Efficient and Specific Generation of MSTN-Edited Hu Sheep Using C-CRISPR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Preparation of Cas9 mRNA and sgRNA
2.3. Manipulation of Sheep Embryos
2.4. Detection of Genome Editing at the Target Sites and POTSs
2.5. Western Blotting and Histological Analysis
2.6. Data Analysis
3. Results
3.1. Efficient Generation of MSTN-Edited Hu Sheep Using C-CRISPR
3.2. Specificity of Genome Editing Using C-CRISPR
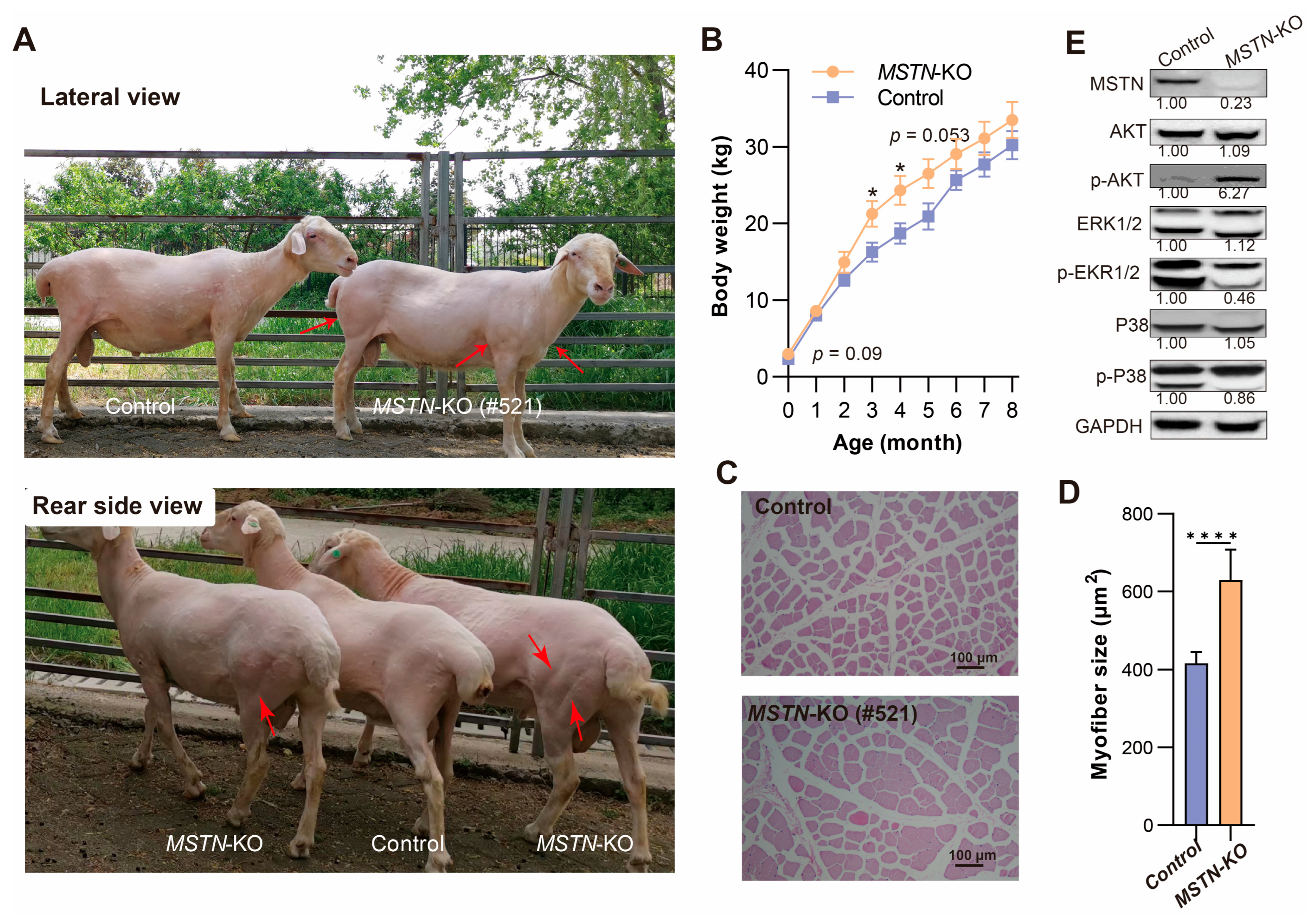
3.3. Phenotype Analysis of MSTN-Edited Hu Sheep
3.4. Molecular Analysis of MSTN-Edited Hu Sheep
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- CRISPR beef cattle get FDA green light. Nat. Biotechnol. 2022, 40, 448. [CrossRef] [PubMed]
- Zhang, Y.; Guo, J.; Yao, X.; Wang, F. Research progress on regulation mechanism of reproductive traits of Hu sheep. J. Nanjing Agric. Univ. 2022, 45, 1032–1040. [Google Scholar]
- Chebo, C.; Betsha, S.; Melesse, A. Chicken genetic diversity, improvement strategies and impacts on egg productivity in Ethiopia: A review. Worlds Poult. Sci. J. 2022, 78, 803–821. [Google Scholar] [CrossRef]
- Cong, L.; Ran, F.A.; Cox, D.; Lin, S.; Barretto, R.; Habib, N.; Hsu, P.D.; Wu, X.; Jiang, W.; Marraffini, L.A.; et al. Multiplex Genome Engineering Using CRISPR/Cas Systems. Science 2013, 339, 819–823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [Green Version]
- Bogdanovich, S.; Krag, T.O.B.; Barton, E.R.; Morris, L.D.; Whittemore, L.-A.; Ahima, R.S.; Khurana, T.S. Functional improvement of dystrophic muscle by myostatin blockade. Nature 2002, 420, 418–421. [Google Scholar] [CrossRef]
- Schuelke, M.; Wagner, K.R.; Stolz, L.E.; Hübner, C.; Riebel, T.; Kömen, W.; Braun, T.; Tobin, J.F.; Lee, S.J. Myostatin mutation associated with gross muscle hypertrophy in a child. N. Engl. J. Med. 2004, 350, 2682–2688. [Google Scholar] [CrossRef] [Green Version]
- Boman, I.A.; Klemetsdal, G.; Nafstad, O.; Blichfeldt, T.; Våge, D.I. Selection based on progeny testing induces rapid changes in myostatin allele frequencies—A case study in sheep. J. Anim. Breed. Genet. Z. Tierz. Zucht. 2011, 128, 52–55. [Google Scholar] [CrossRef]
- Clop, A.; Marcq, F.; Takeda, H.; Pirottin, D.; Tordoir, X.; Bibé, B.; Bouix, J.; Caiment, F.; Elsen, J.M.; Eychenne, F.; et al. A mutation creating a potential illegitimate microRNA target site in the myostatin gene affects muscularity in sheep. Nat. Genet. 2006, 38, 813–818. [Google Scholar] [CrossRef]
- Boman, I.A.; Klemetsdal, G.; Blichfeldt, T.; Nafstad, O.; Våge, D.I. A frameshift mutation in the coding region of the myostatin gene (MSTN) affects carcass conformation and fatness in Norwegian White Sheep (Ovis aries). Anim. Genet. 2009, 40, 418–422. [Google Scholar] [CrossRef]
