High-Resolution Melting (HRM) Analysis for Rapid Molecular Identification of Sparidae Species in the Greek Fish Market
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples Collection and Morphological Identification
2.2. DNA Extraction and PCR
2.3. Sanger Sequencing
2.4. In Silico Analysis and Detection of Polymorphisms
2.5. Restriction Fragment Length Polymorphism (RFLP)
2.6. Multiplex PCR
2.7. HRM Analysis
3. Results
3.1. Morphological Identification and DNA Isolation
3.2. Sequencing-Based Identification—COI Barcoding
3.2.1. P. pagrus
3.2.2. D. dentex
3.2.3. Other Sparidae Species
3.3. Non-Sequencing-Based Techniques
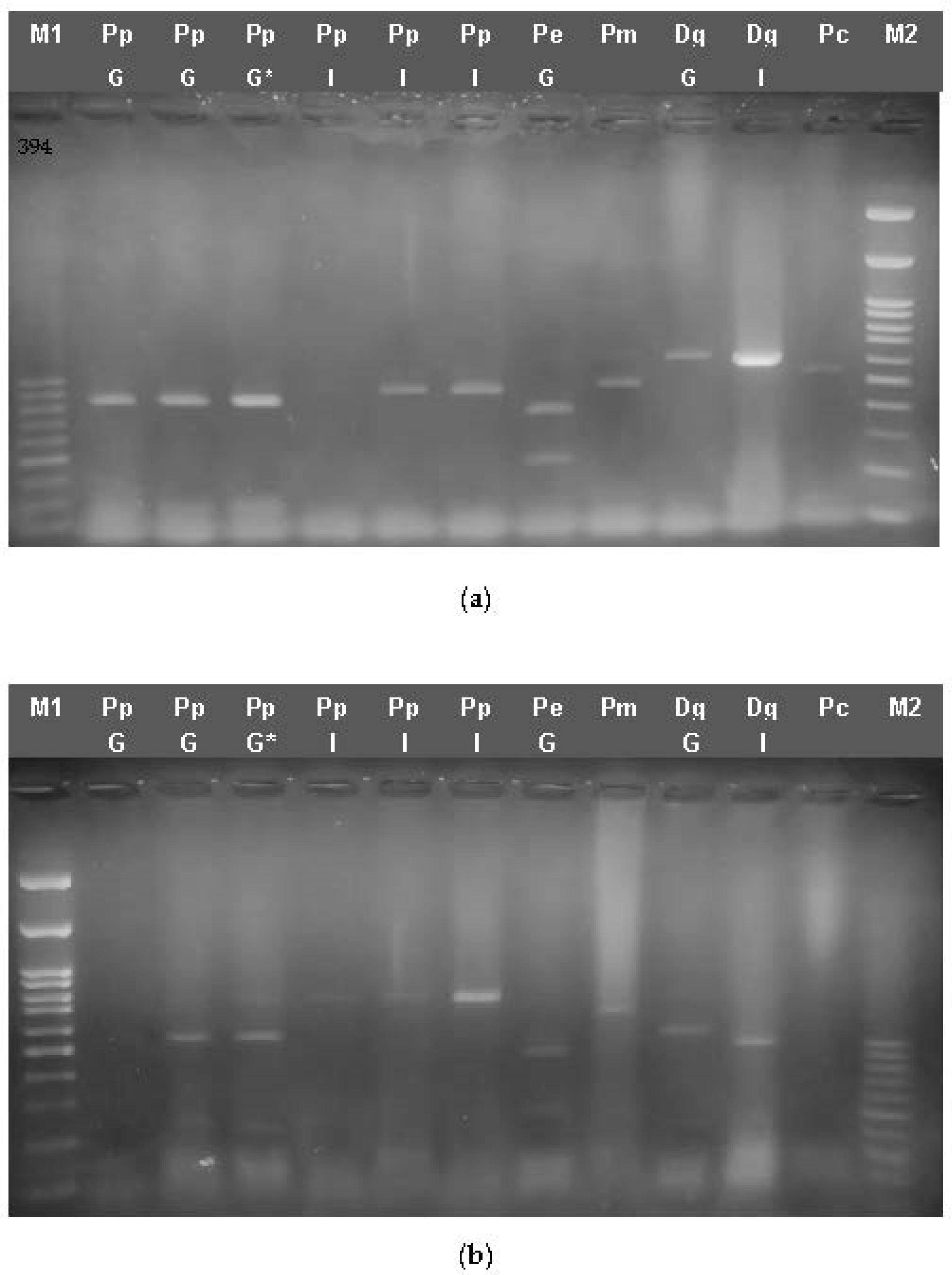
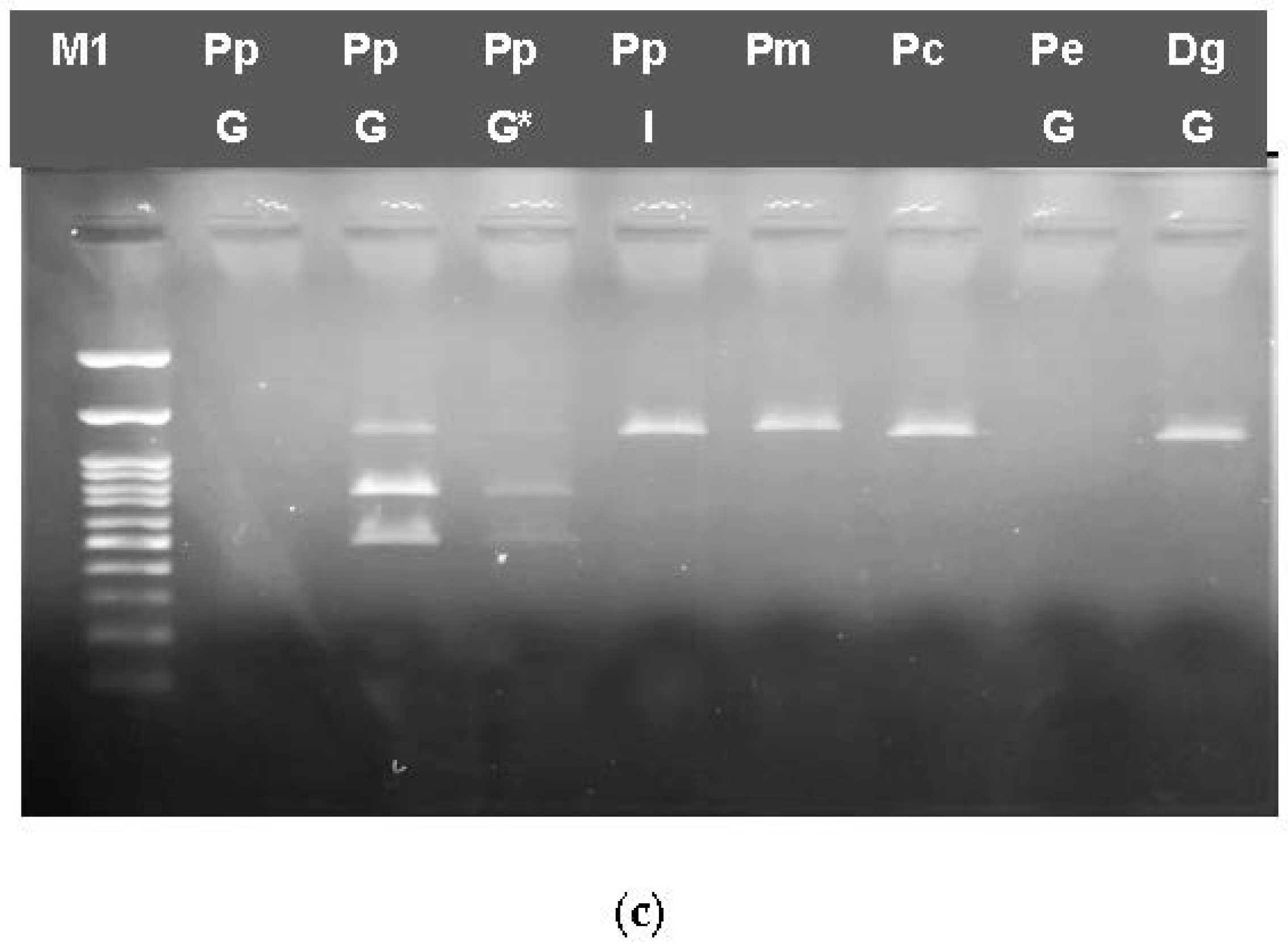
3.3.1. RFLP Analysis
3.3.2. Multiplex PCR
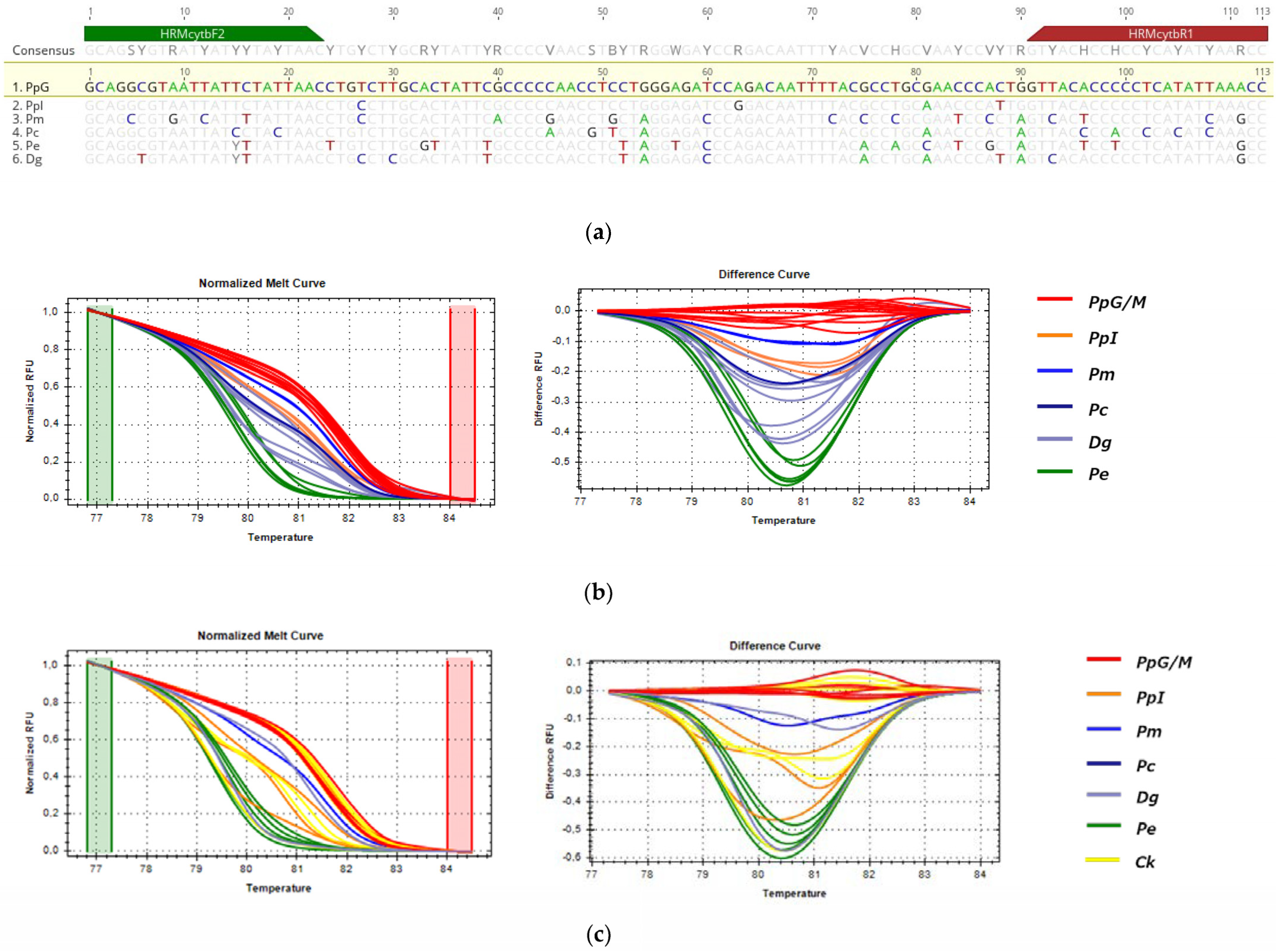
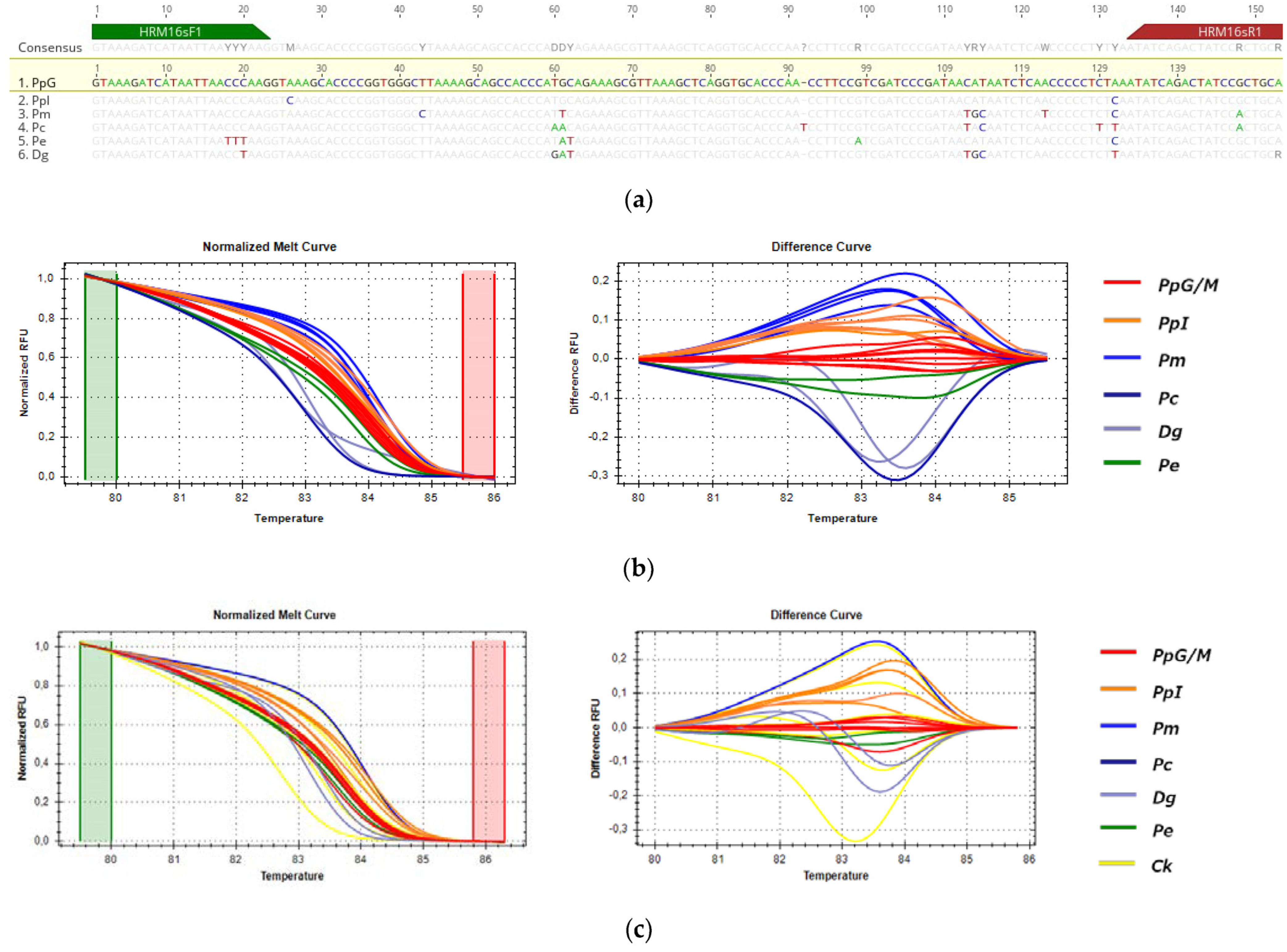
3.3.3. HRM Analysis for the Detection of P. pagrus from Greek Fisheries
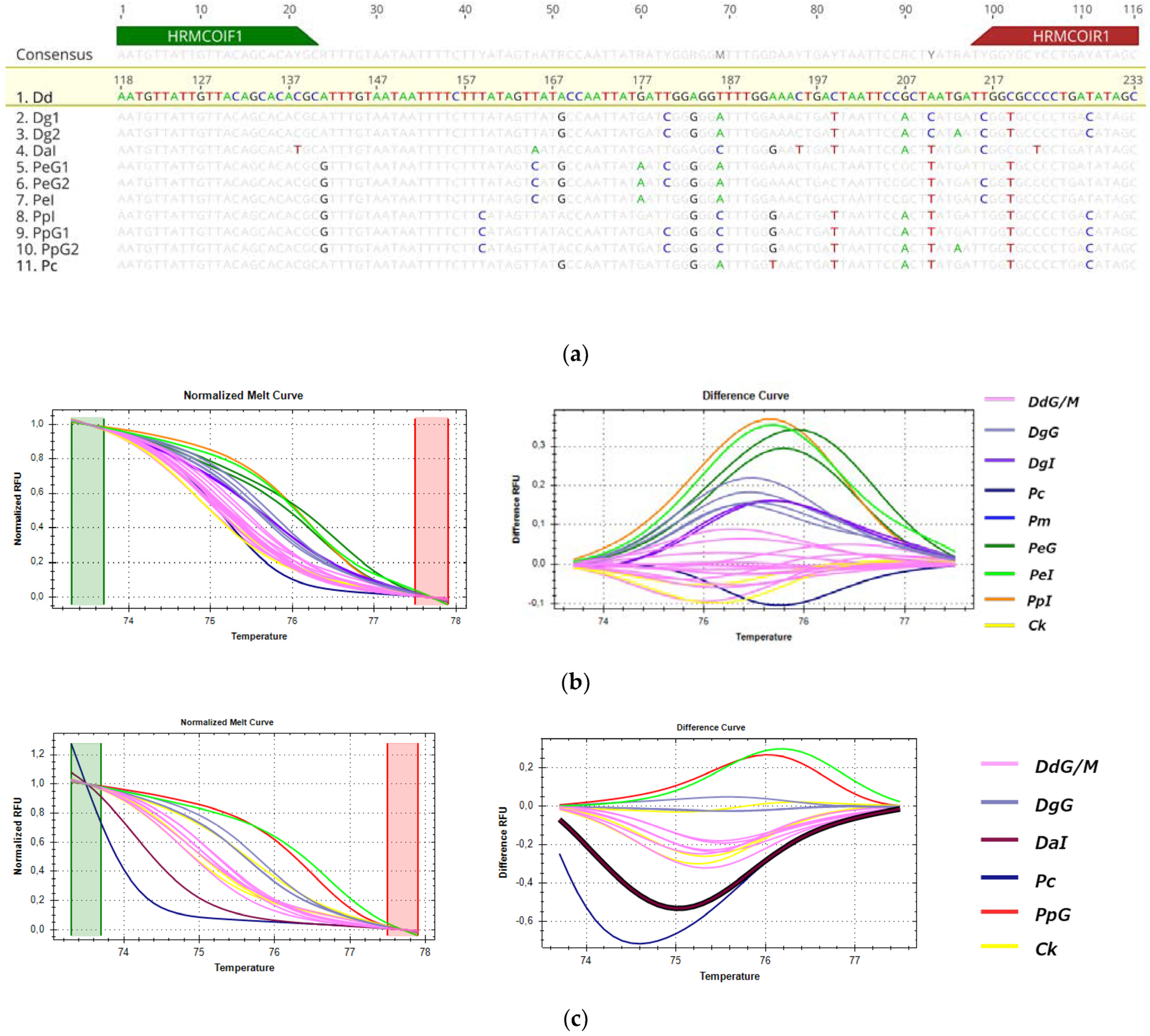
