Genetic Determinants of Atherogenic Indexes
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Population
2.2. Lipid Indexes and Genetic Analyses
2.3. Regression Models
3. Results
3.1. Population and Lipid Characteristics
3.2. Genotype–Phenotype Associations
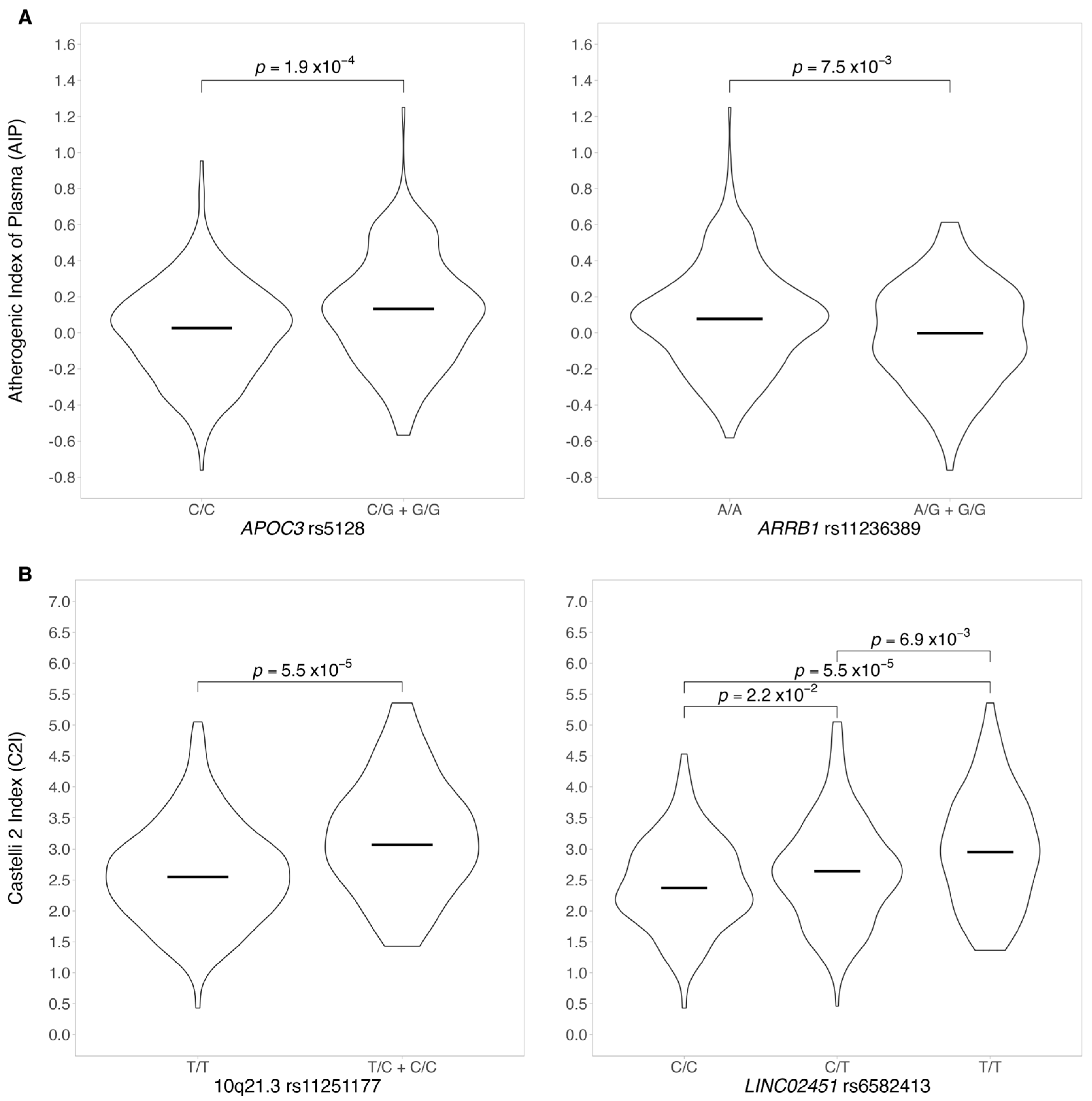
3.2.1. Atherogenic Index of Plasma and AIP Genotype–Phenotype Association
3.2.2. Genotype–Phenotype Association for CI2
4. Discussion
4.1. Genetic Associations with the Atherogenic Index of Plasma, AIP = log(TG/HDLC)
4.2. Genetic Associations with Castelli Index 2, CI2=LDL-C/HDL-C
5. Conclusions
6. Patents
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Lange, L.A.; Lange, E.M.; Bielak, L.F.; Langefeld, C.D.; Kardia, S.L.; Royston, P.; Turner, S.T.; Sheedy, P.F.; Boerwinkle, E.; Peyser, P.A. Autosomal genome-wide scan for coronary artery calcification loci in sibships at high risk for hypertension. Arterioscler. Thromb. Vasc. Biol. 2002, 22, 418–423. [Google Scholar] [CrossRef]
- Herrington, W.; Lacey, B.; Sherliker, P.; Armitage, J.; Lewington, S. Epidemiology of Atherosclerosis and the Potential to Reduce the Global Burden of Atherothrombotic Disease. Circ. Res. 2016, 118, 535–546. [Google Scholar] [CrossRef]
- Araújo, J.A.; Lusis, A.J. Atherosclerosis. In Encyclopedia of Endocrine Diseases; Elsevier: Amsterdam, The Netherlands, 2004; pp. 282–288. [Google Scholar] [CrossRef]
- Scipione, C.A.; Cybulsky, M.I. Early atherogenesis: New insights from new approaches. Curr. Opin. Lipidol. 2022, 33, 271–276. [Google Scholar] [CrossRef]
- Huang, Y.; Gao, L.; Xie, X.; Tan, S.C. Epidemiology of dyslipidemia in Chinese adults: Meta-analysis of prevalence, awareness, treatment, and control. Popul. Health Metr. 2014, 12, 28. [Google Scholar] [CrossRef]
- Bayram, F.; Kocer, D.; Gundogan, K.; Kaya, A.; Demir, O.; Coskun, R.; Sabuncu, T.; Karaman, A.; Cesur, M.; Rizzo, M.; et al. Prevalence of dyslipidemia and associated risk factors in Turkish adults. J. Clin. Lipidol. 2014, 8, 206–216. [Google Scholar] [CrossRef]
- Urina-Jassir, M.; Pacheco-Paez, T.; Paez-Canro, C.; Urina-Triana, M. Statin associated adverse reactions in Latin America: A scoping review. BMJ Open 2021, 11, e050675. [Google Scholar] [CrossRef]
- Pu, J.; Romanelli, R.; Zhao, B.; Azar, K.M.; Hastings, K.G.; Nimbal, V.; Fortmann, S.P.; Palaniappan, L.P. Dyslipidemia in Special Ethnic Populations. Endocrinol. Metab. Clin. N. Am. 2016, 45, 205–216. [Google Scholar] [CrossRef]
- Gao, P.; Wen, X.; Ou, Q.; Zhang, J. Which one of LDL-C /HDL-C ratio and non-HDL-C can better predict the severity of coronary artery disease in STEMI patients. BMC Cardiovasc. Disord. 2022, 22, 318. [Google Scholar] [CrossRef]
