Epigenetic Control of Cell Potency and Fate Determination during Mammalian Gastrulation
Abstract
1. Introduction
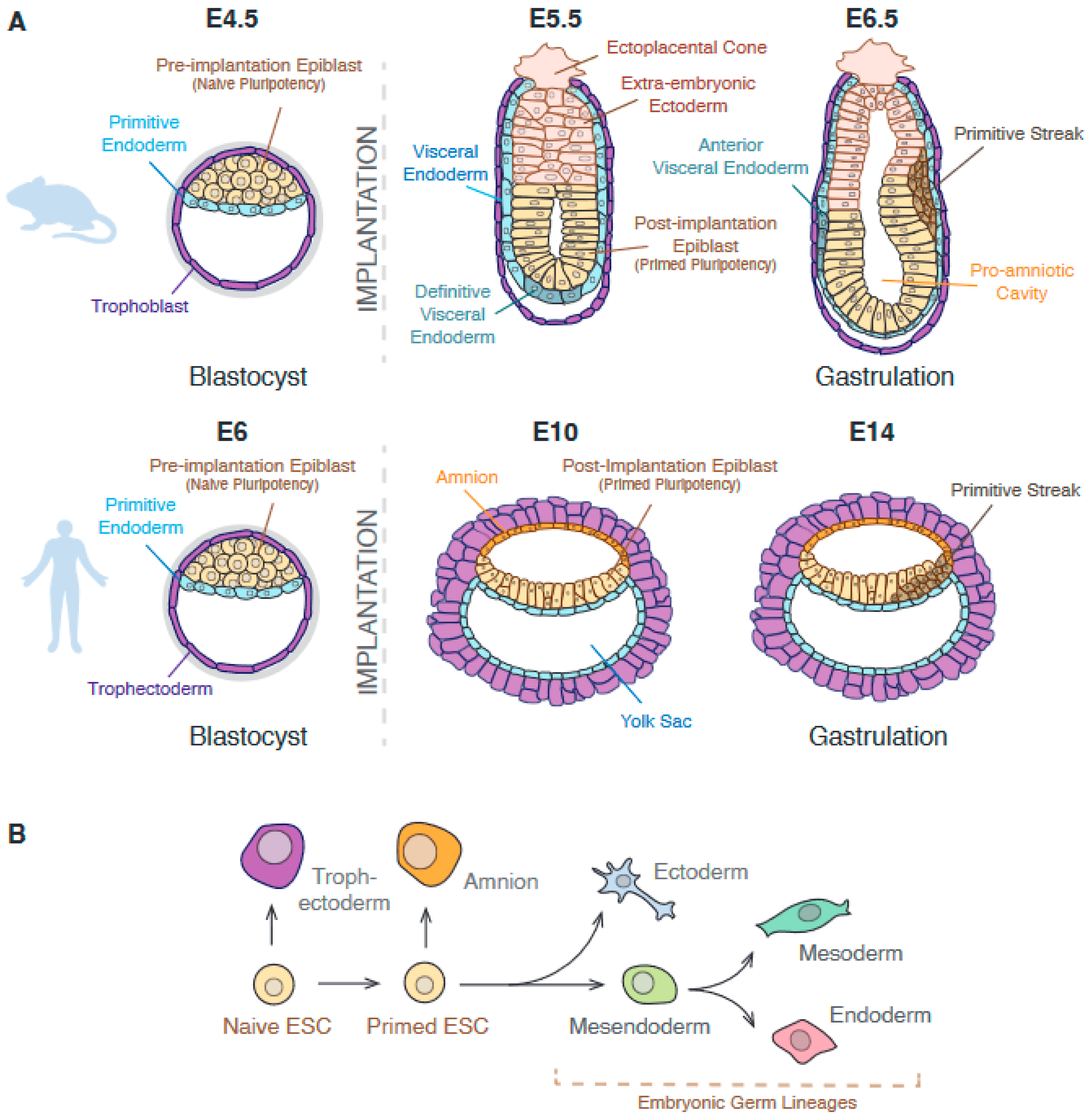
1.1. Embryonic Stem Cells Are a Useful Model to Study Epigenetic Regulation during Gastrulation
1.2. Both Active and Silenced: A State of Gene Regulatory Regions Which Is Critical for Pluripotency
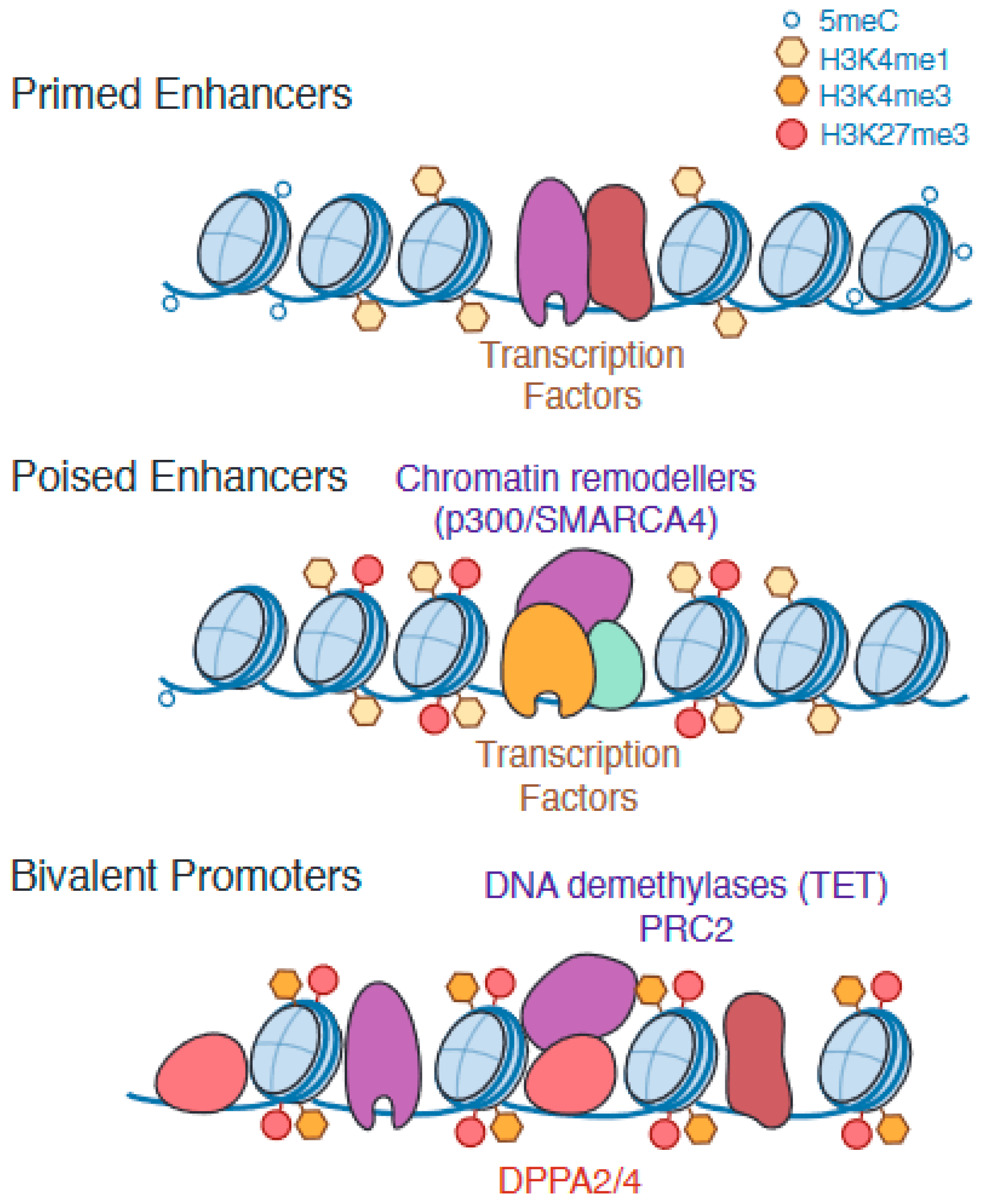
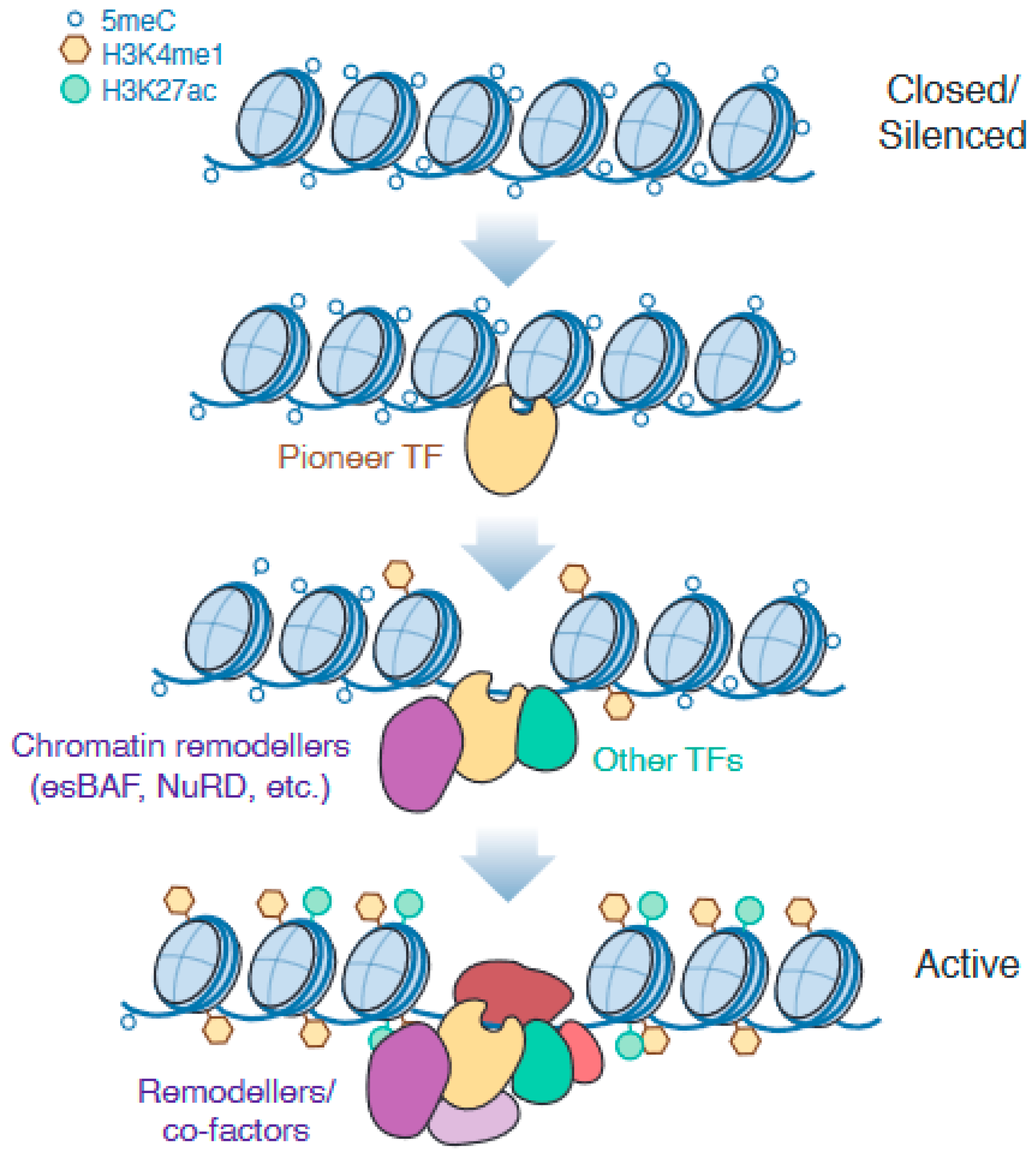
1.3. Epigenetic Remodelling of Enhancers Is Required for Successful Pluripotency Exit and Robust Cell Fate Specification
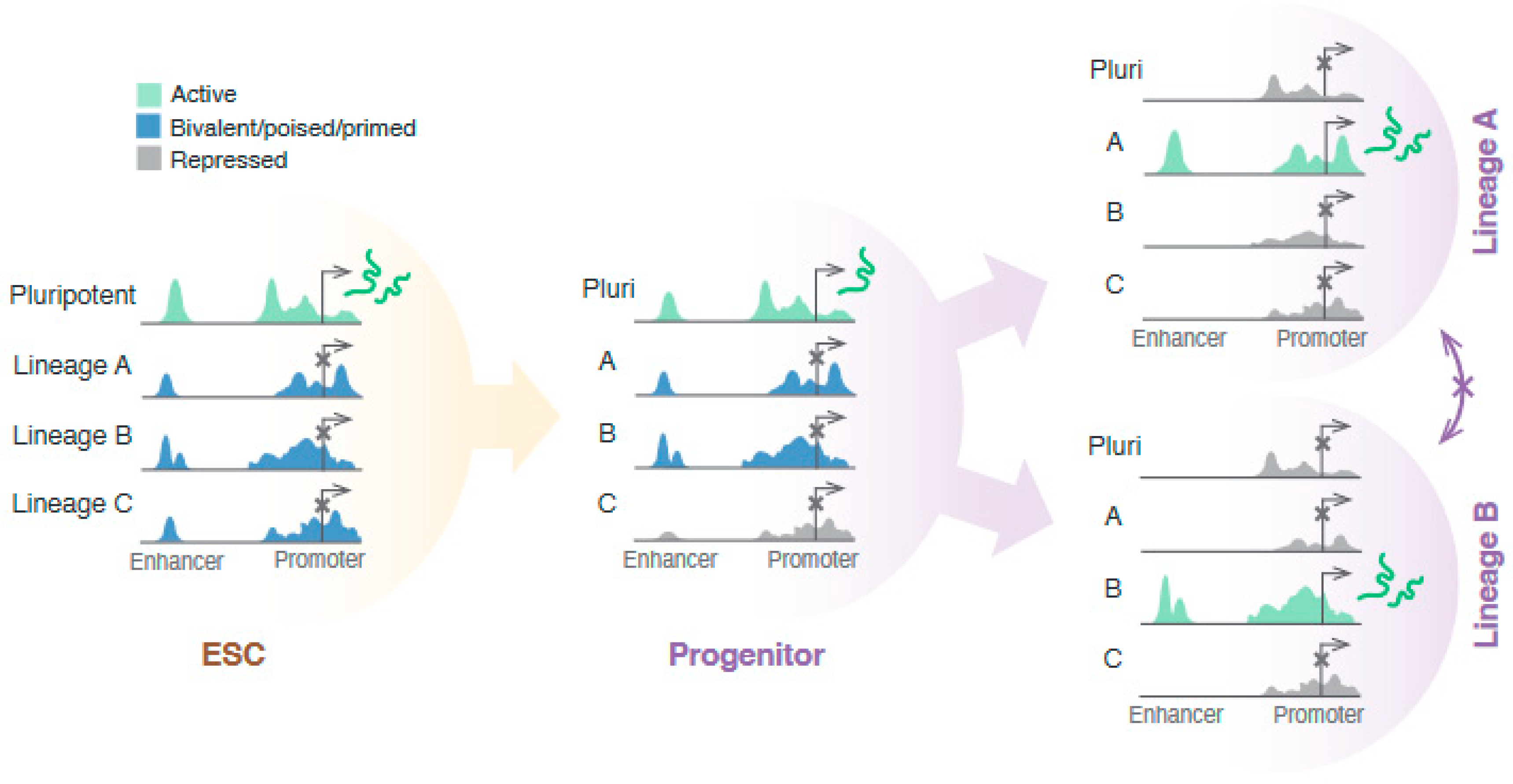
1.4. Redistribution of Repressive Histone Marks Is Important for Fidelity of Lineage Choice
1.5. DNA Methylation Plays a Key but Nuanced Role in Silencing of Gene Regulatory Regions
1.6. How Can Epigenetics Influence Differentiation Trajectories of Multipotent Cells and Vice Versa?
2. Conclusions and Discussion
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Macrae, T.A.; Fothergill-Robinson, J.; Ramalho-Santos, M. Regulation, functions and transmission of bivalent chromatin during mammalian development. Nat. Rev. Mol. Cell Biol. 2023, 24, 6–26. [Google Scholar] [CrossRef] [PubMed]
- Balsalobre, A.; Drouin, J. Pioneer factors as master regulators of the epigenome and cell fate. Nat. Rev. Mol. Cell Biol. 2022, 23, 449–464. [Google Scholar] [CrossRef] [PubMed]
- Smith, Z.D.; Meissner, A. DNA methylation: Roles in mammalian development. Nat. Rev. Genet. 2013, 14, 204–220. [Google Scholar] [CrossRef] [PubMed]
- Gökbuget, D.; Blelloch, R. Epigenetic control of transcriptional regulation in pluripotency and early differentiation. Development 2019, 146, dev164772. [Google Scholar] [CrossRef] [PubMed]
- Atlasi, Y.; Stunnenberg, H.G. The interplay of epigenetic marks during stem cell differentiation and development. Nat. Rev. Genet. 2017, 18, 643–658. [Google Scholar] [CrossRef]
- Argelaguet, R.; Clark, S.J.; Mohammed, H.; Stapel, L.C.; Krueger, C.; Kapourani, C.A.; Imaz-Rosshandler, I.; Lohoff, T.; Xiang, Y.; Hanna, C.W.; et al. Multi-omics profiling of mouse gastrulation at single-cell resolution. Nature 2019, 576, 487–491. [Google Scholar] [CrossRef]
- Pijuan-Sala, B.; Wilson, N.K.; Xia, J.; Hou, X.; Hannah, R.L.; Kinston, S.; Calero-Nieto, F.J.; Poirion, O.; Preissl, S.; Liu, F.; et al. Single-cell chromatin accessibility maps reveal regulatory programs driving early mouse organogenesis. Nat. Cell Biol. 2020, 22, 487–497. [Google Scholar] [CrossRef]
- Barakat, T.S.; Halbritter, F.; Zhang, M.; Rendeiro, A.F.; Perenthaler, E.; Bock, C.; Chambers, I. Functional Dissection of the Enhancer Repertoire in Human Embryonic Stem Cells. Cell Stem Cell 2018, 23, 276–288.e8. [Google Scholar] [CrossRef]
- Zhang, Y.; Xiang, Y.; Yin, Q.; Du, Z.; Peng, X.; Wang, Q.; Fidalgo, M.; Xia, W.; Li, Y.; Zhao, Z.A.; et al. Dynamic epigenomic landscapes during early lineage specification in mouse embryos. Nat. Genet. 2018, 50, 96–105. [Google Scholar] [CrossRef]
- Gifford, C.A.; Ziller, M.J.; Gu, H.; Trapnell, C.; Donaghey, J.; Tsankov, A.; Shalek, A.K.; Kelley, D.R.; Shishkin, A.A.; Issner, R.; et al. Transcriptional and epigenetic dynamics during specification of human embryonic stem cells. Cell 2013, 153, 1149–1163. [Google Scholar] [CrossRef]