3.3.4. HRM Analysis for the Detection of D. dentex from Greek Fisheries
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Molina-Vega, M.; Gómez-Pérez, A.M.; Tinahones, F.J. Fish in the Mediterranean Diet. In The Mediterranean Diet, 2nd ed.; Preedy, V.R., Watson, R.R., Eds.; Elsevier Inc.: Amsterdam, The Netherlands, 2020; pp. 275–284. ISBN 9780128186497. [Google Scholar]
- FAO. World Fisheries and Aquaculture; FAO: Rome, Italy, 2022; pp. 1–11. [Google Scholar] [CrossRef]
- Lawrence, S.; Elliott, C.; Huisman, W.; Dean, M.; van Ruth, S. The 11 sins of seafood: Assessing a decade of food fraud reports in the global supply chain. Compr. Rev. Food Sci. Food Saf. 2022, 21, 3746–3769. [Google Scholar] [CrossRef] [PubMed]
- Ryburn, S.J.; Ballantine, W.M.; Loncan, F.M.; Manning, O.G.; Alston, M.A.; Steinwand, B.; Bruno, J.F. Public awareness of seafood mislabeling. PeerJ 2022, 10, e13486. [Google Scholar] [CrossRef] [PubMed]
- Cermakova, E.; Lencova, S.; Mukherjee, S.; Horka, P.; Vobruba, S.; Demnerova, K.; Zdenkova, K. Identification of Fish Species and Targeted Genetic Modifications Based on DNA Analysis: State of the Art. Foods 2023, 12, 228. [Google Scholar] [CrossRef] [PubMed]
- Warner, K.; Mustain, P.; Lowell, B.; Geren, S.; Talmage, S. Deceptive Dishes: Seafood Swaps Found Worldwide. Oceana 2016. Available online: https://usa.oceana.org/reports/deceptive-dishes-seafood-swaps-found-worldwide/ (accessed on 7 May 2023).
