening panel. World J. Pediatr. 2018, 14, 470–481. [Google Scholar] [CrossRef] [PubMed]
- David, J.; Chrastina, P.; Peskova, K.; Kozich, V.; Friedecky, D.; Adam, T.; Hlidkova, E.; Vinohradska, H.; Novotna, D.; Hedelova, M.; et al. Epidemiology of rare diseases detected by newborn screening in the Czech Republic. Cent. Eur. J. Public Health 2019, 27, 153–159. [Google Scholar] [CrossRef] [PubMed]
- World Medical Association Declaration of Helsinki: Ethical principles for medical research involving human subjects. J. Am. Coll. Dent. 2014, 81, 14–18.
- Ruoppolo, M.; Scolamiero, E.; Caterino, M.; Mirisola, V.; Franconi, F.; Campesi, I. Female and male human babies have distinct blood metabolomic patterns. Mol. Biosyst. 2015, 11, 2483–2492. [Google Scholar] [CrossRef]
- Ruoppolo, M.; Caterino, M.; Albano, L.; Pecce, R.; Di Girolamo, M.G.; Crisci, D.; Costanzo, M.; Milella, L.; Franconi, F.; Campesi, I. Targeted metabolomic profiling in rat tissues reveals sex differences. Sci. Rep. 2018, 8, 4663. [Google Scholar] [CrossRef]
- Ruoppolo, M.; Campesi, I.; Scolamiero, E.; Pecce, R.; Caterino, M.; Cherchi, S.; Mercuro, G.; Tonolo, G.; Franconi, F. Serum metabolomic profiles suggest influence of sex and oral contraceptive use. Am. J. Transl. Res. 2014, 6, 614–624. [Google Scholar]
- Carsana, A.; Frisso, G.; Tremolaterra, M.R.; Ricci, E.; De Rasmo, D.; Salvatore, F. A larger spectrum of intragenic short tandem repeats improves linkage analysis and localization of intragenic recombination detection in the dystrophin gene—An analysis of 93 families from Southern Italy. J. Mol. Diagn. 2007, 9, 64–69. [Google Scholar] [CrossRef]
- Brancaccio, M.; Mennitti, C.; Cesaro, A.; Monda, E.; D’Argenio, V.; Casaburi, G.; Mazzaccara, C.; Ranieri, A.; Fimiani, F.; Barretta, F.; et al. Multidisciplinary In-Depth Investigation in a Young Athlete Suffering from Syncope Caused by Myocardial Bridge. Diagnostics 2021, 11, 2144. [Google Scholar] [CrossRef]
- Limongelli, G.; Nunziato, M.; Mazzaccara, C.; Intrieri, M.; D’Argenio, V.; Esposito, M.V.; Monda, E.; Maggio, F.D.; Frisso, G.; Salvatore, F. Genotype-Phenotype Correlation: A Triple DNA Mutational Event in a Boy Entering Sport Conveys an Additional Pathogenicity Risk. Genes 2020, 11, 524. [Google Scholar] [CrossRef]
- Fioretti, T.; Auricchio, L.; Piccirillo, A.; Vitiello, G.; Ambrosio, A.; Cattaneo, F.; Ammendola, R.; Esposito, G. Multi-Gene Next-Generation Sequencing for Molecular Diagnosis of Autosomal Recessive Congenital Ichthyosis: A Genotype-Phenotype Study of Four Italian Patients. Diagnostics 2020, 10, 995. [Google Scholar] [CrossRef]
- Kopanos, C.; Tsiolkas, V.; Kouris, A.; Chapple, C.E.; Aguilera, M.A.; Meyer, R.; Massouras, A. VarSome: The human genomic variant search engine. Bioinformatics 2019, 35, 1978–1980. [Google Scholar] [CrossRef] [PubMed]
