Histone Deacetylases HD2A and HD2B Undergo Feedback Regulation by ABA and Modulate Drought Tolerance via Mediating ABA-Induced Transcriptional Repression
Abstract
:1. Introduction
2. Results
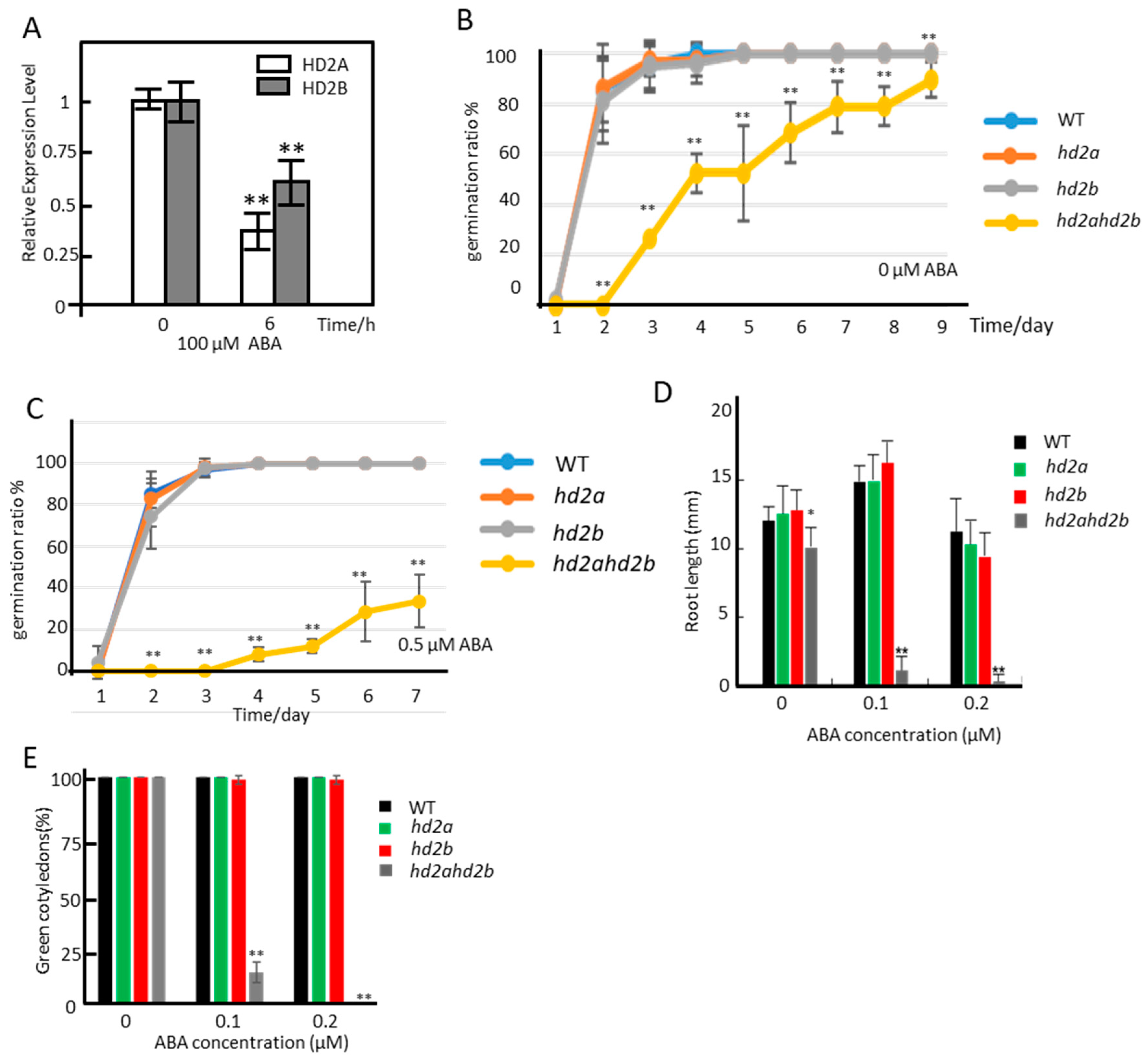
2.1. Loss of HD2A and Reduced Function of HD2B Results in Hypersensitivity towards ABA
2.2. HD2A and HD2B Function Is Essential for the Repression of ABA-Responsive Genes
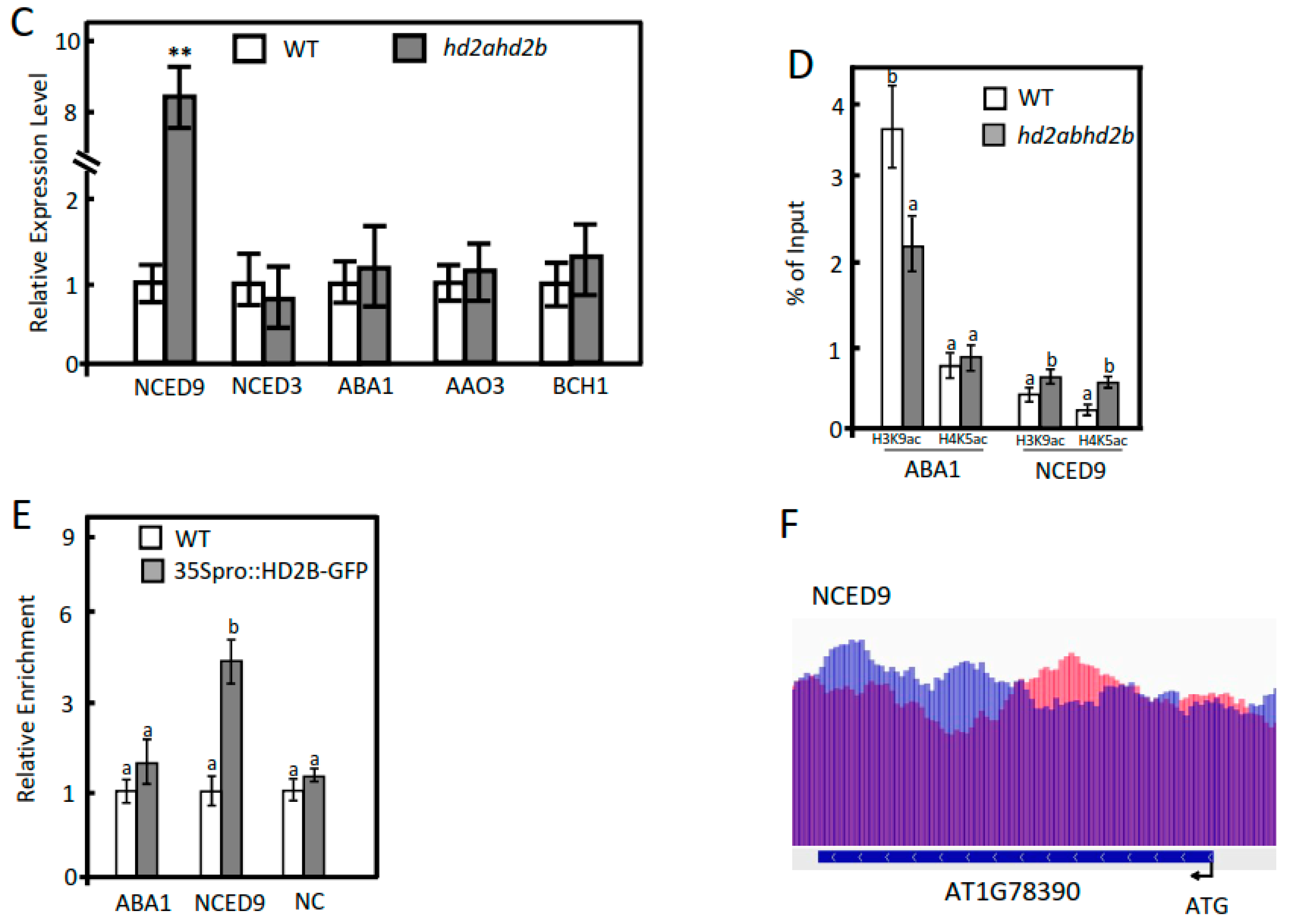
2.3. Under Control Conditions, HD2A and HD2B Repress ABA-Responsive Genes via Histone Deacetylation
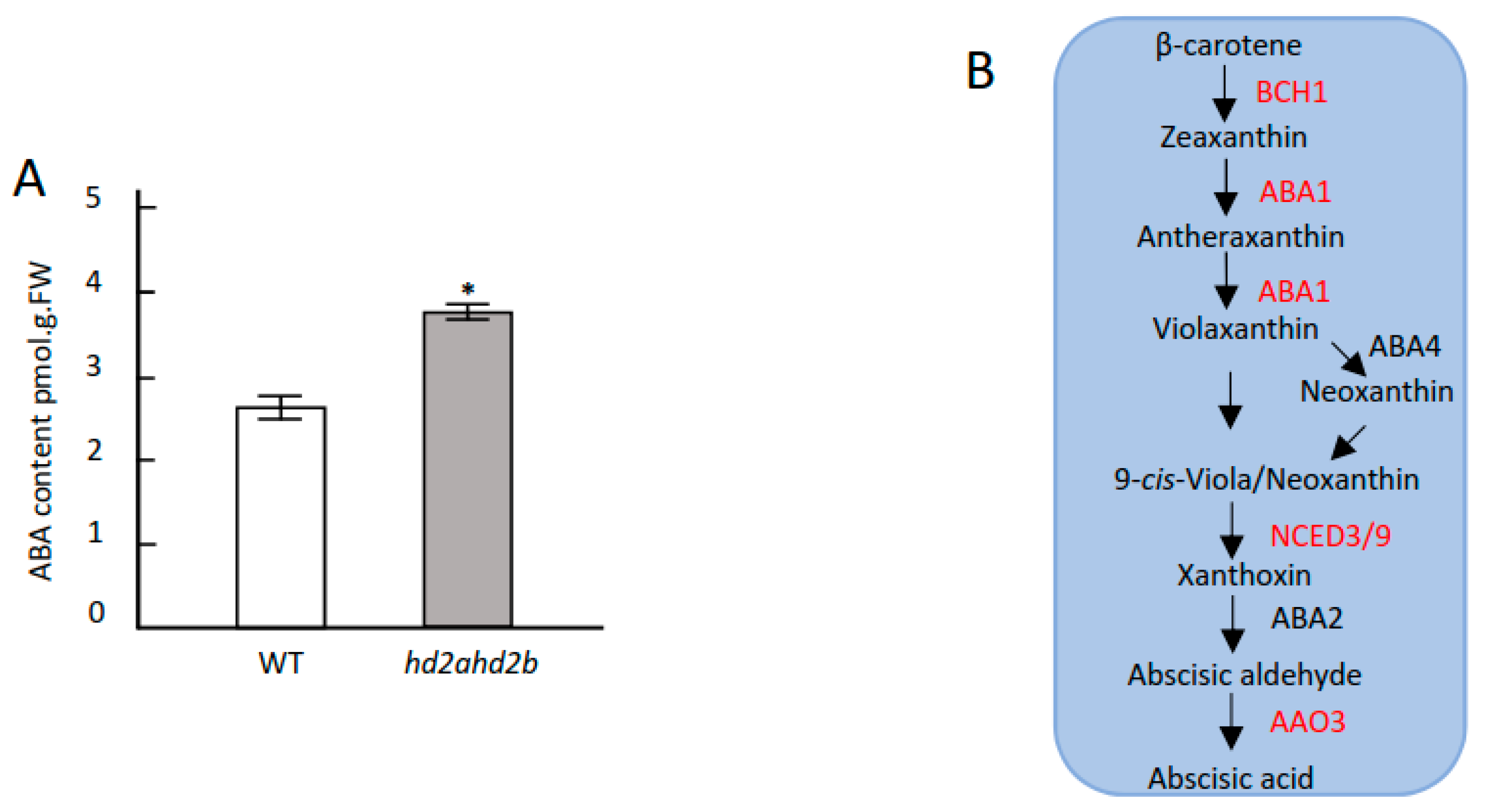
2.4. Loss of HD2A and Reduced Function of HD2B Affects ABA Content, ROS Accumulation, and Stomatal Closure
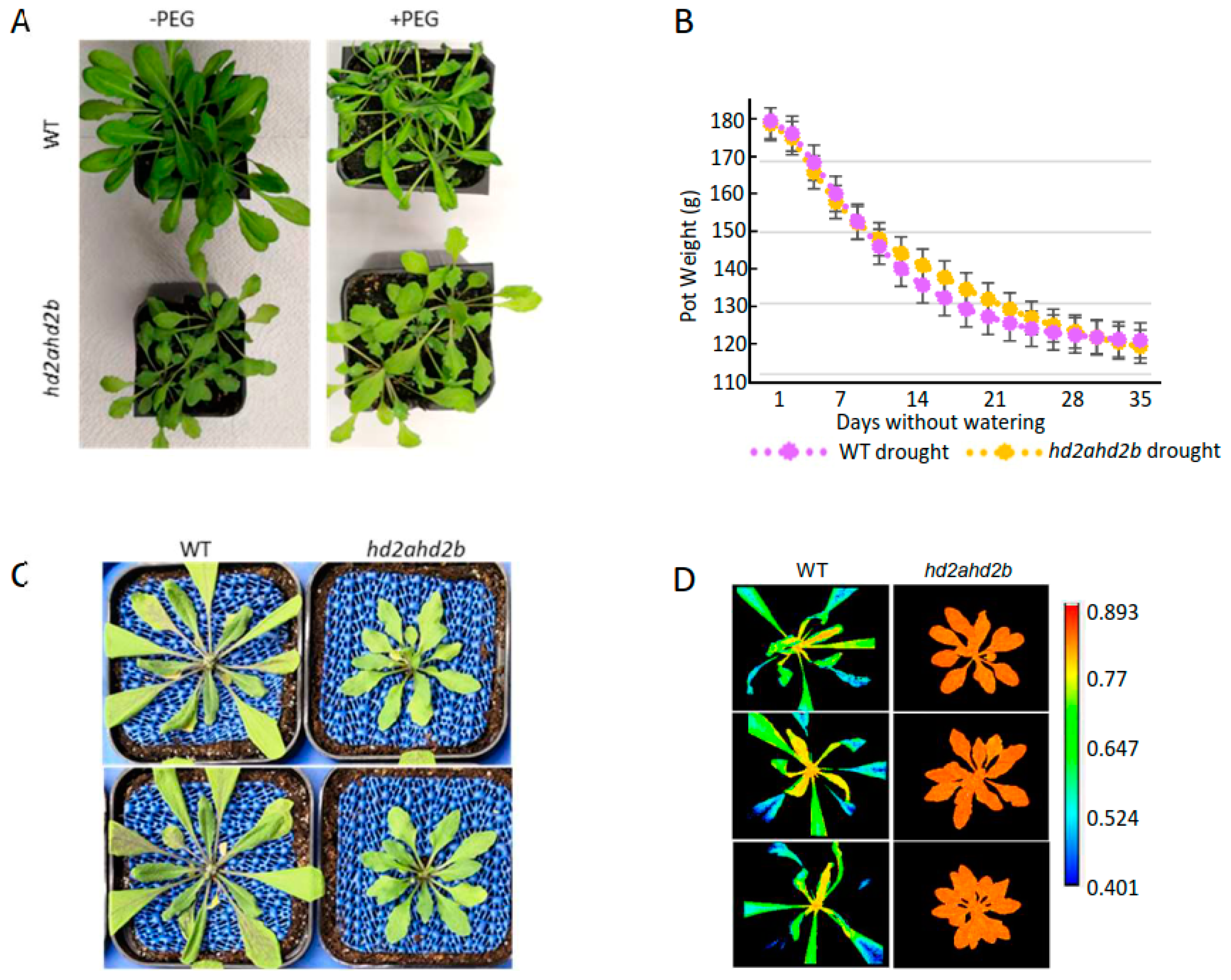
2.5. Loss of HD2A and HD2B Confers Drought Resistance in Arabidopsis
2.6. Loss of HD2A and HD2B Caused a Pleiotropic Phenotype
3. Discussion
3.1. HD2A and HD2B Serve as Negative Regulators of Plant Drought Resistance
3.2. HD2A and HD2B Regulate Drought Stress Responses through an ABA-Dependent Pathway
3.3. HD2A and HD2B Repress the Expression of Certain ABA-Responsive Genes
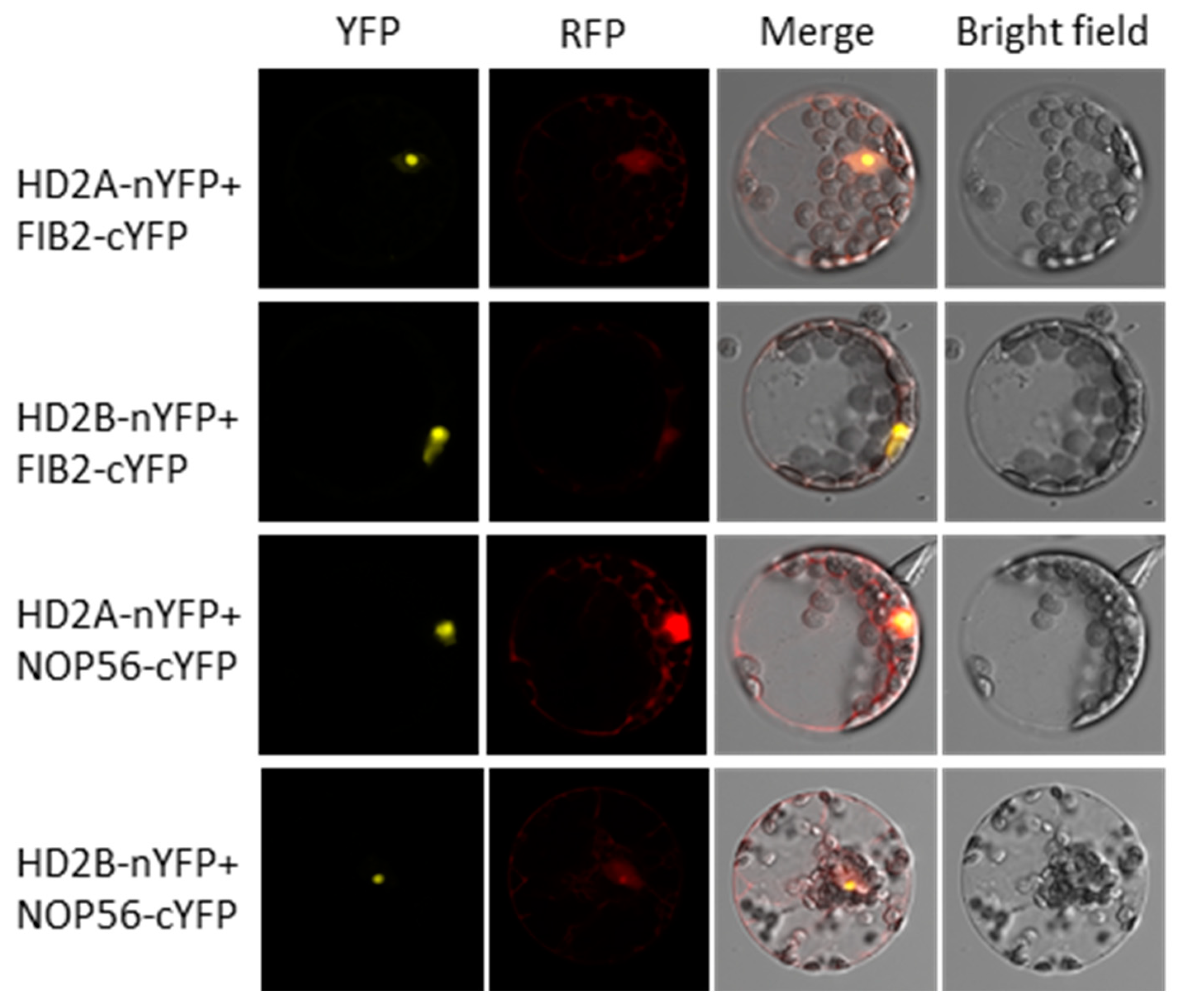
3.4. HD2A and HD2B Function in Drought Tolerance by Affecting ABA-Independent Signaling Pathways and Ribosome Biogenesis
4. Methods
4.1. Plant Materials and Growth Conditions
4.2. Seed Germination, Root Growth, and Green Cotyledon Assay
4.3. Drought Treatment and Data Collection with High-Throughput Automated Non-Invasive Phenotyping Platform
4.4. Determination of ABA, NBT Detection of Superoxide, and Stomatal Aperture Measurement
4.5. DNA Extraction, RNA Extraction, cDNA Synthesis, and Gene Expression Analyses
4.6. RNA-Seq Analysis
4.7. ChIP-qPCR Assays
4.8. Determination of Chl Content
4.9. Bimolecular Fluorescence Complementation Assay
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qin, F.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Achievements and challenges in understanding plant abiotic stress responses and tolerance. Plant Cell Physiol. 2011, 52, 1569–1582. [Google Scholar] [CrossRef] [PubMed]
- Seki, M.; Umezawa, T.; Urano, K.; Shinozaki, K. Regulatory metabolic networks in drought stress responses. Curr. Opin. Plant Biol. 2007, 10, 296–302. [Google Scholar] [CrossRef] [PubMed]
- Chaves, M.M.; Maroco, J.P.; Pereira, J.S. Understanding plant responses to drought—From genes to the whole plant. Funct. Plant Biol. 2003, 30, 239–264. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Rico-Medina, A.; Caño-Delgado, A.I. The physiology of plant responses to drought. Science 2020, 368, 266–269. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Yu, H.; Qu, J.; Cao, Y.; Ding, L.; Feng, W.; Khalid, M.H.B.; Li, W.; Fu, F. Maize ZmBES1/BZR1-5 Decreases ABA Sensitivity and Confers Tolerance to Osmotic Stress in Transgenic Arabidopsis. Int. J. Mol. Sci. 2020, 21, 996. [Google Scholar] [CrossRef]
- López-Cordova, A.; Ramírez-Medina, H.; Silva-Martinez, G.-A.; González-Cruz, L.; Bernardino-Nicanor, A.; Huanca-Mamani, W.; Montero-Tavera, V.; Tovar-Aguilar, A.; Ramírez-Pimentel, J.-G.; Durán-Figueroa, N.-V. LEA13 and LEA30 Are Involved in Tolerance to Water Stress and Stomata Density in Arabidopsis thaliana. Plants 2021, 10, 1694. [Google Scholar] [CrossRef]
- Cao, M.; Liu, X.; Zhang, Y.; Xue, X.; Zhou, X.E.; Melcher, K.; Gao, P.; Wang, F.; Zeng, L.; Zhao, Y.; et al. An ABA-mimicking ligand that reduces water loss and promotes drought resistance in plants. Cell Res. 2013, 23, 1043–1054. [Google Scholar] [CrossRef]
- Chen, K.; Li, G.-J.; Bressan, R.A.; Song, C.-P.; Zhu, J.-K.; Zhao, Y. Abscisic acid dynamics, signaling, and functions in plants. J. Integr. Plant Biol. 2020, 62, 25–54. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef]
- McAdam, S.A.; Brodribb, T.J. Mesophyll cells are the main site of abscisic acid biosynthesis in water-stressed leaves. Plant Physiol. 2018, 177, 911–917. [Google Scholar] [CrossRef]