- Teslovich, T.M.; Musunuru, K.; Smith, A.V.; Edmondson, A.C.; Stylianou, I.M.; Koseki, M.; Pirruccello, J.P.; Ripatti, S.; Chasman, D.I.; Willer, C.J.; et al. Biological, clinical and population relevance of 95 loci for blood lipids. Nature 2010, 466, 707–713. [Google Scholar] [CrossRef]
- Hindy, G.; Dornbos, P.; Chaffin, M.D.; Liu, D.J.; Wang, M.; Selvaraj, M.S.; Zhang, D.; Park, J.; Aguilar-Salinas, C.A.; Antonacci-Fulton, L.; et al. Rare coding variants in 35 genes associate with circulating lipid levels—A multi-ancestry analysis of 170,000 exomes. Am. J. Hum. Genet. 2022, 109, 81–96. [Google Scholar] [CrossRef]
- Weissglas-Volkov, D.; Aguilar-Salinas, C.A.; Nikkola, E.; Deere, K.A.; Cruz-Bautista, I.; Arellano-Campos, O.; Muñoz-Hernandez, L.L.; Gomez-Munguia, L.; Ordoñez-Sánchez, M.L.; Reddy, P.M.V.L.; et al. Genomic study in Mexicans identifies a new locus for triglycerides and refines European lipid loci. J. Med. Genet. 2013, 50, 298–308. [Google Scholar] [CrossRef] [PubMed]
- Weissglas-Volkov, D.; Aguilar-Salinas, C.A.; Sinsheimer, J.S.; Riba, L.; Huertas-Vazquez, A.; Ordoñez-Sánchez, M.L.; Rodriguez-Guillen, R.; Cantor, R.M.; Tusie-Luna, T.; Pajukanta, P. Investigation of Variants Identified in Caucasian Genome-Wide Association Studies for Plasma High-Density Lipoprotein Cholesterol and Triglycerides Levels in Mexican Dyslipidemic Study Samples. Circ. Cardiovasc. Genet. 2010, 3, 31–38. [Google Scholar] [CrossRef] [PubMed]
- Olamoyegun, M.A.; Oluyombo, R.; Asaolu, S.O. Evaluation of dyslipidemia, lipid ratios, and atherogenic index as cardiovascular risk factors among semi-urban dwellers in Nigeria. Ann. Afr. Med. 2016, 15, 194–199. [Google Scholar] [CrossRef]
- Liu, H.; Liu, K.; Pei, L.; Li, S.; Zhao, J.; Zhang, K.; Zong, C.; Zhao, L.; Fang, H.; Wu, J.; et al. Atherogenic Index of Plasma Predicts Outcomes in Acute Ischemic Stroke. Front. Neurol. 2021, 12, 741754. [Google Scholar] [CrossRef]
- Dobiášová, M.; Frohlich, J. The plasma parameter log (TG/HDL-C) as an atherogenic index: Correlation with lipoprotein particle size and esterification rate inapob-lipoprotein-depleted plasma (FERHDL). Clin. Biochem. 2001, 34, 583–588. [Google Scholar] [CrossRef]
- Rivera-Mancía, S.; Colín-Ramírez, E.; Cartas-Rosado, R.; Infante, O.; Vargas-Barrón, J.; Vallejo, M. Indicators of accumulated fat are stronger associated with prehypertension compared with indicators of circulating fat: A cross-sectional study. Medicine 2018, 97, e11869. [Google Scholar] [CrossRef]