- Grobet, L.; Martin, L.J.; Poncelet, D.; Pirottin, D.; Brouwers, B.; Riquet, J.; Schoeberlein, A.; Dunner, S.; Ménissier, F.; Massabanda, J.; et al. A deletion in the bovine myostatin gene causes the double-muscled phenotype in cattle. Nat. Genet. 1997, 17, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Kambadur, R.; Sharma, M.; Smith, T.P.; Bass, J.J. Mutations in myostatin (GDF8) in double-muscled Belgian Blue and Piedmontese cattle. Genome Res. 1997, 7, 910–916. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McPherron, A.C.; Lee, S.J. Double muscling in cattle due to mutations in the myostatin gene. Proc. Natl. Acad. Sci. USA 1997, 94, 12457–12461. [Google Scholar] [CrossRef] [Green Version]
- Matika, O.; Robledo, D.; Pong-Wong, R.; Bishop, S.C.; Riggio, V.; Finlayson, H.; Lowe, N.R.; Hoste, A.E.; Walling, G.A.; Del-Pozo, J.; et al. Balancing selection at a premature stop mutation in the myostatin gene underlies a recessive leg weakness syndrome in pigs. PLoS Genet. 2019, 15, e1007759. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mosher, D.S.; Quignon, P.; Bustamante, C.D.; Sutter, N.B.; Mellersh, C.S.; Parker, H.G.; Ostrander, E.A. A mutation in the myostatin gene increases muscle mass and enhances racing performance in heterozygote dogs. PLoS Genet. 2007, 3, e79. [Google Scholar] [CrossRef]
- Boman, I.A.; Klemetsdal, G.; Nafstad, O.; Blichfeldt, T.; Våge, D.I. Impact of two myostatin (MSTN) mutations on weight gain and lamb carcass classification in Norwegian White Sheep (Ovis aries). Genet. Sel. Evol. GSE 2010, 42, 4. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Yu, H.; Lei, A.; Zhou, J.; Zeng, W.; Zhu, H.; Dong, Z.; Niu, Y.; Shi, B.; Cai, B.; et al. Generation of gene-modified goats targeting MSTN and FGF5 via zygote injection of CRISPR/Cas9 system. Sci. Rep. 2015, 5, 13878. [Google Scholar] [CrossRef] [Green Version]
- Guo, R.; Wan, Y.; Xu, D.; Cui, L.; Deng, M.; Zhang, G.; Jia, R.; Zhou, W.; Wang, Z.; Deng, K.; et al. Generation and evaluation of Myostatin knock-out rabbits and goats using CRISPR/Cas9 system. Sci. Rep. 2016, 6, 29855. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Wang, G.; Hao, Z.; Zhang, G.; Qing, Y.; Liu, S.; Qing, L.; Pan, W.; Chen, L.; Liu, G.; et al. Generation of biallelic knock-out sheep via gene-editing and somatic cell nuclear transfer. Sci. Rep. 2016, 6, 33675. [Google Scholar] [CrossRef] [Green Version]
- Proudfoot, C.; Carlson, D.F.; Huddart, R.; Long, C.R.; Pryor, J.H.; King, T.J.; Lillico, S.G.; Mileham, A.J.; McLaren, D.G.; Whitelaw, C.B.; et al. Genome edited sheep and cattle. Transgenic Res. 2015, 24, 147–153. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Niu, Y.; Zhou, J.; Yu, H.; Kou, Q.; Lei, A.; Zhao, X.; Yan, H.; Cai, B.; Shen, Q.; et al. Multiplex gene editing via CRISPR/Cas9 exhibits desirable muscle hypertrophy without detectable off-target effects in sheep. Sci. Rep. 2016, 6, 32271. [Google Scholar] [CrossRef] [PubMed]