- Iuchi, S.; Kobayashi, M.; Taji, T.; Naramoto, M.; Seki, M.; Kato, T.; Tabata, S.; Kakubari, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Regulation of drought tolerance by gene manipulation of 9-cis-epoxycarotenoid dioxygenase, a key enzyme in abscisic acid biosynthesis in Arabidopsis. Plant J. 2001, 27, 325–333. [Google Scholar] [CrossRef] [PubMed]
- Wan, X.-R.; Li, L. Regulation of ABA level and water-stress tolerance of Arabidopsis by ectopic expression of a peanut 9-cis-epoxycarotenoid dioxygenase gene. Biochem. Biophys. Res. Commun. 2006, 347, 1030–1038. [Google Scholar] [CrossRef]
- Zhao, Y.; Chan, Z.; Gao, J.; Xing, L.; Cao, M.; Yu, C.; Hu, Y.; You, J.; Shi, H.; Zhu, Y.; et al. ABA receptor PYL9 promotes drought resistance and leaf senescence. Proc. Natl. Acad. Sci. USA 2016, 113, 1949–1954. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Liu, J.; Tischer, S.V.; Christmann, A.; Windisch, W.; Schnyder, H.; Grill, E. Leveraging abscisic acid receptors for efficient water use in Arabidopsis. Proc. Natl. Acad. Sci. USA 2016, 113, 6791–6796. [Google Scholar] [CrossRef]
- Kudo, M.; Kidokoro, S.; Yoshida, T.; Mizoi, J.; Kojima, M.; Takebayashi, Y.; Sakakibara, H.; Fernie, A.R.; Shinozaki, K.; Yamaguchi-Shinozaki, K. A gene-stacking approach to overcome the trade-off between drought stress tolerance and growth in Arabidopsis. Plant J. 2019, 97, 240–256. [Google Scholar] [CrossRef] [PubMed]
- Oneto, C.D.; Otegui, M.E.; Baroli, I.; Beznec, A.; Faccio, P.; Bossio, E.; Blumwald, E.; Lewi, D. Water deficit stress tolerance in maize conferred by expression of an isopentenyltransferase (IPT) gene driven by a stress-and maturation-induced promoter. J. Biotechnol. 2016, 220, 66–77. [Google Scholar] [CrossRef]
- Wang, Y.; Ying, J.; Kuzma, M.; Chalifoux, M.; Sample, A.; McArthur, C.; Uchacz, T.; Sarvas, C.; Wan, J.; Dennis, D.T. Molecular tailoring of farnesylation for plant drought tolerance and yield protection. Plant J. 2005, 43, 413–424. [Google Scholar] [CrossRef]
- Lu, F.; Li, W.; Peng, Y.; Cao, Y.; Qu, J.; Sun, F.; Yang, Q.; Lu, Y.; Zhang, X.; Zheng, L.; et al. ZmPP2C26 Alternative Splicing Variants Negatively Regulate Drought Tolerance in Maize. Front. Plant Sci. 2022, 13, 851531. [Google Scholar] [CrossRef]
- Hu, H.; Xiong, L. Genetic engineering and breeding of drought-resistant crops. Annu. Rev. Plant Biol. 2014, 65, 715–741. [Google Scholar] [CrossRef]
- Leung, J.; Merlot, S.; Giraudat, J. The Arabidopsis ABSCISIC ACID-INSENSITIVE2 (ABI2) and ABI1 genes encode homologous protein phosphatases 2C involved in abscisic acid signal transduction. Plant Cell 1997, 9, 759–771. [Google Scholar]
- Bailey-Serres, J.; Parker, J.E.; Ainsworth, E.A.; Oldroyd, G.E.D.; Schroeder, J.I. Genetic strategies for improving crop yields. Nature 2019, 575, 109–118. [Google Scholar] [CrossRef] [PubMed]
- Murata, Y.; Pei, Z.-M.; Mori, I.C.; Schroeder, J. Abscisic Acid Activation of Plasma Membrane Ca2+ Channels in Guard Cells Requires Cytosolic NAD(P)H and Is Differentially Disrupted Upstream and Downstream of Reactive Oxygen Species Production in abi1-1 and abi2-1 Protein Phosphatase 2C Mutants. Plant Cell 2001, 13, 2513–2523. [Google Scholar] [CrossRef] [PubMed]
- Vartanian, N.; Marcotte, L.; Giraudat, J. Drought rhizogenesis in Arabidopsis thaliana (differential responses of hormonal mutants). Plant Physiol. 1994, 104, 761–767. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Zhang, Y.; Li, Q.; Liu, S.; He, F.; An, Y.; Zhou, Y.; Liu, C.; Yin, W.; Xia, X. PdGNC confers drought tolerance by mediating stomatal closure resulting from NO and H2O2 production via the direct regulation of PdHXK1 expression in Populus. New Phytol. 2021, 230, 1868–1882. [Google Scholar] [CrossRef]
- Hollender, C.; Liu, Z. Histone deacetylase genes in Arabidopsis development. J. Integr. Plant Biol. 2008, 50, 875–885. [Google Scholar] [CrossRef]
- Wolffe, A.P. Histone deacetylase—A regulator of transcription. Science 1996, 272, 371. [Google Scholar] [CrossRef]