- Xie, W.; Schultz, M.D.; Lister, R.; Hou, Z.; Rajagopal, N.; Ray, P.; Whitaker, J.W.; Tian, S.; Hawkins, R.D.; Leung, D.; et al. Epigenomic Analysis of Multilineage Differentiation of Human Embryonic Stem Cells. Cell 2013, 153, 1134–1148. [Google Scholar] [CrossRef] [PubMed]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic Stem Cell Lines Derived from Human Blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef] [PubMed]
- Bryja, V.; Bonilla, S.; Arenas, E. Derivation of mouse embryonic stem cells. Nat. Protoc. 2006, 1, 2082–2087. [Google Scholar] [CrossRef] [PubMed]
- Kumar, B.; Navarro, C.; Winblad, N.; Schell, J.P.; Zhao, C.; Weltner, J.; Baqué-Vidal, L.; Mantero, A.S.; Petropoulos, S.; Lanner, F.; et al. Polycomb repressive complex 2 shields naïve human pluripotent cells from trophectoderm differentiation. Nat. Cell Biol. 2022, 24, 845–857. [Google Scholar] [CrossRef]
- Zijlmans, D.W.; Talon, I.; Verhelst, S.; Bendall, A.; Van Nerum, K.; Javali, A.; Malcolm, A.A.; van Knippenberg, S.S.F.A.; Biggins, L.; To, S.K.; et al. Integrated multi-omics reveal polycomb repressive complex 2 restricts human trophoblast induction. Nat. Cell Biol. 2022, 24, 858–871. [Google Scholar] [CrossRef]
- Tesar, P.J.; Chenoweth, J.G.; Brook, F.A.; Davies, T.J.; Evans, E.P.; Mack, D.L.; Gardner, R.L.; McKay, R.D.G. New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 2007, 448, 196–199. [Google Scholar] [CrossRef]
- Guo, G.; Stirparo, G.G.; Strawbridge, S.E.; Spindlow, D.; Yang, J.; Clarke, J.; Dattani, A.; Yanagida, A.; Li, M.A.; Myers, S.; et al. Human naive epiblast cells possess unrestricted lineage potential. Cell Stem Cell 2021, 28, 1040–1056.e6. [Google Scholar] [CrossRef]
- Bernstein, B.E.; Mikkelsen, T.S.; Xie, X.; Kamal, M.; Huebert, D.J.; Cuff, J.; Fry, B.; Meissner, A.; Wernig, M.; Plath, K.; et al. A Bivalent Chromatin Structure Marks Key Developmental Genes in Embryonic Stem Cells. Cell 2006, 125, 315–326. [Google Scholar] [CrossRef]
- Azuara, V.; Perry, P.; Sauer, S.; Spivakov, M.; Jørgensen, H.F.; John, R.M.; Gouti, M.; Casanova, M.; Warnes, G.; Merkenschlager, M.; et al. Chromatin signatures of pluripotent cell lines. Nat. Cell Biol. 2006, 8, 532–538. [Google Scholar] [CrossRef]
- Glaser, S.; Schaft, J.; Lubitz, S.; Vintersten, K.; van der Hoeven, F.; Tufteland, K.R.; Aasland, R.; Anastassiadis, K.; Ang, S.; Stewart, A.F. Multiple epigenetic maintenance factors implicated by the loss of Mll2 in mouse development. Development 2006, 133, 1423–1432. [Google Scholar] [CrossRef]
- Mas, G.; Blanco, E.; Ballaré, C.; Sansó, M.; Spill, Y.G.; Hu, D.; Aoi, Y.; Le Dily, F.; Shilatifard, A.; Marti-Renom, M.A.; et al. Promoter bivalency favors an open chromatin architecture in embryonic stem cells. Nat. Genet. 2018, 50, 1452–1462. [Google Scholar] [CrossRef] [PubMed]
- Lubitz, S.; Glaser, S.; Schaft, J.; Stewart, A.F.; Anastassiadis, K. Increased apoptosis and skewed differentiation in mouse embryonic stem cells lacking the histone methyltransferase Mll2. Mol. Biol. Cell 2007, 18, 2356–2366. [Google Scholar] [CrossRef] [PubMed]
- Eckersley-Maslin, M.A.; Parry, A.; Blotenburg, M.; Krueger, C.; Ito, Y.; Franklin, V.N.R.; Narita, M.; D’Santos, C.S.; Reik, W. Epigenetic priming by Dppa2 and 4 in pluripotency facilitates multi-lineage commitment. Nat. Struct. Mol. Biol. 2020, 27, 696–705. [Google Scholar] [CrossRef] [PubMed]