- Bi, Y.; Hua, Z.; Liu, X.; Hua, W.; Ren, H.; Xiao, H.; Zhang, L.; Li, L.; Wang, Z.; Laible, G.; et al. Isozygous and selectable marker-free MSTN knockout cloned pigs generated by the combined use of CRISPR/Cas9 and Cre/LoxP. Sci. Rep. 2016, 6, 31729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ren, H.; Xiao, W.; Qin, X.; Cai, G.; Chen, H.; Hua, Z.; Cheng, C.; Li, X.; Hua, W.; Xiao, H.; et al. Myostatin regulates fatty acid desaturation and fat deposition through MEF2C/miR222/SCD5 cascade in pigs. Commun. Biol. 2020, 3, 612. [Google Scholar] [CrossRef] [PubMed]
- Fan, Z.; Liu, Z.; Xu, K.; Wu, T.; Ruan, J.; Zheng, X.; Bao, S.; Mu, Y.; Sonstegard, T.; Li, K. Long-term, multidomain analyses to identify the breed and allelic effects in MSTN-edited pigs to overcome lameness and sustainably improve nutritional meat production. Sci. China Life Sci. 2022, 65, 362–375. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Kalds, P.; Luo, Q.; Sun, K.; Zhao, X.; Gao, Y.; Cai, B.; Huang, S.; Kou, Q.; Petersen, B.; et al. Optimized Cas9:sgRNA delivery efficiently generates biallelic MSTN knockout sheep without affecting meat quality. BMC Genom. 2022, 23, 348. [Google Scholar] [CrossRef]
- Tait-Burkard, C.; Doeschl-Wilson, A.; McGrew, M.J.; Archibald, A.L.; Sang, H.M.; Houston, R.D.; Whitelaw, C.B.; Watson, M. Livestock 2.0—Genome editing for fitter, healthier, and more productive farmed animals. Genome Biol. 2018, 19, 204. [Google Scholar] [CrossRef] [Green Version]
- Kalds, P.; Gao, Y.; Zhou, S.; Cai, B.; Huang, X.; Wang, X.; Chen, Y. Redesigning small ruminant genomes with CRISPR toolkit: Overview and perspectives. Theriogenology 2020, 147, 25–33. [Google Scholar] [CrossRef]
- Niu, Y.; Shen, B.; Cui, Y.; Chen, Y.; Wang, J.; Wang, L.; Kang, Y.; Zhao, X.; Si, W.; Li, W.; et al. Generation of gene-modified cynomolgus monkey via Cas9/RNA-mediated gene targeting in one-cell embryos. Cell 2014, 156, 836–843. [Google Scholar] [CrossRef] [Green Version]
- Dingwei, P.; Ruiqiang, L.; Wu, Z.; Min, W.; Xuan, S.; Zeng, J.; Liu, X.; Chen, Y.; He, Z. Editing the cystine knot motif of MSTN enhances muscle development of Liang Guang Small Spotted pigs. Hereditas 2021, 43, 261–270. [Google Scholar] [CrossRef]
- Li, R.; Zeng, W.; Ma, M.; Wei, Z.; Liu, H.; Liu, X.; Wang, M.; Shi, X.; Zeng, J.; Yang, L.; et al. Precise editing of myostatin signal peptide by CRISPR/Cas9 increases the muscle mass of Liang Guang Small Spotted pigs. Transgenic Res. 2020, 29, 149–163. [Google Scholar] [CrossRef]
- Zhu, X.X.; Zhan, Q.M.; Wei, Y.Y.; Yan, A.F.; Feng, J.; Liu, L.; Lu, S.S.; Tang, D.S. CRISPR/Cas9-mediated MSTN disruption accelerates the growth of Chinese Bama pigs. Reprod. Domest. Anim. Zuchthyg. 2020, 55, 1314–1327. [Google Scholar] [CrossRef] [PubMed]