- Yang, X.-J.; Seto, E. Collaborative spirit of histone deacetylases in regulating chromatin structure and gene expression. Curr. Opin. Genet. Dev. 2003, 13, 143–153. [Google Scholar] [CrossRef]
- Demetriou, K.; Kapazoglou, A.; Tondelli, A.; Francia, E.; Stanca, M.A.; Bladenopoulos, K.; Tsaftaris, A.S.J.P.P. Epigenetic chromatin modifiers in barley: I. Cloning, mapping and expression analysis of the plant specific HD2 family of histone deacetylases from barley, during seed development and after hormonal treatment. Physiol. Plant 2009, 136, 358–368. [Google Scholar] [CrossRef]
- Lagacé, M.; Chantha, S.-C.; Major, G.; Matton, D.P. Fertilization induces strong accumulation of a histone deacetylase (HD2) and of other chromatin-remodeling proteins in restricted areas of the ovules. Plant Mol. Biol. 2003, 53, 759–769. [Google Scholar] [CrossRef]
- Aquea, F.; Timmermann, T.; Arce-Johnson, P. Analysis of histone acetyltransferase and deacetylase families of Vitis vinifera. Plant Physiol. Biochem. 2010, 48, 194–199. [Google Scholar] [CrossRef]
- Bourque, S.; Dutartre, A.; Hammoudi, V.; Blanc, S.; Dahan, J.; Jeandroz, S.; Pichereaux, C.; Rossignol, M.; Wendehenne, D. Type-2 histone deacetylases as new regulators of elicitor-induced cell death in plants. New Phytol. 2011, 192, 127–139. [Google Scholar] [CrossRef] [PubMed]
- Sokol, A.; Kwiatkowska, A.; Jerzmanowski, A.; Prymakowska-Bosak, M. Up-regulation of stress-inducible genes in tobacco and Arabidopsis cells in response to abiotic stresses and ABA treatment correlates with dynamic changes in histone H3 and H4 modifications. Planta 2007, 227, 245–254. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.M.; To, T.K.; Ishida, J.; Morosawa, T.; Kawashima, M.; Matsui, A.; Toyoda, T.; Kimura, H.; Shinozaki, K.; Seki, M. Alterations of lysine modifications on the histone H3 N-tail under drought stress conditions in Arabidopsis thaliana. Plant Cell Physiol. 2008, 49, 1580–1588. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Liu, X.; Thorn, G.; Duan, J.; Tian, L. Expression analysis of histone acetyltransferases in rice under drought stress. Biochem. Biophys. Res. Commun. 2014, 443, 400–405. [Google Scholar] [CrossRef]
- Shen, Y.; Lei, T.; Cui, X.; Liu, X.; Zhou, S.; Zheng, Y.; Guérard, F.; Issakidis-Bourguet, E.; Zhou, D.X. Arabidopsis histone deacetylase HDA 15 directly represses plant response to elevated ambient temperature. Plant J. 2019, 100, 991–1006. [Google Scholar] [CrossRef]
- Baek, D.; Shin, G.; Kim, M.C.; Shen, M.; Lee, S.Y.; Yun, D.J. Histone Deacetylase HDA9 With ABI4 Contributes to Abscisic Acid Homeostasis in Drought Stress Response. Front. Plant Sci. 2020, 11, 143. [Google Scholar] [CrossRef]
- Tasset, C.; Singh Yadav, A.; Sureshkumar, S.; Singh, R.; van der Woude, L.; Nekrasov, M.; Tremethick, D.; van Zanten, M.; Balasubramanian, S. POWERDRESS-mediated histone deacetylation is essential for thermomorphogenesis in Arabidopsis thaliana. PLoS Genet. 2018, 14, e1007280. [Google Scholar] [CrossRef]
- Lee, H.G.; Seo, P.J. MYB96 recruits the HDA15 protein to suppress negative regulators of ABA signaling in Arabidopsis. Nat. Commun. 2019, 10, 1713. [Google Scholar] [CrossRef]
- Chen, L.T.; Wu, K. Role of histone deacetylases HDA6 and HDA19 in ABA and abiotic stress response. Plant Signal. Behav. 2010, 5, 1318–1320. [Google Scholar] [CrossRef]
- Tanaka, M.; Kikuchi, A.; Kamada, H. The Arabidopsis histone deacetylases HDA6 and HDA19 contribute to the repression of embryonic properties after germination. Plant Physiol. 2008, 146, 149–161. [Google Scholar] [CrossRef]
- Zhou, Y.; Tan, B.; Luo, M.; Li, Y.; Liu, C.; Chen, C.; Yu, C.W.; Yang, S.; Dong, S.; Ruan, J.; et al. Histone Deacetylase19 interacts with HSL1 and participates in the repression of seed maturation genes in Arabidopsis seedlings. Plant Cell 2013, 25, 134–148. [Google Scholar] [CrossRef] [PubMed]