- Blanco-Fernandez, C.; Ardura, A.; Masiá, P.; Rodriguez, N.; Voces, L.; Fernandez-Raigoso, M.; Roca, A.; Machado-Schiaffino, G.; Dopico, E.; Garcia-Vazquez, E. Fraud in highly appreciated fish detected from DNA in Europe may undermine the Development Goal of sustainable fishing in Africa. Sci. Rep. 2021, 11, 11423. [Google Scholar] [CrossRef]
- Verrez-Bagnis, V.; Sotelo, C.G.; Mendes, R.; Silva, H.; Kappel, K.; Schröder, U. Methods for Seafood Authenticity Testing in Europe. In Bioactive Molecules in Food; Mérillon, J.M., Ramawat, K., Eds.; Springer: Berlin/Heidelberg, Germany, 2019; pp. 2063–2117. ISBN 9783319780306. [Google Scholar] [CrossRef]
- Rasmussen, R.S.; Morrissey, M.T. Application of DNA-Based Methods to Identify Fish and Seafood Substitution on the Commercial Market. Compr. Rev. Food Sci. Food Saf. 2009, 8, 118–154. [Google Scholar] [CrossRef]
- EUMOFA. EU Consumer Habits Regarding Fishery and Aquaculture Products—Annex 1 Mapping and analysis of existing studies on consumer habits. Eur. Mark. Obs. Fish. Aquac. Prod. 2017, 46. Available online: https://data.europa.e (accessed on 1 March 2023).
- Manooch, C.S.; Hassler, W.W. Synopsis of Biological Data on the Red Porgy, Pagrus Pagrus (Linnaeus); Penn State University: State College, PA, USA, 2013. [Google Scholar] [CrossRef]
- Froese, R.D.P. (Ed.) FishBase. Version (02/2023); World Wide Web Electronic Publication 2023. Available online: www.fishbase.org (accessed on 1 March 2023).
- European Commission Council Regulation (EC). No 1967/2006 of 21 December 2006 Concerning Management Measures for the Sustainable Exploitation of Fishery Resources in the Mediterranean Sea. Off. J. Eur. Union. 2019. Available online: http://data.europa.eu/eli/reg/2006/1967/2019-08-14 (accessed on 7 May 2023).
- Hellenic Aquaculture Producers Organization. Greek Aquaculture 2022 Annual Report. Available online: https://fishfromgreece.com/en/kathgoria/exhibitions/greek-aquaculture-annual-report-2022/ (accessed on 21 April 2023).