- Moro, L.N.; Viale, D.L.; Bastón, J.I.; Arnold, V.; Suvá, M.; Wiedenmann, E.; Olguín, M.; Miriuka, S.; Vichera, G. Generation of myostatin edited horse embryos using CRISPR/Cas9 technology and somatic cell nuclear transfer. Sci. Rep. 2020, 10, 15587. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Ouyang, H.; Xie, Z.; Yao, C.; Guo, N.; Li, M.; Jiao, H.; Pang, D. Efficient Generation of Myostatin Mutations in Pigs Using the CRISPR/Cas9 System. Sci. Rep. 2015, 5, 16623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zuo, E.; Cai, Y.J.; Li, K.; Wei, Y.; Wang, B.A.; Sun, Y.; Liu, Z.; Liu, J.; Hu, X.; Wei, W.; et al. One-step generation of complete gene knockout mice and monkeys by CRISPR/Cas9-mediated gene editing with multiple sgRNAs. Cell Res. 2017, 27, 933–945. [Google Scholar] [CrossRef]
- Bae, S.; Park, J.; Kim, J.S. Cas-OFFinder: A fast and versatile algorithm that searches for potential off-target sites of Cas9 RNA-guided endonucleases. Bioinformatics 2014, 30, 1473–1475. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Guo, R.; Deng, M.; Liu, Z.; Pang, J.; Zhang, G.; Wang, Z.; Wang, F. Efficient generation of CLPG1-edited rabbits using the CRISPR/Cas9 system. Reprod. Domest. Anim. Zuchthyg. 2019, 54, 538–544. [Google Scholar] [CrossRef] [PubMed]
- Kilian, N.; Goryaynov, A.; Lessard, M.D.; Hooker, G.; Toomre, D.; Rothman, J.E.; Bewersdorf, J. Assessing photodamage in live-cell STED microscopy. Nat. Methods 2018, 15, 755–756. [Google Scholar] [CrossRef]
- Iyer, V.; Boroviak, K.; Thomas, M.; Doe, B.; Riva, L.; Ryder, E.; Adams, D.J. No unexpected CRISPR-Cas9 off-target activity revealed by trio sequencing of gene-edited mice. PLoS Genet. 2018, 14, e1007503. [Google Scholar] [CrossRef]
- Wilson, C.J.; Fennell, T.; Bothmer, A.; Maeder, M.L.; Reyon, D.; Cotta-Ramusino, C.; Fernandez, C.A.; Marco, E.; Barrera, L.A.; Jayaram, H.; et al. Response to “Unexpected mutations after CRISPR-Cas9 editing in vivo”. Nat. Methods 2018, 15, 236–237. [Google Scholar] [CrossRef]
- Smith, C.; Gore, A.; Yan, W.; Abalde-Atristain, L.; Li, Z.; He, C.; Wang, Y.; Brodsky, R.A.; Zhang, K.; Cheng, L.; et al. Whole-genome sequencing analysis reveals high specificity of CRISPR/Cas9 and TALEN-based genome editing in human iPSCs. Cell Stem Cell 2014, 15, 12–13. [Google Scholar] [CrossRef] [Green Version]
- Suzuki, K.; Yu, C.; Qu, J.; Li, M.; Yao, X.; Yuan, T.; Goebl, A.; Tang, S.; Ren, R.; Aizawa, E.; et al. Targeted gene correction minimally impacts whole-genome mutational load in human-disease-specific induced pluripotent stem cell clones. Cell Stem Cell 2014, 15, 31–36. [Google Scholar] [CrossRef] [Green Version]