- Gu, D.; Chen, C.Y.; Zhao, M.; Zhao, L.; Duan, X.; Duan, J.; Wu, K.; Liu, X. Identification of HDA15-PIF1 as a key repression module directing the transcriptional network of seed germination in the dark. Nucleic Acids Res. 2017, 45, 7137–7150. [Google Scholar] [CrossRef]
- Zhou, Y.; Yang, P.; Zhang, F.; Luo, X.; Xie, J. Histone deacetylase HDA19 interacts with histone methyltransferase SUVH5 to regulate seed dormancy in Arabidopsis. Plant Biol. 2020, 22, 1062–1071. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Yu, C.W.; Chen, F.F.; Zhao, L.; Tian, G.; Liu, X.; Cui, Y.; Yang, J.Y.; Wu, K. Histone deacetylase HDA6 is functionally associated with AS1 in repression of KNOX genes in arabidopsis. PLoS Genet. 2012, 8, e1003114. [Google Scholar] [CrossRef]
- Kim, W.; Latrasse, D.; Servet, C.; Zhou, D.X. Arabidopsis histone deacetylase HDA9 regulates flowering time through repression of AGL19. Biochem. Biophys. Res. Commun. 2013, 432, 394–398. [Google Scholar] [CrossRef]
- Luo, M.; Wang, Y.-Y.; Liu, X.; Yang, S.; Lu, Q.; Cui, Y.; Wu, K. HD2C interacts with HDA6 and is involved in ABA and salt stress response in Arabidopsis. J. Exp. Bot. 2012, 63, 3297–3306. [Google Scholar] [CrossRef] [PubMed]
- Sridha, S.; Wu, K. Identification of AtHD2C as a novel regulator of abscisic acid responses in Arabidopsis. Plant J. 2006, 46, 124–133. [Google Scholar] [CrossRef]
- Tahir, M.S.; Karagiannis, J.; Tian, L. HD2A and HD2C co-regulate drought stress response by modulating stomatal closure and root growth in Arabidopsis. Front. Plant Sci. 2022, 13, 1062722. [Google Scholar] [CrossRef] [PubMed]
- Buszewicz, D.; Archacki, R.; Palusiński, A.; Kotliński, M.; Fogtman, A.; Iwanicka-Nowicka, R.; Sosnowska, K.; Kuciński, J.; Pupel, P.; Olędzki, J.; et al. HD2C histone deacetylase and a SWI/SNF chromatin remodelling complex interact and both are involved in mediating the heat stress response in Arabidopsis. Plant Cell Environ. 2016, 39, 2108–2122. [Google Scholar] [CrossRef]
- Han, Z.; Yu, H.; Zhao, Z.; Hunter, D.; Luo, X.; Duan, J.; Tian, L. AtHD2D Gene Plays a Role in Plant Growth, Development, and Response to Abiotic Stresses in Arabidopsis thaliana. Front. Plant Sci. 2016, 7, 310. [Google Scholar] [CrossRef]
- Guo, Z.; Li, Z.; Liu, Y.; An, Z.; Peng, M.; Shen, W.H.; Dong, A.; Yu, Y. MRG1/2 histone methylation readers and HD2C histone deacetylase associate in repression of the florigen gene FT to set a proper flowering time in response to day-length changes. New Phytol. 2020, 227, 1453–1466. [Google Scholar] [CrossRef] [PubMed]
- Luo, Y.; Shi, D.-Q.; Jia, P.-F.; Bao, Y.; Li, H.-J.; Yang, W.-C. Nucleolar histone deacetylases HDT1, HDT2 and HDT3 regulate plant reproductive development. J. Genet. Genom. 2021, 49, 30–39. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Torres-Garcia, J.; Latrasse, D.; Benhamed, M.; Schilderink, S.; Zhou, W.; Kulikova, O.; Hirt, H.; Bisseling, T. Plant-specific histone deacetylases HDT1/2 regulate GIBBERELLIN 2-OXIDASE2 expression to control Arabidopsis root meristem cell number. Plant Cell 2017, 29, 2183–2196. [Google Scholar] [CrossRef] [PubMed]
- Yano, R.; Takebayashi, Y.; Nambara, E.; Kamiya, Y.; Seo, M. Combining association mapping and transcriptomics identify HD2B histone deacetylase as a genetic factor associated with seed dormancy in Arabidopsis thaliana. Plant J. 2013, 74, 815–828. [Google Scholar] [CrossRef]
- Yongtao, H.; Elisabeth, G.; Christoph, J.W.; Patrick, H.; Gregor, J.H.; Robert, K.; Claude, B.; Joerg, D.; Christian, L. Arabidopsis Histone Deacetylase HD2A and HD2B Regulate Seed Dormancy by Repressing Delay of Germination 1. Front. Plant Sci. 2023, 14, 1431. [Google Scholar]
- Bourque, S.; Jeandroz, S.; Grandperret, V.; Lehotai, N.; Aimé, S.; Soltis, D.E.; Miles, N.W.; Melkonian, M.; Deyholos, M.K.; Leebens-Mack, J.H.; et al. The Evolution of HD2 Proteins in Green Plants. Trends Plant Sci. 2016, 21, 1008–1016. [Google Scholar] [CrossRef]
- Wu, K.; Tian, L.; Malik, K.; Brown, D.; Miki, B. Functional analysis of HD2 histone deacetylase homologues in Arabidopsis thaliana. Plant J. 2000, 22, 19–27. [Google Scholar] [CrossRef]