- Eckersley-Maslin, M.A. Keeping your options open: Insights from Dppa2/4 into how epigenetic priming factors promote cell plasticity. Biochem. Soc. Trans. 2020, 48, 2891–2902. [Google Scholar] [CrossRef]
- Creyghton, M.P.; Cheng, A.W.; Welstead, G.G.; Kooistra, T.; Carey, B.W.; Steine, E.J.; Hanna, J.; Lodato, M.A.; Frampton, G.M.; Sharp, P.A.; et al. Histone H3K27ac separates active from poised enhancers and predicts developmental state. Proc. Natl. Acad. Sci. USA 2010, 107, 21931–21936. [Google Scholar] [CrossRef]
- Rada-Iglesias, A.; Bajpai, R.; Swigut, T.; Brugmann, S.A.; Flynn, R.A.; Wysocka, J. A unique chromatin signature uncovers early developmental enhancers in humans. Nature 2011, 470, 279–285. [Google Scholar] [CrossRef]
- Cruz-Molina, S.; Respuela, P.; Tebartz, C.; Kolovos, P.; Nikolic, M.; Fueyo, R.; van Ijcken, W.F.J.; Grosveld, F.; Frommolt, P.; Bazzi, H.; et al. PRC2 Facilitates the Regulatory Topology Required for Poised Enhancer Function during Pluripotent Stem Cell Differentiation. Cell Stem Cell 2017, 20, 689–705.e9. [Google Scholar] [CrossRef]
- Crispatzu, G.; Rehimi, R.; Pachano, T.; Bleckwehl, T.; Cruz-Molina, S.; Xiao, C.; Mahabir, E.; Bazzi, H.; Rada-Iglesias, A. The chromatin, topological and regulatory properties of pluripotency-associated poised enhancers are conserved in vivo. Nat. Commun. 2021, 12, 4344. [Google Scholar] [CrossRef]
- Kim, H.S.; Tan, Y.; Ma, W.; Merkurjev, D.; Destici, E.; Ma, Q.; Suter, T.; Ohgi, K.; Friedman, M.; Skowronska-Krawczyk, D.; et al. Pluripotency factors functionally premark cell-type-restricted enhancers in ES cells. Nature 2018, 556, 510–514. [Google Scholar] [CrossRef]
- Stock, J.K.; Giadrossi, S.; Casanova, M.; Brookes, E.; Vidal, M.; Koseki, H.; Brockdorff, N.; Fisher, A.G.; Pombo, A. Ring1-mediated ubiquitination of H2A restrains poised RNA polymerase II at bivalent genes in mouse ES cells. Nat. Cell Biol. 2007, 9, 1428–1435. [Google Scholar] [CrossRef]
- Adelman, K.; Lis, J.T. Promoter-proximal pausing of RNA polymerase II: Emerging roles in metazoans. Nat. Rev. Genet. 2012, 13, 720–731. [Google Scholar] [CrossRef] [PubMed]
- Kumar, D.; Cinghu, S.; Oldfield, A.J.; Yang, P.; Jothi, R. Decoding the function of bivalent chromatin in development and cancer. Genome Res. 2021, 31, 2170–2184. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wang, L.; Li, J.; Ding, Z.; Xiao, J.; Yin, X.; He, S.; Shi, P.; Dong, L.; Li, G.; et al. Structural insight into autoinhibition and histone H3-induced activation of DNMT3A. Nature 2015, 517, 640–644. [Google Scholar] [CrossRef] [PubMed]
- Ooi, S.K.T.; Qiu, C.; Bernstein, E.; Li, K.; Jia, D.; Yang, Z.; Erdjument-Bromage, H.; Tempst, P.; Lin, S.; Allis, C.D.; et al. DNMT3L connects unmethylated lysine 4 of histone H3 to de novo methylation of DNA. Nature 2007, 448, 714–717. [Google Scholar] [CrossRef] [PubMed]
- Douillet, D.; Sze, C.C.; Ryan, C.; Piunti, A.; Shah, A.P.; Ugarenko, M.; Marshall, S.A.; Rendleman, E.J.; Zha, D.; Helmin, K.A.; et al. Uncoupling histone H3K4 trimethylation from developmental gene expression via an equilibrium of COMPASS, Polycomb and DNA methylation. Nat. Genet. 2020, 52, 615–625. [Google Scholar] [CrossRef]
- Yoon, S.J.; Foley, J.W.; Baker, J.C. HEB associates with PRC2 and SMAD2/3 to regulate developmental fates. Nat. Commun. 2015, 6, 6546. [Google Scholar] [CrossRef]