- Colín-Ramírez, E.; Rivera-Mancía, S.; Infante-Vázquez, O.; Cartas-Rosado, R.; Vargas-Barrón, J.; Madero, M.; Vallejo, M. Protocol for a prospective longitudinal study of risk factors for hypertension incidence in a Mexico City population: The Tlalpan 2020 cohort. BMJ Open 2017, 7, e016773. [Google Scholar] [CrossRef]
- Liu, T.; Liu, J.; Wu, Z.; Lv, Y.; Li, W. Predictive value of the atherogenic index of plasma for chronic total occlusion before coronary angiography. Clin. Cardiol. 2021, 44, 518–525. [Google Scholar] [CrossRef]
- Fernández-Macías, J.C.; Ochoa-Martínez, A.C.; Varela-Silva, J.A.; Pérez-Maldonado, I.N. Atherogenic Index of Plasma: Novel Predictive Biomarker for Cardiovascular Illnesses. Arch. Med. Res. 2019, 50, 285–294. [Google Scholar] [CrossRef]
- Koca, T.T.; Tugan, C.B.; Seyithanoglu, M.; Kocyigit, B.F. The Clinical Importance of the Plasma Atherogenic Index, Other Lipid Indexes, and Urinary Sodium and Potassium Excretion in Patients with Stroke. Eurasian J. Med. 2019, 51, 172–176. [Google Scholar] [CrossRef] [PubMed]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. PLINK: A Tool Set for Whole-Genome Association and Population-Based Linkage Analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2015; ISBN 3-900051-07-0. [Google Scholar]
- Turner, S.; Armstrong, L.L.; Bradford, Y.; Carlson, C.S.; Crawford, D.C.; Crenshaw, A.T.; De Andrade, M.; Doheny, K.F.; Haines, J.L.; Hayes, G.; et al. Quality Control Procedures for Genome-Wide Association Studies. Curr. Protoc. Hum. Genet. 2011, 68, 1.19.1–1.19.18. [Google Scholar] [CrossRef]
- Alexander, D.H.; Lange, K. Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinform. 2011, 12, 246. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.; Lee, S.; Min, J.-Y.; Min, K.-B. Association between Serum Lipid Parameters and Cognitive Performance in Older Adults. J. Clin. Med. 2021, 10, 5405. [Google Scholar] [CrossRef] [PubMed]
- Uffelmann, E.; Huang, Q.Q.; Munung, N.S.; de Vries, J.; Okada, Y.; Martin, A.R.; Martin, H.C.; Lappalainen, T.; Posthuma, D. Genome-wide association studies. Nat. Rev. Methods Prim. 2021, 1, 59. [Google Scholar] [CrossRef]
- Jurado-Camacho, P.A.; Cid-Soto, M.A.; Barajas-Olmos, F.; García-Ortíz, H.; Baca-Peynado, P.; Martínez-Hernández, A.; Centeno-Cruz, F.; Contreras-Cubas, C.; González-Villalpando, M.E.; Saldaña-Álvarez, Y.; et al. Exome Sequencing Data Analysis and a Case-Control Study in Mexican Population Reveals Lipid Trait Associations of New and Known Genetic Variants in Dyslipidemia-Associated Loci. Front. Genet. 2022, 13, 807381. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez-Covarrubias, V. Lipidomics in longevity and healthy aging. Biogerontology 2013, 14, 663–672. [Google Scholar] [CrossRef]