- Muhammad, I.I.; Kong, S.L.; Akmar Abdullah, S.N.; Munusamy, U. RNA-seq and ChIP-seq as Complementary Approaches for Comprehension of Plant Transcriptional Regulatory Mechanism. Int. J. Mol. Sci. 2020, 21, 167. [Google Scholar] [CrossRef]
- Pepke, S.; Wold, B.; Mortazavi, A. Computation for ChIP-seq and RNA-seq studies. Nat. Methods 2009, 6, S22–S32. [Google Scholar] [CrossRef]
- Latrasse, D.; Jégu, T.; Li, H.; de Zelicourt, A.; Raynaud, C.; Legras, S.; Gust, A.; Samajova, O.; Veluchamy, A.; Rayapuram, N.; et al. MAPK-triggered chromatin reprogramming by histone deacetylase in plant innate immunity. Genome Biol. 2017, 18, 131. [Google Scholar] [CrossRef]
- Zhang, D.P.; Zhou, Y.; Yin, J.F.; Yan, X.J.; Lin, S.; Xu, W.F.; Baluška, F.; Wang, Y.P.; Xia, Y.J.; Liang, G.H.; et al. Rice G-protein subunits qPE9-1 and RGB1 play distinct roles in abscisic acid responses and drought adaptation. J. Exp. Bot. 2015, 66, 6371–6384. [Google Scholar] [CrossRef] [PubMed]
- Soon, F.-F.; Ng, L.-M.; Zhou, X.E.; West, G.M.; Kovach, A.; Tan, M.E.; Suino-Powell, K.M.; He, Y.; Xu, Y.; Chalmers, M.J. Molecular mimicry regulates ABA signaling by SnRK2 kinases and PP2C phosphatases. Science 2012, 335, 85–88. [Google Scholar] [CrossRef] [PubMed]
- Song, X.-J.; Matsuoka, M. Bar the windows: An optimized strategy to survive drought and salt adversities. Genes Dev. 2009, 23, 1709–1713. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Ma, W.; Niu, J.F.; Li, B.; Zhou, W.; Liu, S.; Yan, Y.P.; Ma, J.; Wang, Z.Z. Systematic analysis of SmWD40s, and responding of SmWD40-170 to drought stress by regulation of ABA- and H(2)O(2)-induced stomal movement in Salvia miltiorrhiza bunge. Plant Physiol. Biochem. 2020, 153, 131–140. [Google Scholar] [CrossRef]
- Hogewoning, S.W.; Wientjes, E.; Douwstra, P.; Trouwborst, G.; van Ieperen, W.; Croce, R.; Harbinson, J. Photosynthetic quantum yield dynamics: From photosystems to leaves. Plant Cell 2012, 24, 1921–1935. [Google Scholar] [CrossRef]
- Barbagallo, R.P.; Oxborough, K.; Pallett, K.E.; Baker, N.R. Rapid, Noninvasive Screening for Perturbations of Metabolism and Plant Growth Using Chlorophyll Fluorescence Imaging. Plant Physiol. 2003, 132, 485–493. [Google Scholar] [CrossRef]
- Huang, B.; Fry, J.; Wang, B. Water Relations and Canopy Characteristics of Tall Fescue Cultivars during and after Drought Stress. HortSci. A Publ. Am. Soc. Hortic. Sci. 1998, 33, 837–840. [Google Scholar] [CrossRef]
- Paknejad, F.; Nasri, M.; Moghadam, H.T.; Zahedi, H.; Alahmadi, M.J. Effects of drought stress on chlorophyll fluorescence parameters, chlorophyll content and grain yield of wheat cultivars. J. Biol. Sci. 2007, 7, 841–847. [Google Scholar] [CrossRef]
- Sánchez-García, A.B.; Aguilera, V.; Micol-Ponce, R.; Jover-Gil, S.; Ponce, M.R. Arabidopsis MAS2, an essential gene that encodes a homolog of animal NF-κ B activating protein, is involved in 45S ribosomal DNA silencing. Plant Cell 2015, 27, 1999–2015. [Google Scholar] [CrossRef]
- Wieckowski, Y.; Schiefelbein, J. Nuclear ribosome biogenesis mediated by the DIM1A rRNA dimethylase is required for organized root growth and epidermal patterning in Arabidopsis. Plant Cell 2012, 24, 2839–2856. [Google Scholar] [CrossRef]
- Chen, X.; Lu, L.; Qian, S.; Scalf, M.; Smith, L.M.; Zhong, X. Canonical and noncanonical actions of Arabidopsis histone deacetylases in ribosomal RNA processing. Plant Cell 2018, 30, 134–152. [Google Scholar] [CrossRef] [PubMed]
- Lechertier, T.; Grob, A.; Hernandez-Verdun, D.; Roussel, P. Fibrillarin and Nop56 interact before being co-assembled in box C/D snoRNPs. Exp. Cell Res. 2009, 315, 928–942. [Google Scholar] [CrossRef] [PubMed]
- Zhao, T.; Zhan, Z.; Jiang, D. Histone modifications and their regulatory roles in plant development and environmental memory. J. Genet. Genom. 2019, 46, 467–476. [Google Scholar] [CrossRef]