- Metzis, V.; Steinhauser, S.; Pakanavicius, E.; Gouti, M.; Stamataki, D.; Ivanovitch, K.; Watson, T.; Rayon, T.; Mousavy Gharavy, S.N.; Lovell-Badge, R.; et al. Nervous System Regionalization Entails Axial Allocation before Neural Differentiation. Cell 2018, 175, 1105–1118.e17. [Google Scholar] [CrossRef]
- Tang, W.W.C.; Castillo-Venzor, A.; Gruhn, W.H.; Kobayashi, T.; Penfold, C.A.; Morgan, M.D.; Sun, D.; Irie, N.; Surani, M.A. Sequential enhancer state remodelling defines human germline competence and specification. Nat. Cell Biol. 2022, 24, 448–460. [Google Scholar] [CrossRef]
- Bleckwehl, T.; Crispatzu, G.; Schaaf, K.; Respuela, P.; Bartusel, M.; Benson, L.; Clark, S.J.; Dorighi, K.M.; Barral, A.; Laugsch, M.; et al. Enhancer-associated H3K4 methylation safeguards in vitro germline competence. Nat. Commun. 2021, 12, 1–19. [Google Scholar] [CrossRef]
- Wang, A.; Yue, F.; Li, Y.; Xie, R.; Harper, T.; Patel, N.A.; Muth, K.; Palmer, J.; Qiu, Y.; Wang, J.; et al. Epigenetic priming of enhancers predicts developmental competence of hESC-derived endodermal lineage intermediates. Cell Stem Cell 2015, 16, 386–399. [Google Scholar] [CrossRef]
- Iwafuchi-Doi, M.; Donahue, G.; Kakumanu, A.; Watts, J.A.; Mahony, S.; Pugh, B.F.; Lee, D.; Kaestner, K.H.; Zaret, K.S. The Pioneer Transcription Factor FoxA Maintains an Accessible Nucleosome Configuration at Enhancers for Tissue-Specific Gene Activation. Mol. Cell 2016, 62, 79–91. [Google Scholar] [CrossRef] [PubMed]
- Tsankov, A.M.; Gu, H.; Akopian, V.; Ziller, M.J.; Donaghey, J.; Amit, I.; Gnirke, A.; Meissner, A. Transcription factor binding dynamics during human ES cell differentiation. Nature 2015, 518, 344–349. [Google Scholar] [CrossRef] [PubMed]
- Donaghey, J.; Thakurela, S.; Charlton, J.; Chen, J.S.; Smith, Z.D.; Gu, H.; Pop, R.; Clement, K.; Stamenova, E.K.; Karnik, R.; et al. Genetic determinants and epigenetic effects of pioneer-factor occupancy. Nat. Genet. 2018, 50, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Blassberg, R.; Patel, H.; Watson, T.; Gouti, M.; Metzis, V.; Delás, M.J.; Briscoe, J. Sox2 levels regulate the chromatin occupancy of WNT mediators in epiblast progenitors responsible for vertebrate body formation. Nat. Cell Biol. 2022, 24, 633–644. [Google Scholar] [CrossRef]
- Bunina, D.; Abazova, N.; Diaz, N.; Noh, K.M.; Krijgsveld, J.; Zaugg, J.B. Genomic Rewiring of SOX2 Chromatin Interaction Network during Differentiation of ESCs to Postmitotic Neurons. Cell Syst. 2020, 10, 480–494.e8. [Google Scholar] [CrossRef]
- Tosic, J.; Kim, G.J.; Pavlovic, M.; Schröder, C.M.; Mersiowsky, S.L.; Barg, M.; Hofherr, A.; Probst, S.; Köttgen, M.; Hein, L.; et al. Eomes and Brachyury control pluripotency exit and germ-layer segregation by changing the chromatin state. Nat. Cell Biol. 2019, 21, 1518–1531. [Google Scholar] [CrossRef]
- Ho, L.; Ronan, J.L.; Wu, J.; Staahl, B.T.; Chen, L.; Kuo, A.; Lessard, J.; Nesvizhskii, A.I.; Ranish, J.; Crabtree, G.R. An embryonic stem cell chromatin remodeling complex, esBAF, is essential for embryonic stem cell self-renewal and pluripotency. Proc. Natl. Acad. Sci. USA 2009, 106, 5181–5186. [Google Scholar] [CrossRef]
- Iurlaro, M.; Stadler, M.B.; Masoni, F.; Jagani, Z.; Galli, G.G.; Schübeler, D. Mammalian SWI / SNF continuously restores local accessibility to chromatin. Nat. Genet. 2021, 53, 279–287. [Google Scholar] [CrossRef]