- Muñoz, A.; Millán, J.; Pintó, X.; Zúñiga, M.; Rubiés-Prat, J.; Pallardo, L.F.; Masana, L.; Mangas, A.; Hernández-Mijares, A.; González-Santos, P.; et al. Lipoprotein ratios: Physiological significance and clinical usefulness in cardiovascular prevention. Vasc. Health Risk Manag. 2009, 5, 757–765. [Google Scholar] [CrossRef]
- Kammar-García, A.; López-Moreno, P.; Hernández-Hernández, M.E.; Ortíz-Bueno, A.M.; Martínez-Montaño, M.D.L.C. Atherogenic index of plasma as a marker of cardiovascular risk factors in Mexicans aged 18 to 22 years. Proc. (Bayl. Univ. Med. Cent.) 2020, 34, 22–27. [Google Scholar] [CrossRef]
- Afsin, A.; Kaya, H.; Suner, A.; Uzel, K.E.; Bursa, N.; Hosoglu, Y.; Yavuz, F.; Asoglu, R. Plasma atherogenic indices are independent predictors of slow coronary flow. BMC Cardiovasc. Disord. 2021, 21, 608. [Google Scholar] [CrossRef]
- Du, R.; Li, M.; Wang, X.; Wang, S.; Li, S.; Tian, H.; Wu, Y.; Zhang, C. LDL-C/HDL-C ratio associated with carotid intima-media thickness and carotid plaques in male but not female patients with type 2 diabetes. Clin. Chim. Acta 2020, 511, 215–220. [Google Scholar] [CrossRef]
- Ahn, S.S.; Lee, L.E.; Pyo, J.Y.; Song, J.J.; Park, Y.-B.; Lee, S.-W. Atherogenic index of plasma predicts cerebrovascular accident occurrence in antineutrophil cytoplasmic antibody-associated vasculitis. Lipids Health Dis. 2020, 19, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Enomoto, M.; Adachi, H.; Hirai, Y.; Fukami, A.; Satoh, A.; Otsuka, M.; Kumagae, S.-I.; Nanjo, Y.; Yoshikawa, K.; Esaki, E.; et al. LDL-C/HDL-C Ratio Predicts Carotid Intima-Media Thickness Progression Better Than HDL-C or LDL-C Alone. J. Lipids 2011, 2011, 549137. [Google Scholar] [CrossRef] [PubMed]
- Song, Y.; Zhu, L.; Richa, M.; Li, P.; Yang, Y.; Li, S. Associations of the APOC3 rs5128 polymorphism with plasma APOC3 and lipid levels: A meta-analysis. Lipids Health Dis. 2015, 14, 32. [Google Scholar] [CrossRef]
- Wu, J.; Zhou, Q.; Wei, Z.; Wei, J.; Cui, M. Atherogenic Index of Plasma and Coronary Artery Disease in the Adult Population: A Meta-Analysis. Front. Cardiovasc. Med. 2021, 8, 817441. [Google Scholar] [CrossRef]
- Rocco, C.A.; Mecikovsky, D.; Aulicino, P.; Bologna, R.; Sen, L.; Mangano, A. Hypercholesterolemia Is Associated with the Apolipoprotein C-III (APOC3) Genotype in Children Receiving HAART: An Eight-Year Retrospective Study. PLoS ONE 2012, 7, e39678. [Google Scholar] [CrossRef]