- Girolami, F.; Frisso, G.; Benelli, M.; Crotti, L.; Iascone, M.; Mango, R.; Mazzaccara, C.; Pilichou, K.; Arbustini, E.; Tomberli, B.; et al. Contemporary genetic testing in inherited cardiac disease: Tools, ethical issues, and clinical applications. J. Cardiovasc. Med. 2018, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Frisso, G.; Detta, N.; Coppola, P.; Mazzaccara, C.; Pricolo, M.R.; D’Onofrio, A.; Limongelli, G.; Calabro, R.; Salvatore, F. Functional Studies and In Silico Analyses to Evaluate Non-Coding Variants in Inherited Cardiomyopathies. Int. J. Mol. Sci. 2016, 17, 1883. [Google Scholar] [CrossRef]
- Richards, S.; Aziz, N.; Bale, S.; Bick, D.; Das, S.; Gastier-Foster, J.; Grody, W.W.; Hegde, M.; Lyon, E.; Spector, E.; et al. Standards and guidelines for the interpretation of sequence variants: A joint consensus recommendation of the American College of Medical Genetics and Genomics and the Association for Molecular Pathology. Genet. Med. 2015, 17, 405–424. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y. I-TASSER server for protein 3D structure prediction. BMC Bioinform. 2008, 9, 40. [Google Scholar] [CrossRef] [PubMed]
- Pricolo, M.R.; Herrero-Galan, E.; Mazzaccara, C.; Losi, M.A.; Alegre-Cebollada, J.; Frisso, G. Protein Thermodynamic Destabilization in the Assessment of Pathogenicity of a Variant of Uncertain Significance in Cardiac Myosin Binding Protein C. J. Cardiovasc. Transl. Res. 2020, 13, 867–877. [Google Scholar] [CrossRef] [PubMed]
- Stenson, P.D.; Mort, M.; Ball, E.V.; Shaw, K.; Phillips, A.; Cooper, D.N. The Human Gene Mutation Database: Building a comprehensive mutation repository for clinical and molecular genetics, diagnostic testing and personalized genomic medicine. Hum. Genet. 2014, 133, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Landrum, M.J.; Lee, J.M.; Riley, G.R.; Jang, W.; Rubinstein, W.S.; Church, D.M.; Maglott, D.R. ClinVar: Public archive of relationships among sequence variation and human phenotype. Nucleic Acids Res. 2014, 42, D980–D985. [Google Scholar] [CrossRef]
- Sherry, S.T.; Ward, M.H.; Kholodov, M.; Baker, J.; Phan, L.; Smigielski, E.M.; Sirotkin, K. dbSNP: The NCBI database of genetic variation. Nucleic Acids Res. 2001, 29, 308–311. [Google Scholar] [CrossRef]
- Abecasis, G.R.; Auton, A.; Brooks, L.D.; DePristo, M.A.; Durbin, R.M.; Handsaker, R.E.; Kang, H.M.; Marth, G.T.; McVean, G.A. An integrated map of genetic variation from 1092 human genomes. Nature 2012, 491, 56–65. [Google Scholar] [CrossRef]
- Fu, W.Q.; O’Connor, T.D.; Jun, G.; Kang, H.M.; Abecasis, G.; Leal, S.M.; Gabriel, S.; Rieder, M.J.; Altshuler, D.; Shendure, J.; et al. Analysis of 6,515 exomes reveals the recent origin of most human protein-coding variants. Nature 2013, 493, 216, Correction in Nature 2013, 495, 270. [Google Scholar] [CrossRef] [PubMed]
- Diekman, E.F.; de Koning, T.J.; Verhoeven-Duif, N.M.; Rovers, M.M.; van Hasselt, P.M. Survival and psychomotor development with early betaine treatment in patients with severe methylenetetrahydrofolate reductase deficiency. JAMA Neurol. 2014, 71, 188–194. [Google Scholar] [CrossRef] [PubMed]
- Huemer, M.; Diodato, D.; Schwahn, B.; Schiff, M.; Bandeira, A.; Benoist, J.F.; Burlina, A.; Cerone, R.; Couce, M.L.; Garcia-Cazorla, A.; et al. Guidelines for diagnosis and management of the cobalamin-related remethylation disorders cblC, cblD, cblE, cblF, cblG, cblJ and MTHFR deficiency. J. Inherit. Metab. Dis. 2017, 40, 21–48. [Google Scholar] [CrossRef]