- Schick, S.; Grosche, S.; Kohl, K.E.; Drpic, D.; Jaeger, M.G.; Marella, N.C.; Imrichova, H.; Lin, J.M.G.; Hofstätter, G.; Schuster, M.; et al. Acute BAF perturbation causes immediate changes in chromatin accessibility. Nat. Genet. 2021, 53, 269–278. [Google Scholar] [CrossRef]
- King, H.W.; Klose, R.J. The pioneer factor OCT4 requires the chromatin remodeller BRG1 to support gene regulatory element function in mouse embryonic stem cells. Elife 2017, 6, 1–24. [Google Scholar] [CrossRef]
- Takaku, M.; Grimm, S.A.; Shimbo, T.; Perera, L.; Menafra, R.; Stunnenberg, H.G.; Archer, T.K.; Machida, S.; Kurumizaka, H.; Wade, P.A. GATA3-dependent cellular reprogramming requires activation-domain dependent recruitment of a chromatin remodeler. Genome Biol. 2016, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Whyte, W.A.; Bilodeau, S.; Orlando, D.A.; Hoke, H.A.; Frampton, G.M.; Foster, C.T.; Cowley, S.M.; Young, R.A. Enhancer decommissioning by LSD1 during embryonic stem cell differentiation. Nature 2012, 482, 221–225. [Google Scholar] [CrossRef] [PubMed]
- Bornelöv, S.; Reynolds, N.; Xenophontos, M.; Gharbi, S.; Johnstone, E.; Floyd, R.; Ralser, M.; Signolet, J.; Loos, R.; Dietmann, S.; et al. The Nucleosome Remodeling and Deacetylation Complex Modulates Chromatin Structure at Sites of Active Transcription to Fine-Tune Gene Expression. Mol. Cell 2018, 71, 56–72.e4. [Google Scholar] [CrossRef] [PubMed]
- O’Carroll, D.; Erhardt, S.; Pagani, M.; Barton, S.C.; Surani, M.A.; Jenuwein, T. The Polycomb -Group Gene Ezh2 Is Required for Early Mouse Development. Mol. Cell. Biol. 2001, 21, 4330–4336. [Google Scholar] [CrossRef]
- Pasini, D.; Bracken, A.P.; Jensen, M.R.; Denchi, E.L.; Helin, K. Suz12 is essential for mouse development and for EZH2 histone methyltransferase activity. EMBO J. 2004, 23, 4061–4071. [Google Scholar] [CrossRef]
- Faust, C.; Lawson, K.A.; Schork, N.J.; Thiel, B.; Magnuson, T. The Polycomb -group gene eed is required for normal morphogenetic movements during gastrulation in the mouse embryo. Development 1998, 125, 4495–4506. [Google Scholar] [CrossRef]
- Collinson, A.; Collier, A.J.; Morgan, N.P.; Sienerth, A.R.; Chandra, T.; Andrews, S.; Rugg-Gunn, P.J. Deletion of the Polycomb-Group Protein EZH2 Leads to Compromised Self-Renewal and Differentiation Defects in Human Embryonic Stem Cells. Cell Rep. 2016, 17, 2700–2714. [Google Scholar] [CrossRef]
- Shan, Y.; Liang, Z.; Xing, Q.; Zhang, T.; Wang, B.; Tian, S.; Huang, W.; Zhang, Y.; Yao, J.; Zhu, Y.; et al. PRC2 specifies ectoderm lineages and maintains pluripotency in primed but not naïve ESCs. Nat. Commun. 2017, 8, 672. [Google Scholar] [CrossRef]
- Riising, E.M.; Comet, I.; Leblanc, B.; Wu, X.; Johansen, J.V.; Helin, K. Gene Silencing Triggers Polycomb Repressive Complex 2 Recruitment to CpG Islands Genome Wide. Mol. Cell 2014, 55, 347–360. [Google Scholar] [CrossRef]
- Petracovici, A.; Bonasio, R. Distinct PRC2 subunits regulate maintenance and establishment of Polycomb repression during differentiation. Mol. Cell 2021, 81, 2625–2639.e5. [Google Scholar] [CrossRef]
- Smith, Z.D.; Chan, M.M.; Mikkelsen, T.S.; Gu, H.; Gnirke, A.; Regev, A.; Meissner, A. A unique regulatory phase of DNA methylation in the early mammalian embryo. Nature 2012, 484, 339–344. [Google Scholar] [CrossRef] [PubMed]