- Zhou, C.; Labbe, H.; Sridha, S.; Wang, L.; Tian, L.; Latoszek-Green, M.; Yang, Z.; Brown, D.; Miki, B.; Wu, K. Expression and function of HD2-type histone deacetylases in Arabidopsis development. Plant J. 2004, 38, 715–724. [Google Scholar] [CrossRef] [PubMed]
- Luo, M.; Cheng, K.; Xu, Y.; Yang, S.; Wu, K. Plant Responses to Abiotic Stress Regulated by Histone Deacetylases. Front. Plant Sci. 2017, 8, 2147. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; He, X.; Gao, Y.; Zhou, C.; Chiang, V.L.; Li, W. Histone Acetylation Changes in Plant Response to Drought Stress. Genes 2021, 12, 1409. [Google Scholar] [CrossRef] [PubMed]
- Chinnusamy, V.; Zhu, J.-K. Epigenetic regulation of stress responses in plants. J. Integr. Plant Biol. 2009, 12, 133–139. [Google Scholar] [CrossRef]
- Kim, J.M.; To, T.K.; Nishioka, T.; Seki, M. Chromatin regulation functions in plant abiotic stress responses. J. Plant Cell Environ. 2010, 33, 604–611. [Google Scholar] [CrossRef]
- Ueda, M.; Matsui, A.; Nakamura, T.; Abe, T.; Sunaoshi, Y.; Shimada, H.; Seki, M. Versatility of HDA19-deficiency in increasing the tolerance of Arabidopsis to different environmental stresses. Plant Signal. Behav. 2018, 13, e1475808. [Google Scholar] [CrossRef]
- Zhao, B.; Shao, Z.; Wang, L.; Zhang, F.; Chakravarty, D.; Zong, W. MYB44-ENAP1/2 restricts HDT4 to regulate drought tolerance in Arabidopsis. PLoS Genet. 2022, 18, e1010473. [Google Scholar] [CrossRef]
- Liu, S.; Lv, Z.; Liu, Y.; Li, L.; Zhang, L. Network analysis of ABA-dependent and ABA-independent drought responsive genes in Arabidopsis thaliana. Genet. Mol. Biol. 2018, 41, 624–637. [Google Scholar] [CrossRef] [PubMed]
- Leung, J.; Giraudat, J. Abscisic Acid Signal Transduction. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998, 49, 199–222. [Google Scholar] [CrossRef] [PubMed]
- Pei, Z.M.; Kuchitsu, K.; Ward, J.M.; Schwarz, M.; Schroeder, J.I. Differential abscisic acid regulation of guard cell slow anion channels in Arabidopsis wild-type and abi1 and abi2 mutants. Plant Cell 1997, 9, 409–423. [Google Scholar] [PubMed]
- Pei, Z.-M.; Murata, Y.; Benning, G.; Thomine, S.; Klüsener, B.; Allen, G.J.; Grill, E.; Schroeder, J.I. Calcium channels activated by hydrogen peroxide mediate abscisic acid signalling in guard cells. Nature 2000, 406, 731–734. [Google Scholar] [CrossRef]
- Qi, J.; Wang, J.; Gong, Z.; Zhou, J.M. Apoplastic ROS signaling in plant immunity. Curr. Opin. Plant Biol. 2017, 38, 92–100. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef]
- Qi, J.; Song, C.-P.; Wang, B.; Zhou, J.; Kangasjärvi, J.; Zhu, J.-K.; Gong, Z. Reactive oxygen species signaling and stomatal movement in plant responses to drought stress and pathogen attack. J. Integr. Plant Biol. 2018, 60, 805–826. [Google Scholar] [CrossRef]
- Chang, Y.N.; Zhu, C.; Jiang, J.; Zhang, H.; Zhu, J.K.; Duan, C.G. Epigenetic regulation in plant abiotic stress responses. J. Integr. Plant Biol. 2020, 62, 563–580. [Google Scholar] [CrossRef]
- Song, L.; Huang, S.C.; Wise, A.; Castanon, R.; Nery, J.R.; Chen, H.; Watanabe, M.; Thomas, J.; Bar-Joseph, Z.; Ecker, J.R. A transcription factor hierarchy defines an environmental stress response network. Science 2016, 354, aag1550. [Google Scholar] [CrossRef]
- Ueda, M.; Seki, M. Histone Modifications Form Epigenetic Regulatory Networks to Regulate Abiotic Stress Response. Plant Physiol. 2020, 182, 15–26. [Google Scholar] [CrossRef]
- Chen, L.-T.; Luo, M.; Wang, Y.-Y.; Wu, K. Involvement of Arabidopsis histone deacetylase HDA6 in ABA and salt stress response. J. Exp. Bot. 2010, 61, 3345–3353. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, T.; Mogami, J.; Yamaguchi-Shinozaki, K. ABA-dependent and ABA-independent signaling in response to osmotic stress in plants. Curr. Opin. Plant Biol. 2014, 21, 133–139. [Google Scholar] [CrossRef] [PubMed]