- Veres, A.; Gosis, B.S.; Ding, Q.; Collins, R.; Ragavendran, A.; Brand, H.; Erdin, S.; Cowan, C.A.; Talkowski, M.E.; Musunuru, K. Low incidence of off-target mutations in individual CRISPR-Cas9 and TALEN targeted human stem cell clones detected by whole-genome sequencing. Cell Stem Cell 2014, 15, 27–30. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aryal, N.K.; Wasylishen, A.R.; Lozano, G. CRISPR/Cas9 can mediate high-efficiency off-target mutations in mice in vivo. Cell Death Dis. 2018, 9, 1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iyer, V.; Shen, B.; Zhang, W.; Hodgkins, A.; Keane, T.; Huang, X.; Skarnes, W.C. Off-target mutations are rare in Cas9-modified mice. Nat. Methods 2015, 12, 479. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Liu, J.; Niu, Y.; Li, Y.; Zhou, S.; Li, C.; Ma, B.; Kou, Q.; Petersen, B.; Sonstegard, T.; et al. Low incidence of SNVs and indels in trio genomes of Cas9-mediated multiplex edited sheep. BMC Genom. 2018, 19, 397. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Zhou, H.; Hu, J.; Li, S.; Luo, Y.; Hickford, J.G. Two single nucleotide polymorphisms in the promoter of the ovine myostatin gene (MSTN) and their effect on growth and carcass muscle traits in New Zealand Romney sheep. J. Anim. Breed. Genet. Z. Tierz. Zucht. 2016, 133, 219–226. [Google Scholar] [CrossRef]
- Crispo, M.; Mulet, A.P.; Tesson, L.; Barrera, N.; Cuadro, F.; dos Santos-Neto, P.C.; Nguyen, T.H.; Creneguy, A.; Brusselle, L.; Anegon, I.; et al. Efficient Generation of Myostatin Knock-Out Sheep Using CRISPR/Cas9 Technology and Microinjection into Zygotes. PLoS ONE 2015, 10, e0136690. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Petersen, B. More abundant and healthier meat: Will the MSTN editing epitome empower the commercialization of gene editing in livestock? Sci. China Life Sci. 2022, 65, 448–450. [Google Scholar] [CrossRef]
- McCroskery, S.; Thomas, M.; Maxwell, L.; Sharma, M.; Kambadur, R. Myostatin negatively regulates satellite cell activation and self-renewal. J. Cell Biol. 2003, 162, 1135–1147. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.; Langley, B.; Berry, C.; Sharma, M.; Kirk, S.; Bass, J.; Kambadur, R. Myostatin, a negative regulator of muscle growth, functions by inhibiting myoblast proliferation. J. Biol. Chem. 2000, 275, 40235–40243. [Google Scholar] [CrossRef] [Green Version]
- Sartori, R.; Milan, G.; Patron, M.; Mammucari, C.; Blaauw, B.; Abraham, R.; Sandri, M. Smad2 and 3 transcription factors control muscle mass in adulthood. Am. J. Physiol. Cell Physiol. 2009, 296, C1248–C1257. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Amirouche, A.; Durieux, A.C.; Banzet, S.; Koulmann, N.; Bonnefoy, R.; Mouret, C.; Bigard, X.; Peinnequin, A.; Freyssenet, D. Down-regulation of Akt/mammalian target of rapamycin signaling pathway in response to myostatin overexpression in skeletal muscle. Endocrinology 2009, 150, 286–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trendelenburg, A.U.; Meyer, A.; Rohner, D.; Boyle, J.; Hatakeyama, S.; Glass, D.J. Myostatin reduces Akt/TORC1/p70S6K signaling, inhibiting myoblast differentiation and myotube size. Am. J. Physiol. Cell Physiol. 2009, 296, C1258–C1270. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morissette, M.R.; Cook, S.A.; Buranasombati, C.; Rosenberg, M.A.; Rosenzweig, A. Myostatin inhibits IGF-I-induced myotube hypertrophy through Akt. Am. J. Physiol. Cell Physiol. 2009, 297, C1124–C1132. [Google Scholar] [CrossRef] [Green Version]