- Guo, H.; Zhu, P.; Yan, L.; Li, R.; Hu, B.; Lian, Y.; Yan, J.; Ren, X.; Lin, S.; Li, J.; et al. The DNA methylation landscape of human early embryos. Nature 2014, 511, 606–610. [Google Scholar] [CrossRef] [PubMed]
- Dai, H.; Wang, B.; Yang, L.; Chen, J.; Zhu, G.; Sun, M.; Ge, H.; Wang, R.; Chapman, D.L.; Tang, F.; et al. TET-mediated DNA demethylation controls gastrulation by regulating Lefty—Nodal signalling. Nat. Publ. Gr. 2016, 538, 528–532. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Yue, X.; Pastor, W.A.; Lin, L.; Georges, R.; Chavez, L.; Evans, S.M.; Rao, A. Tet proteins influence the balance between neuroectodermal and mesodermal fate choice by inhibiting Wnt signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E8267–E8276. [Google Scholar] [CrossRef] [PubMed]
- Cheng, S.; Mittnenzweig, M.; Mayshar, Y.; Lifshitz, A.; Dunjić, M.; Rais, Y.; Ben-Yair, R.; Gehrs, S.; Chomsky, E.; Mukamel, Z.; et al. The intrinsic and extrinsic effects of TET proteins during gastrulation. Cell 2022, 185, 3169–3185.e20. [Google Scholar] [CrossRef] [PubMed]
- Kreibich, E.; Kleinendorst, R.; Barzaghi, G.; Kaspar, S.; Krebs, A.R. Single-molecule footprinting identifies context-dependent regulation of enhancers by DNA methylation. Mol. Cell 2023, 83, 787–802.e9. [Google Scholar] [CrossRef]
- Kaluscha, S.; Domcke, S.; Wirbelauer, C.; Stadler, M.B.; Durdu, S.; Burger, L.; Schübeler, D. Evidence that direct inhibition of transcription factor binding is the prevailing mode of gene and repeat repression by DNA methylation. Nat. Genet. 2022, 54, 1895–1906. [Google Scholar] [CrossRef]
- Kribelbauer, J.F.; Laptenko, O.; Chen, S.; Martini, G.D.; Freed-Pastor, W.A.; Prives, C.; Mann, R.S.; Bussemaker, H.J. Quantitative Analysis of the DNA Methylation Sensitivity of Transcription Factor Complexes. Cell Rep. 2017, 19, 2383–2395. [Google Scholar] [CrossRef]
- Smukler, S.R.; Runciman, S.B.; Xu, S.; Van Der Kooy, D. Embryonic stem cells assume a primitive neural stem cell fate in the absence of extrinsic influences. J. Cell Biol. 2006, 172, 79–90. [Google Scholar] [CrossRef]
- Chambers, S.M.; Fasano, C.A.; Papapetrou, E.P.; Tomishima, M.; Sadelain, M.; Studer, L. Highly efficient neural conversion of human ES and iPS cells by dual inhibition of SMAD signaling. Nat. Biotechnol. 2009, 27, 275–280. [Google Scholar] [CrossRef]
- Winnier, G.; Blessing, M.; Labosky, P.A.; Hogan, B.L.M. Bone morphogenetic protein-4 is required for mesoderm formation and patterning in the mouse. Genes Dev. 1995, 9, 2105–2116. [Google Scholar] [CrossRef] [PubMed]
- Di-Gregorio, A.; Sancho, M.; Stuckey, D.W.; Crompton, L.A.; Godwin, J.; Mishina, Y.; Rodriguez, T.A. BMP signalling inhibits premature neural differentiation in the mouse embryo. Development 2007, 134, 3359–3369. [Google Scholar] [CrossRef] [PubMed]
- Lauria, A.; Meng, G.; Proserpio, V.; Rapelli, S.; Maldotti, M.; Polignano, I.L.; Anselmi, F.; Incarnato, D.; Krepelova, A.; Donna, D.; et al. DNMT3B supports meso-endoderm differentiation from mouse embryonic stem cells. Nat. Commun. 2023, 14, 367. [Google Scholar] [CrossRef]
- Loh, C.H.; van Genesen, S.; Perino, M.; Bark, M.R.; Veenstra, G.J.C. Loss of PRC2 subunits primes lineage choice during exit of pluripotency. Nat. Commun. 2021, 12, 6985. [Google Scholar] [CrossRef] [PubMed]