- Lei, L.; Shi, J.; Chen, J.; Zhang, M.; Sun, S.; Xie, S.; Li, X.; Zeng, B.; Peng, L.; Hauck, A.; et al. Ribosome profiling reveals dynamic translational landscape in maize seedlings under drought stress. Plant J. 2015, 84, 1206–1218. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Liang, C.; Zheng, M.; Liu, L.; An, Y.; Xu, H.; Xiao, S.; Nie, L. Ribosome Hibernation as a Stress Response of Bacteria. Protein Pept. Lett. 2020, 27, 1082–1091. [Google Scholar] [CrossRef]
- Steffen, K.K.; McCormick, M.A.; Pham, K.M.; MacKay, V.L.; Delaney, J.R.; Murakami, C.J.; Kaeberlein, M.; Kennedy, B.K. Ribosome Deficiency Protects Against ER Stress in Saccharomyces cerevisiae. Genetics 2012, 191, 107–118. [Google Scholar] [CrossRef]
- Huang, K.-C.; Lin, W.-C.; Cheng, W.-H. Salt hypersensitive mutant 9, a nucleolar APUM23 protein, is essential for salt sensitivity in association with the ABA signaling pathway in Arabidopsis. BMC Plant Biol. 2018, 18, 40. [Google Scholar] [CrossRef]
- Micol-Ponce, R.; Sarmiento-Mañús, R.; Ruiz-Bayón, A.; Montacié, C.; Sáez-Vasquez, J.; Ponce, M.R. Arabidopsis RIBOSOMAL RNA PROCESSING7 is required for 18S rRNA maturation. Plant Cell 2018, 30, 2855–2872. [Google Scholar] [CrossRef]
- Abbasi, N.; Kim, H.B.; Park, N.-I.; Kim, H.-S.; Kim, Y.-K.; Park, Y.-I.; Choi, S.-B. APUM23, a nucleolar Puf domain protein, is involved in pre-ribosomal RNA processing and normal growth patterning in Arabidopsis. Plant J. 2010, 64, 960–976. [Google Scholar] [CrossRef]
- Okajima, Y.; Taneda, H.; Noguchi, K.; Terashima, I. Optimum leaf size predicted by a novel leaf energy balance model incorporating dependencies of photosynthesis on light and temperature. Ecol. Res. 2012, 27, 333–346. [Google Scholar] [CrossRef]
- Kurdistani, S.K.; Tavazoie, S.; Grunstein, M. Mapping Global Histone Acetylation Patterns to Gene Expression. Cell 2004, 117, 721–733. [Google Scholar] [CrossRef]
- Wang, D.; Urisman, A.; Liu, Y.; Springer, M.; Ksiazek, T.; Erdman, D.; Mardis, E.; Hickenbotham, M.; Magrini, V.; Eldred, J. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003, 1, E2. [Google Scholar] [CrossRef] [PubMed]
- De Nadal, E.; Zapater, M.; Alepuz, P.M.; Sumoy, L.; Mas, G.; Posas, F.J.N. The MAPK Hog1 recruits Rpd3 histone deacetylase to activate osmoresponsive genes. Nature 2004, 427, 370–374. [Google Scholar] [CrossRef] [PubMed]
- Wu, K.; Tian, L.; Zhou, C.; Brown, D.; Miki, B. Repression of gene expression by Arabidopsis HD2 histone deacetylases. Plant J. 2003, 34, 241–247. [Google Scholar] [CrossRef] [PubMed]
- Sharma, N.; Bender, Y.; Boyle, K.; Fobert, P.R. High-level expression of sugar inducible gene2 (HSI2) is a negative regulator of drought stress tolerance in Arabidopsis. BMC Plant Biol. 2013, 13, 170. [Google Scholar] [CrossRef]
- Suzuki, M.; Wang, H.H.-Y.; McCarty, D.R. Repression of the LEAFY COTYLEDON 1/B3 Regulatory Network in Plant Embryo Development by VP1/ABSCISIC ACID INSENSITIVE 3-LIKE B3 Genes. Plant Physiol. 2007, 143, 902–911. [Google Scholar] [CrossRef]
- Parsons, G.G.; Spenser, C.A. Mitotic repression of RNA polymerase II transcription is accompanied by release of transcription elongation complexes. Mol. Cell. Biol. 1997, 17, 5791–5802. [Google Scholar] [CrossRef]
- Attieh, Y.; Geng, Q.R.; DiNardo, C.D.; Zheng, H.; Jia, Y.; Fang, Z.-H.; Gañán-Gómez, I.; Yang, H.; Wei, Y.; Kantarjian, H.; et al. Low frequency of H3.3 mutations and upregulated DAXX expression in MDS. Blood 2013, 121, 4009–4011. [Google Scholar] [CrossRef]
- Ginno, P.A.; Burger, L.; Seebacher, J.; Iesmantavicius, V.; Schübeler, D. Cell cycle-resolved chromatin proteomics reveals the extent of mitotic preservation of the genomic regulatory landscape. Nat. Commun. 2018, 9, 4048. [Google Scholar] [CrossRef]
- Alabert, C.; Barth, T.K.; Reverón-Gómez, N.; Sidoli, S.; Schmidt, A.; Jensen, O.N.; Imhof, A.; Groth, A. Two distinct modes for propagation of histone PTMs across the cell cycle. Genes Dev. 2015, 29, 585–590. [Google Scholar] [CrossRef]
- Cimini, D.; Mattiuzzo, M.; Torosantucci, L.; Degrassi, F. Histone hyperacetylation in mitosis prevents sister chromatid separation and produces chromosome segregation defects. Mol. Biol. Cell 2003, 14, 3821–3833. [Google Scholar] [CrossRef]