- Huemer, M.; Baumgartner, M.R. The clinical presentation of cobalamin-related disorders: From acquired deficiencies to inborn errors of absorption and intracellular pathways. J. Inherit. Metab. Dis. 2019, 42, 686–705. [Google Scholar] [CrossRef]
- Maruotti, G.M.; Frisso, G.; Calcagno, G.; Fortunato, G.; Castaldo, G.; Martinelli, P.; Sacchetti, L.; Salvatore, F. Prenatal diagnosis of inherited diseases: 20 years’ experience of an Italian Regional Reference Centre. Clin. Chem. Lab. Med. 2013, 51, 2211–2217. [Google Scholar] [CrossRef] [PubMed]
- Esposito, G.; Ruggiero, R.; Savarese, M.; Savarese, G.; Tremolaterra, M.R.; Salvatore, F.; Carsana, A. Prenatal molecular diagnosis of inherited neuromuscular diseases: Duchenne/Becker muscular dystrophy, myotonic dystrophy type 1 and spinal muscular atrophy. Clin. Chem. Lab. Med. 2013, 51, 2239–2245. [Google Scholar] [CrossRef] [PubMed]
- Lombardo, B.; D’Argenio, V.; Monda, E.; Vitale, A.; Caiazza, M.; Sacchetti, L.; Pastore, L.; Limongelli, G.; Frisso, G.; Mazzaccara, C. Genetic analysis resolves differential diagnosis of a familial syndromic dilated cardiomyopathy: A new case of Alstrom syndrome. Mol. Genet. Genom. Med. 2020, 8, e1260. [Google Scholar] [CrossRef]
- Sumner, J.; Jencks, D.A.; Khani, S.; Matthews, R.G. Photoaffinity labeling of methylenetetrahydrofolate reductase with 8-azido-S-adenosylmethionine. J. Biol. Chem. 1986, 261, 7697–7700. [Google Scholar] [CrossRef]
- Mazzaccara, C.; Limongelli, G.; Petretta, M.; Vastarella, R.; Pacileo, G.; Bonaduce, D.; Salvatore, F.; Frisso, G. A common polymorphism in the SCN5A gene is associated with dilated cardiomyopathy. J. Cardiovasc. Med. 2018, 19, 344–350. [Google Scholar] [CrossRef]
- Yamada, K.; Chen, Z.; Rozen, R.; Matthews, R.G. Effects of common polymorphisms on the properties of recombinant human methylenetetrahydrofolate reductase. Proc. Natl. Acad. Sci. USA 2001, 98, 14853–14858. [Google Scholar] [CrossRef]
- Guenther, B.D.; Sheppard, C.A.; Tran, P.; Rozen, R.; Matthews, R.G.; Ludwig, M.L. The structure and properties of methylenetetrahydrofolate reductase from Escherichia coli suggest how folate ameliorates human hyperhomocysteinemia. Nat. Struct. Biol. 1999, 6, 359–365. [Google Scholar] [CrossRef] [PubMed]
- Knowles, L.; Morris, A.A.; Walter, J.H. Treatment with Mefolinate (5-Methyltetrahydrofolate), but Not Folic Acid or Folinic Acid, Leads to Measurable 5-Methyltetrahydrofolate in Cerebrospinal Fluid in Methylenetetrahydrofolate Reductase Deficiency. JIMD Rep. 2016, 29, 103–107. [Google Scholar] [CrossRef] [PubMed]
- Yverneau, M.; Leroux, S.; Imbard, A.; Gleich, F.; Arion, A.; Moreau, C.; Nassogne, M.C.; Szymanowski, M.; Tardieu, M.; Touati, G.; et al. Influence of early identification and therapy on long-term outcomes in early-onset MTHFR deficiency. J. Inherit. Metab. Dis. 2022, 45, 848–861. [Google Scholar] [CrossRef] [PubMed]

 Carrier male;
Carrier male;  Carrier female;
Carrier female;  Affected male;
Affected male;  Affected female;
Affected female;  Proband.