- Hsueh, W.-C.; Nair, A.K.; Kobes, S.; Chen, P.; Göring, H.H.; Pollin, T.I.; Malhotra, A.; Knowler, W.C.; Baier, L.J.; Hanson, R.L. Identity-by-Descent Mapping Identifies Major Locus for Serum Triglycerides in Amerindians Largely Explained by an APOC3 Founder Mutation. Circ. Cardiovasc. Genet. 2017, 10, e001809. [Google Scholar] [CrossRef]
- Berg, M.E.v.D.; Warren, H.R.; Cabrera, C.P.; Verweij, N.; Mifsud, B.; Haessler, J.; Bihlmeyer, N.A.; Fu, Y.-P.; Weiss, S.; Lin, H.J.; et al. Discovery of novel heart rate-associated loci using the Exome Chip. Hum. Mol. Genet. 2017, 26, 2346–2363. [Google Scholar] [CrossRef]
- Savarese, M.; Maggi, L.; Vihola, A.; Jonson, P.H.; Tasca, G.; Ruggiero, L.; Bello, L.; Magri, F.; Giugliano, T.; Torella, A.; et al. Interpreting Genetic Variants in Titin in Patients With Muscle Disorders. JAMA Neurol. 2018, 75, 557–565. [Google Scholar] [CrossRef] [PubMed]
- Tharp, C.A.; Haywood, M.E.; Sbaizero, O.; Taylor, M.R.G.; Mestroni, L. The Giant Protein Titin’s Role in Cardiomyopathy: Genetic, Transcriptional, and Post-translational Modifications of TTN and Their Contribution to Cardiac Disease. Front. Physiol. 2019, 10, 1436. [Google Scholar] [CrossRef]
- Ye, D.; Zhou, W.; Hamrick, S.K.; Tester, D.J.; Kim, C.J.; Barajas-Martinez, H.; Hu, D.; Giudicessi, J.R.; Antzelevitch, C.; Ackerman, M.J. Acacetin, a Potent Transient Outward Current Blocker, May Be a Novel Therapeutic for KCND3 -Encoded Kv4.3 Gain-of-Function-Associated J-Wave Syndromes. Circ. Genom. Precis. Med. 2022, 15, 419–428. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Guo, Y.; Shi, H.; Liu, C.-L.; Panganiban, R.A.; Chung, W.; O’Connor, L.J.; Himes, B.E.; Gazal, S.; Hasegawa, K.; et al. Shared genetic and experimental links between obesity-related traits and asthma subtypes in UK Biobank. J. Allergy Clin. Immunol. 2020, 145, 537–549. [Google Scholar] [CrossRef]
- Beigi, F.; Schmeckpeper, J.; Pow-Anpongkul, P.; Payne, J.A.; Zhang, L.; Zhang, Z.; Huang, J.; Mirotsou, M.; Dzau, V.J. C3orf58, a Novel Paracrine Protein, Stimulates Cardiomyocyte Cell-Cycle Progression Through the PI3K–AKT–CDK7 Pathway. Circ. Res. 2013, 113, 372–380. [Google Scholar] [CrossRef] [PubMed]
- Yano, Y.; Butler, K.R.; Hall, M.E.; Schwartz, G.L.; Knopman, D.S.; Lirette, S.T.; Jones, D.W.; Wilson, J.G.; Hall, J.E.; Correa, A.; et al. Associations of Nocturnal Blood Pressure With Cognition by Self-Identified Race in Middle-Aged and Older Adults: The GENOA (Genetic Epidemiology Network of Arteriopathy) Study. J. Am. Heart Assoc. 2017, 6, e007022. [Google Scholar] [CrossRef] [PubMed]