- Ueno, Y.; Ishikawa, T.; Watanabe, K.; Terakura, S.; Iwakawa, H.; Okada, K.; Machida, C.; Machida, Y. Histone Deacetylases and ASYMMETRIC LEAVES2 Are Involved in the Establishment of Polarity in Leaves of Arabidopsis. Plant Cell 2007, 19, 445–457. [Google Scholar] [CrossRef] [PubMed]
- Meng, L.; Feldman, L. A rapid TRIzol-based two-step method for DNA-free RNA extraction from Arabidopsis siliques and dry seeds. Biotechnol. J. 2010, 5, 183–186. [Google Scholar] [CrossRef] [PubMed]
- Križman, M.; Jakše, J.; Baričevič, D.; Javornik, B.; Prošek, M. Robust CTAB-activated charcoal protocol for plant DNA extraction. J. Acta Agric. Slov. 2006, 87, 427–433. [Google Scholar]
- Berardini, T.Z.; Reiser, L.; Li, D.; Mezheritsky, Y.; Muller, R.; Strait, E.; Huala, E. The Arabidopsis information resource: Making and mining the “gold standard” annotated reference plant genome. Genesis 2015, 53, 474–485. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N.; Marth, G.; Abecasis, G.; Durbin, R. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Pertea, M.; Pertea, G.M.; Antonescu, C.M.; Chang, T.C.; Mendell, J.T.; Salzberg, S.L. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 2015, 33, 290–295. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Ageeva-Kieferle, A.; Georgii, E.; Winkler, B.; Ghirardo, A.; Albert, A.; Hüther, P.; Mengel, A.; Becker, C.; Schnitzler, J.-P.; Durner, J.; et al. Nitric oxide coordinates growth, development, and stress response via histone modification and gene expression. Plant Physiol. 2021, 187, 336–360. [Google Scholar] [CrossRef]
- Bowler, C.; Benvenuto, G.; Laflamme, P.; Molino, D.; Probst, A.V.; Tariq, M.; Paszkowski, J. Chromatin techniques for plant cells. Plant J. 2004, 39, 776–789. [Google Scholar] [CrossRef]
- Liang, Y.; Urano, D.; Liao, K.-L.; Hedrick, T.L.; Gao, Y.; Jones, A.M. A nondestructive method to estimate the chlorophyll content of Arabidopsis seedlings. Plant Methods 2017, 13, 26. [Google Scholar] [CrossRef] [PubMed]
- Grefen, C.; Blatt, M.R. A 2in1 cloning system enables ratiometric bimolecular fluorescence complementation (rBiFC). Biotechniques 2012, 53, 311–314. [Google Scholar] [CrossRef] [PubMed]
- Yoo, S.D.; Cho, Y.H.; Sheen, J. Arabidopsis mesophyll protoplasts: A versatile cell system for transient gene expression analysis. Nat. Protoc. 2007, 2, 1565–1572. [Google Scholar] [CrossRef] [PubMed]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Haouel, A.; Georgii, E.; Priego-Cubero, S.; Wurm, C.J.; Hemmler, D.; Schmitt-Kopplin, P.; Becker, C.; Durner, J.; Lindermayr, C. Histone Deacetylases HD2A and HD2B Undergo Feedback Regulation by ABA and Modulate Drought Tolerance via Mediating ABA-Induced Transcriptional Repression. Genes 2023, 14, 1199. https://doi.org/10.3390/genes14061199
Han Y, Haouel A, Georgii E, Priego-Cubero S, Wurm CJ, Hemmler D, Schmitt-Kopplin P, Becker C, Durner J, Lindermayr C. Histone Deacetylases HD2A and HD2B Undergo Feedback Regulation by ABA and Modulate Drought Tolerance via Mediating ABA-Induced Transcriptional Repression. Genes. 2023; 14(6):1199. https://doi.org/10.3390/genes14061199
Chicago/Turabian StyleHan, Yongtao, Amira Haouel, Elisabeth Georgii, Santiago Priego-Cubero, Christoph J. Wurm, Daniel Hemmler, Philippe Schmitt-Kopplin, Claude Becker, Jörg Durner, and Christian Lindermayr. 2023. "Histone Deacetylases HD2A and HD2B Undergo Feedback Regulation by ABA and Modulate Drought Tolerance via Mediating ABA-Induced Transcriptional Repression" Genes 14, no. 6: 1199. https://doi.org/10.3390/genes14061199
APA StyleHan, Y., Haouel, A., Georgii, E., Priego-Cubero, S., Wurm, C. J., Hemmler, D., Schmitt-Kopplin, P., Becker, C., Durner, J., & Lindermayr, C. (2023). Histone Deacetylases HD2A and HD2B Undergo Feedback Regulation by ABA and Modulate Drought Tolerance via Mediating ABA-Induced Transcriptional Repression. Genes, 14(6), 1199. https://doi.org/10.3390/genes14061199