- Philip, B.; Lu, Z.; Gao, Y. Regulation of GDF-8 signaling by the p38 MAPK. Cell Signal. 2005, 17, 365–375. [Google Scholar] [CrossRef]
- Yang, W.; Chen, Y.; Zhang, Y.; Wang, X.; Yang, N.; Zhu, D. Extracellular signal-regulated kinase 1/2 mitogen-activated protein kinase pathway is involved in myostatin-regulated differentiation repression. Cancer Res. 2006, 66, 1320–1326. [Google Scholar] [CrossRef] [Green Version]
- Lipina, C.; Kendall, H.; McPherron, A.C.; Taylor, P.M.; Hundal, H.S. Mechanisms involved in the enhancement of mammalian target of rapamycin signalling and hypertrophy in skeletal muscle of myostatin-deficient mice. FEBS Lett. 2010, 584, 2403–2408. [Google Scholar] [CrossRef] [Green Version]
- Fang, L.; Chang, H.M.; Cheng, J.C.; Yu, Y.; Leung, P.C.; Sun, Y.P. Growth Differentiation Factor-8 Decreases StAR Expression Through ALK5-Mediated Smad3 and ERK1/2 Signaling Pathways in Luteinized Human Granulosa Cells. Endocrinology 2015, 156, 4684–4694. [Google Scholar] [CrossRef] [Green Version]
- Pan, S.; Zhang, L.; Liu, Z.; Xing, H. Myostatin suppresses adipogenic differentiation and lipid accumulation by activating crosstalk between ERK1/2 and PKA signaling pathways in porcine subcutaneous preadipocytes. J. Anim. Sci. 2021, 99, skab287. [Google Scholar] [CrossRef]
- Li, F.; Yang, H.; Duan, Y.; Yin, Y. Myostatin regulates preadipocyte differentiation and lipid metabolism of adipocyte via ERK1/2. Cell Biol. Int. 2011, 35, 1141–1146. [Google Scholar] [CrossRef]

| sgRNAs | Donors Superovulated | Embryo Injected/Embryos Transferred | Delivered Recipients /Total Recipients | Edited Lambs /Total Lambs |
|---|---|---|---|---|
| sgC1–C4 | 5 | 83/70 | 5/13 (38.46%) | 9/10 (90.00%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, R.; Wang, H.; Meng, C.; Gui, H.; Li, Y.; Chen, F.; Zhang, C.; Zhang, H.; Ding, Q.; Zhang, J.; et al. Efficient and Specific Generation of MSTN-Edited Hu Sheep Using C-CRISPR. Genes 2023, 14, 1216. https://doi.org/10.3390/genes14061216
Guo R, Wang H, Meng C, Gui H, Li Y, Chen F, Zhang C, Zhang H, Ding Q, Zhang J, et al. Efficient and Specific Generation of MSTN-Edited Hu Sheep Using C-CRISPR. Genes. 2023; 14(6):1216. https://doi.org/10.3390/genes14061216
Chicago/Turabian StyleGuo, Rihong, Huili Wang, Chunhua Meng, Hongbing Gui, Yinxia Li, Fang Chen, Chenjian Zhang, Han Zhang, Qiang Ding, Jianli Zhang, and et al. 2023. "Efficient and Specific Generation of MSTN-Edited Hu Sheep Using C-CRISPR" Genes 14, no. 6: 1216. https://doi.org/10.3390/genes14061216