- Zanni, G.; Hsiao, C.-T.; Fu, S.-J.; Tang, C.-Y.; Capuano, A.; Bosco, L.; Graziola, F.; Bellacchio, E.; Servidei, S.; Primiano, G.; et al. Novel KCND3 Variant Underlying Nonprogressive Congenital Ataxia or SCA19/22 Disrupt KV4.3 Protein Expression and K+ Currents with Variable Effects on Channel Properties. Int. J. Mol. Sci. 2021, 22, 4986. [Google Scholar] [CrossRef]
- Pollini, L.; Galosi, S.; Tolve, M.; Caputi, C.; Carducci, C.; Angeloni, A.; Leuzzi, V. KCND3-Related Neurological Disorders: From Old to Emerging Clinical Phenotypes. Int. J. Mol. Sci. 2020, 21, 5802. [Google Scholar] [CrossRef]
- Feitosa, M.F.; Myers, R.H.; Pankow, J.S.; Province, M.A.; Borecki, I.B. LIPC variants in the promoter and intron 1 modify HDL-C levels in a sex-specific fashion. Atherosclerosis 2009, 204, 171–177. [Google Scholar] [CrossRef]
- Talmud, P.J.; Drenos, F.; Shah, S.; Shah, T.; Palmen, J.; Verzilli, C.; Gaunt, T.R.; Pallas, J.; Lovering, R.; Li, K.; et al. Gene-centric Association Signals for Lipids and Apolipoproteins Identified via the HumanCVD BeadChip. Am. J. Hum. Genet. 2009, 85, 628–642. [Google Scholar] [CrossRef]
- Boyle, A.P.; Hong, E.L.; Hariharan, M.; Cheng, Y.; Schaub, M.A.; Kasowski, M.; Karczewski, K.J.; Park, J.; Hitz, B.C.; Weng, S.; et al. Annotation of functional variation in personal genomes using RegulomeDB. Genome Res. 2012, 22, 1790–1797. [Google Scholar] [CrossRef]
| Males N = 170 | Females N = 256 | All N = 426 | |
|---|---|---|---|
| Age, y | 38 (18–53) | 40 (17–52) | 39 (17–53) |
| Weight, kg | 78.1 (51.2–125) | 63.1 (41.7–119) | 70 (41.7–125) |
| Height, m | 1.70 (1.50–1.99) | 1.57 (1.36–1.72) | 1.61 (1.36–1.99) |
| BMI, kg/m2 | 26.8 (16.9–40.3) | 26.2 (16.9–47.1) | 26.4 (16.8–47.1) |
| Waist circumference, cm | 94.0 (63.0–130) | 85.0 (54.0–126) | 89.0 (54.0–130) |
| Glucose, mg/dL | 94.0 (72.0–166) | 90.0 (74.0–241) | 92.0 (72.0–241) |
| Uric acid, mg/dL | 6.34 (1.82–10.0) | 4.62 (2.30–7.58) | 5.31 (1.82–10.0) |
| Creatinine, mg/dL | 0.95 (0.62–1.40) | 0.69 (0.44–1.19) | 0.77 (0.44–1.40) |
| Cholesterol, mmol/dL | 4.62 (2.96–8.30) | 4.39 (2.16–7.06) | 4.49 (2.16–8.30) |
| HDL-C, mmol/dL | 1.10 (0.60–2.12) | 1.22 (0.73–2.27) | 1.16 (0.60–2.27) |
| LDL-C, mmol/dL | 3.09 (0.98–6.87) | 2.93 (0.54–5.38) | 3.00 (0.54–6.87) |
| Triglycerides (TG), mmol/dL | 1.48 (0.47–15.4) | 1.22 (0.22–5.86) | 1.32 (0.22–15.34) |
| Dyslipidemia, n (%) | 119 (70%) | 85 (72%) | 304 (71%) |
| Castelli risk index 2 (CI2) 1 | 2.91 (1.25–5.36) | 2.45 (0.43–4.92) | 2.60 (0.43–5.36) |
| Atherogenic index of plasma (AIP) 2 | 0.48 (−0.18–1.61) | 0.38 (−0.40–1.17) | 0.42 (−0.40–1.61) |