- Moran, S.; Martínez-Cardús, A.; Sayols, S.; Musulén, E.; Balañá, C.; Estival-Gonzalez, A.; Moutinho, C.; Heyn, H.; Diaz-Lagares, A.; de Moura, M.C.; et al. Epigenetic profiling to classify cancer of unknown primary: A multicentre, retrospective analysis. Lancet Oncol. 2016, 17, 1386–1395. [Google Scholar] [CrossRef]
- Lo Riso, P.; Villa, C.E.; Gasparoni, G.; Vingiani, A.; Luongo, R.; Manfredi, A.; Jungmann, A.; Bertolotti, A.; Borgo, F.; Garbi, A.; et al. A cell-of-origin epigenetic tracer reveals clinically distinct subtypes of high-grade serous ovarian cancer. Genome Med. 2020, 12, 94. [Google Scholar] [CrossRef]
- Ho, Y.-T.; Shimbo, T.; Wijaya, E.; Ouchi, Y.; Takaki, E.; Yamamoto, R.; Kikuchi, Y.; Kaneda, Y.; Tamai, K. Chromatin accessibility identifies diversity in mesenchymal stem cells from different tissue origins. Sci. Rep. 2018, 8, 17765. [Google Scholar] [CrossRef]
- Polo, J.M.; Liu, S.; Figueroa, M.E.; Kulalert, W.; Eminli, S.; Tan, K.Y.; Apostolou, E.; Stadtfeld, M.; Li, Y.; Shioda, T.; et al. Cell type of origin influences the molecular and functional properties of mouse induced pluripotent stem cells. Nat. Biotechnol. 2010, 28, 848–855. [Google Scholar] [CrossRef]
- Jadhav, U.; Cavazza, A.; Banerjee, K.K.; Xie, H.; O’Neill, N.K.; Saenz-Vash, V.; Herbert, Z.; Madha, S.; Orkin, S.H.; Zhai, H.; et al. Extensive Recovery of Embryonic Enhancer and Gene Memory Stored in Hypomethylated Enhancer DNA. Mol. Cell 2019, 74, 542–554.e5. [Google Scholar] [CrossRef]
- Wong, Y.F.; Kumar, Y.; Proks, M.; Herrera, J.A.R.; Rothová, M.M.; Monteiro, R.S.; Pozzi, S.; Jennings, R.E.; Hanley, N.A.; Bickmore, W.A.; et al. Expansion of ventral foregut is linked to changes in the enhancer landscape for organ-specific differentiation. Nat. Cell Biol. 2023, 25, 481–492. [Google Scholar] [CrossRef]
- Theunissen, T.W.; Powell, B.E.; Wang, H.; Mitalipova, M.; Faddah, D.A.; Reddy, J.; Fan, Z.P.; Maetzel, D.; Ganz, K.; Shi, L.; et al. Systematic identification of culture conditions for induction and maintenance of naive human pluripotency. Cell Stem Cell 2014, 15, 471–487. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, S.; Penfold, C.A.; Slatery, E.; Siriwardena, D.; Drummer, C.; Clark, S.; Strawbridge, S.E.; Kishimoto, K.; Vickers, A.; Tewary, M.; et al. Spatial profiling of early primate gastrulation in utero. Nature 2022, 609, 136–143. [Google Scholar] [CrossRef] [PubMed]
- Rostovskaya, M.; Andrews, S.; Reik, W.; Rugg-Gunn, P.J. Amniogenesis occurs in two independent waves in primates. Cell Stem Cell 2022, 29, 744–759.e6. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.N.; Dobreva, M.P.; Graham, L.; Huylebroeck, D.; Lawson, K.A.; Zwijsen, A. Amnion formation in the mouse embryo: The single amniochorionic fold model. BMC Dev. Biol. 2011, 11, 48. [Google Scholar] [CrossRef]
- van den Brink, S.C.; van Oudenaarden, A. 3D gastruloids: A novel frontier in stem cell-based in vitro modeling of mammalian gastrulation. Trends Cell Biol. 2021, 31, 747–759. [Google Scholar] [CrossRef]
- Zhu, C.; Zhang, Y.; Li, Y.E.; Lucero, J.; Behrens, M.M.; Ren, B. Joint profiling of histone modifications and transcriptome in single cells from mouse brain. Nat. Methods 2021, 18, 283–292. [Google Scholar] [CrossRef]