Proband.
 Carrier male;
Carrier male;  Carrier female;
Carrier female;  Affected male;
Affected male;  Affected female;
Affected female;  Proband.
Proband.

| Biochemical Markers | Molecular Analysis | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Patient (Date of Birth) | DBS (I Spot) | DBS (II Spot) | Serum Tests | HGVS § cDNA | HGVS § Protein | Variant Classification | |||||
| Met * | Hcy # | Met * | Hcy # | Met ** | Hcy # | ClinVar | HGMD §§ | ACMG §§§ | |||
| Patient 1 (20 February 2018) | 4 | 53.7 | 6 | 42.3 | 10 | 106.7 | c.176G>C c.665C>T c.1286A>C c.1769T>G | p.Trp59Ser p.Ala222Val p.Glu429Ala p.Leu590Arg | P LB (DR) CI NR | CM155512 CM950819 CM981315 NR | LP B B LP |
| Patient 2 (7 February 2020) | 5.5 | 4.4 | 9 | 5.11 | 31 | 14.7 | c.665C>T c.1286A>C | p.Ala222Val p.Glu429Ala | LB (DR) CI | CM950819 CM981315 | B B |
| Patient 3 (20 October 2020) | 5.07 | 11 | 8.6 | 16.3 | 29 | 53.1 | c.1320G>A c.1683G>A | p.Ser440 = (splicing) p.Trp561Ter | P/LP P | CM155527 CM155520 | LP P |
| Patient 4 (24 July 2022) | 5.6 | 9.2 | 8 | 7 | 23 | 54.5 | c.665C>T c.1160G>A | p.Ala222Val p.Gly387Asp | LB (DR) NR | CM950819 CM000527 | B VUS |
| Patient born before the introduction of NBS | |||||||||||
| Patient 5 (1 August 2009) | NP | NP | NP | NP | NP | 423 pre therapy 27.9 during therapy | c.665C>T c.973C>T c.1970G>C | p.Ala222Val p.Arg325Cys p.Ter657Serext*50 | LB (DR) VUS LB | CM950819 CM950822 CM035841 | B LP LP |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barretta, F.; Uomo, F.; Fecarotta, S.; Albano, L.; Crisci, D.; Verde, A.; Fisco, M.G.; Gallo, G.; Dottore Stagna, D.; Pricolo, M.R.; et al. Contribution of Genetic Test to Early Diagnosis of Methylenetetrahydrofolate Reductase (MTHFR) Deficiency: The Experience of a Reference Center in Southern Italy. Genes 2023, 14, 980. https://doi.org/10.3390/genes14050980
Barretta F, Uomo F, Fecarotta S, Albano L, Crisci D, Verde A, Fisco MG, Gallo G, Dottore Stagna D, Pricolo MR, et al. Contribution of Genetic Test to Early Diagnosis of Methylenetetrahydrofolate Reductase (MTHFR) Deficiency: The Experience of a Reference Center in Southern Italy. Genes. 2023; 14(5):980. https://doi.org/10.3390/genes14050980
Chicago/Turabian StyleBarretta, Ferdinando, Fabiana Uomo, Simona Fecarotta, Lucia Albano, Daniela Crisci, Alessandra Verde, Maria Grazia Fisco, Giovanna Gallo, Daniela Dottore Stagna, Maria Rosaria Pricolo, and et al. 2023. "Contribution of Genetic Test to Early Diagnosis of Methylenetetrahydrofolate Reductase (MTHFR) Deficiency: The Experience of a Reference Center in Southern Italy" Genes 14, no. 5: 980. https://doi.org/10.3390/genes14050980