| High TG > 1.9 mmol/L | High Cholesterol > 5 mmol/L | High LDL-C > 3.9 mmol/L | Low HDL-C < 1.04 mmol/L | |
|---|---|---|---|---|
| 1 Males % | 30%, n = 51 | 35.3%, n = 60 | 16.5%, n = 28 | 38%, n = 65 |
| 1 Females % | 16%, n = 41 | 22.3%, n = 59 | 10.9%, n = 28 | 26%, n = 67 |
| p-value 2 | 8.74 × 10−6 | 2.32 × 10−3 | 1.80 × 10−3 | 2.59 × 10−5 |
| Gene | Chr | rs Identifier | Coefficient | p-Value |
|---|---|---|---|---|
| Variants associated with AIP | ||||
| APOA1/APOC3 | 11 | rs5128, C > G | 0.094 | 2.61 × 10−6 |
| CYBA | 16 | rs12709102, T > C | 0.078 | 3.91 × 10−6 |
| ARRIB1 | 11 | rs11236389, A > G | −0.102 | 6.63 × 10−6 |
| TTN/CCDC141 | 2 | rs10497528, A > C | 0.089 | 8.29 × 10−6 |
| KCND3 | 1 | rs6703437 | −0.177 | 0.90 × 10−6 |
| APOA1/APOC3 | 11 | rs5072, G > A | 0.091 | 8.94 × 10−6 |
| Variants associated with CI2 | ||||
| Intergenic | 10q21.3 | rs11251177, T > C | 0.606 | 1.07 × 10−7 |
| LINC02451 | 12 | rs6582413, T > C | 0.259 | 5.19 × 10−7 |
| LINC02451 | 12 | rs12817366, C > T | 0.254 | 1.88 × 10−6 |
| Intergenic | 12 | rs34115639, C > T | 0.244 | 7.06 × 10−6 |
| Intergenic | 12 | rs10880344, T > C | −0.233 | 7.10 × 10−6 |
| Intergenic | 6 | rs7762658, C > T | −0.247 | 2.03 × 10−6 |
| LIPC/ALDH1A2 | 15 | rs261342, C > G | 0.227 | 1.10 × 10−6 |
| DIPK2B | 23 | rs4294309, A > G | 0.306 | 1.18 × 10−5 |
| KCND3 | 1 | rs6703437, G > A | −0.234 | 1.76 × 10−5 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Texis, T.; Rivera-Mancía, S.; Colín-Ramírez, E.; Cartas-Rosado, R.; Koepsell, D.; Rubio-Carrasco, K.; Rodríguez-Dorantes, M.; Gonzalez-Covarrubias, V. Genetic Determinants of Atherogenic Indexes. Genes 2023, 14, 1214. https://doi.org/10.3390/genes14061214
Texis T, Rivera-Mancía S, Colín-Ramírez E, Cartas-Rosado R, Koepsell D, Rubio-Carrasco K, Rodríguez-Dorantes M, Gonzalez-Covarrubias V. Genetic Determinants of Atherogenic Indexes. Genes. 2023; 14(6):1214. https://doi.org/10.3390/genes14061214
Chicago/Turabian StyleTexis, Tomas, Susana Rivera-Mancía, Eloisa Colín-Ramírez, Raul Cartas-Rosado, David Koepsell, Kenneth Rubio-Carrasco, Mauricio Rodríguez-Dorantes, and Vanessa Gonzalez-Covarrubias. 2023. "Genetic Determinants of Atherogenic Indexes" Genes 14, no. 6: 1214. https://doi.org/10.3390/genes14061214
APA StyleTexis, T., Rivera-Mancía, S., Colín-Ramírez, E., Cartas-Rosado, R., Koepsell, D., Rubio-Carrasco, K., Rodríguez-Dorantes, M., & Gonzalez-Covarrubias, V. (2023). Genetic Determinants of Atherogenic Indexes. Genes, 14(6), 1214. https://doi.org/10.3390/genes14061214