- Zhu, C.; Yu, M.; Huang, H.; Juric, I.; Abnousi, A.; Hu, R.; Lucero, J.; Behrens, M.M.; Hu, M.; Ren, B. An ultra high-throughput method for single-cell joint analysis of open chromatin and transcriptome. Nat. Struct. Mol. Biol. 2019, 26, 1063–1070. [Google Scholar] [CrossRef]
- Chen, S.; Lake, B.B.; Zhang, K. High-throughput sequencing of the transcriptome and chromatin accessibility in the same cell. Nat. Biotechnol. 2019, 37, 1452–1457. [Google Scholar] [CrossRef]
- Clark, S.J.; Argelaguet, R.; Kapourani, C.A.; Stubbs, T.M.; Lee, H.J.; Alda-Catalinas, C.; Krueger, F.; Sanguinetti, G.; Kelsey, G.; Marioni, J.C.; et al. ScNMT-seq enables joint profiling of chromatin accessibility DNA methylation and transcription in single cells e. Nat. Commun. 2018, 9, 1–9. [Google Scholar] [CrossRef]
- Dimitriu, M.A.; Lazar-Contes, I.; Roszkowski, M.; Mansuy, I.M. Single-Cell Multiomics Techniques: From Conception to Applications. Front. Cell Dev. Biol. 2022, 10, 1–16. [Google Scholar] [CrossRef]
- Pulecio, J.; Verma, N.; Mejía-Ramírez, E.; Huangfu, D.; Raya, A. CRISPR/Cas9-Based Engineering of the Epigenome. Cell Stem Cell 2017, 21, 431–447. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, Y.; Chen, Y.; Li, M.; Shao, Z.; Zhang, M.Q.; Xu, J. CAPTURE: In Situ Analysis of Chromatin Composition of Endogenous Genomic Loci by Biotinylated dCas9. Curr. Protoc. Mol. Biol. 2018, 123, e64. [Google Scholar] [CrossRef] [PubMed]
- Knaupp, A.S.; Mohenska, M.; Larcombe, M.R.; Ford, E.; Lim, S.M.; Wong, K.; Chen, J.; Firas, J.; Huang, C.; Liu, X.; et al. TINC—A Method to Dissect Regulatory Complexes at Single-Locus Resolution—Reveals an Extensive Protein Complex at the Nanog Promoter. Stem Cell Reports 2020, 15, 1246–1259. [Google Scholar] [CrossRef]
- Sönmezer, C.; Kleinendorst, R.; Imanci, D.; Barzaghi, G.; Villacorta, L.; Schübeler, D.; Benes, V.; Molina, N.; Krebs, A.R. Molecular Co-occupancy Identifies Transcription Factor Binding Cooperativity In Vivo. Mol. Cell 2021, 81, 255–267.e6. [Google Scholar] [CrossRef] [PubMed]
| Epigenetic Modification | Writers | Erasers | Location | Role |
|---|---|---|---|---|
| DNA methylation (5meC) | DNMT3A, DNMT3B (maintenance: DNMT1) | TET1, TET2, TET3 | CpG | Repression |
| H3K27ac | HATs, e.g., CBP/p300 | HDACs, SIRTs | Active promoters and enhancers | Activation |
| H3K27me3 | PRC2.1, PRC2.2 | KDM6A, KDM6B | Repressed and bivalent promoters, poised enhancers | Repression |
| H3K4me1 | COMPASS-like (KMT2C, KMT2D) | KDM1A, KDM1B | Active and poised/primed enhancers | Priming, activating |
| H3K4me3 | COMPASS-like (KMT2A, KMT2B) | KDM5A, KDM5B, KDM5C, KDM5D, KDM2B | Mostly promoters (active and bivalent) | Priming, activating |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sullivan, A.E. Epigenetic Control of Cell Potency and Fate Determination during Mammalian Gastrulation. Genes 2023, 14, 1143. https://doi.org/10.3390/genes14061143
Sullivan AE. Epigenetic Control of Cell Potency and Fate Determination during Mammalian Gastrulation. Genes. 2023; 14(6):1143. https://doi.org/10.3390/genes14061143
Chicago/Turabian StyleSullivan, Adrienne E. 2023. "Epigenetic Control of Cell Potency and Fate Determination during Mammalian Gastrulation" Genes 14, no. 6: 1143. https://doi.org/10.3390/genes14061143
APA StyleSullivan, A. E. (2023). Epigenetic Control of Cell Potency and Fate Determination during Mammalian Gastrulation. Genes, 14(6), 1143. https://doi.org/10.3390/genes14061143