- Cladas, Y.; Spalla, K.; Doudoumis, V.; Ketsilis-Rinos, V.; Batargias, C.; Koutsikopoulos, C. Presence Confirmation of Non-Native Species Pagrus major (Temminck And Schlegel, 1843) in the Eastern Mediterranean. Oceanogr. Fish. Open Access J. 2019, 9, 555756. [Google Scholar] [CrossRef]
- European Commission Council Regulation (EC). No. 535/2008 Laying down Detailed Rules for the Implementation of Council Regulation (EC) No. 708/2007 Concerning Use of Alien and Locally Absent Species in Aquaculture. Off. J. Eur. Union 2008, 2008, 6–9. [Google Scholar]
- Cocolin, L.; D’Agaro, E.; Manzano, M.; Lanari, D.; Comi, G. Rapid PCR-RFLP Method for the Identification of Marine Fish Fillets (Seabass, Seabream, Umbrine, and Dentex). J. Food Sci. 2000, 65, 1315–1317. [Google Scholar] [CrossRef]
- Schiefenhövel, K.; Rehbein, H. Differentiation of Sparidae species by DNA sequence analysis, PCR-SSCP and IEF of sarcoplasmic proteins. Food Chem. 2013, 138, 154–160. [Google Scholar] [CrossRef]
- Marengo, M.; Durieux, E.D.; Marchand, B.; Francour, P. A review of biology, fisheries and population structure of Dentex dentex (Sparidae). Rev. Fish Biol. Fish. 2014, 24, 1065–1088. [Google Scholar] [CrossRef]
- Carpenter, K.E.; Russel, B. Dentex Dentex. The IUCN Red List of Threatened Species 2014. 2014; 8235, e.T170245A1300534(accessed on 19 April 2023). [Google Scholar] [CrossRef]
- Koumoundouros, G.; Carrillo, J.; Divanach, P.; Kentouri, M. The rearing of common dentex Dentex dentex (L.) during the hatchery and on-growing phases. Aquaculture 2004, 240, 165–173. [Google Scholar] [CrossRef]
- Rueda, F.; Martinez, F. A review on the biology and potential aquaculture of Dentex dentex. Rev. Fish Biol. Fish. 2001, 11, 57–70. [Google Scholar] [CrossRef]
- Katavic, I.; Grubiŝíc, L.; Skakelja, N. Growth performance of pink dentex as compared to four other sparids reared in marine cages in Croatia. Aquac. Int. 2000, 8, 455–461. [Google Scholar] [CrossRef]
- European Commission Council Regulation (EU). No 1379/2013 on the Common Organization of the Markets in Fishery and Aquaculture Products, Amending Council Regulations (EC) No 1184/2006 and (EC) No 1224/2009 and Repealin. Off. J. Eur. Union 2013, L354, 1–21. [Google Scholar]
- Teletchea, F. Molecular identification methods of fish species: Reassessment and possible applications. Rev. Fish Biol. Fish. 2009, 19, 265–293. [Google Scholar] [CrossRef]
- Angers, A.; Ballin, N.; Hofherr, J.; Kagkli, D.; Lievens, A.; Maquet, A.; Martinsohn, J.; Paracchini, V.; Petrillo, M.; Puertas Gallardo, A. Enhancing Fish Species Identification Using Novel Markers and Emerging Technologies; EUR 28505 EN; Publications Office of the European Union: Luxembourg, 2017; ISBN 978-92-79-66293-5. [Google Scholar]
- Ward, R.D.; Hanner, R.; Hebert, P.D.N. The campaign to DNA barcode all fishes, FISH-BOL. J. Fish Biol. 2009, 74, 329–356. [Google Scholar] [CrossRef]
- Di Pinto, A.; Marchetti, P.; Mottola, A.; Bozzo, G.; Bonerba, E.; Ceci, E.; Bottaro, M.; Tantillo, G. Species identification in fish fillet products using DNA barcoding. Fish. Res. 2015, 170, 9–13. [Google Scholar] [CrossRef]
- Rasmussen, R.S.; Morrissey, M.T. DNA-Based Methods for the Identification of Commercial Fish and Seafood Species. Compr. Rev. Food Sci. Food Saf. 2008, 7, 280–295. [Google Scholar] [CrossRef] [PubMed]
- Tomás, C.; Ferreira, I.; Faria, M. Codfish authentication by a fast Short Amplicon High Resolution Melting Analysis (SA-HRMA) method. Food Control 2017, 71, 255–263. [Google Scholar] [CrossRef]
- Chen, C.; Ding, Y.; Wang, Y.; Jiang, Q.; Wang, F.; Lu, C.; Zhang, L.; Zhu, C. High-Resolution Melting Analysis of COI Sequences Distinguishes Pufferfish Species (Takifugu spp.) in China. J. Agric. Food Chem. 2021, 69, 794–804. [Google Scholar] [CrossRef]
- Fitzcharles, E. Rapid discrimination between four Antarctic fish species, genus Macrourus, using HRM analysis. Fish. Res. 2012, 127–128, 166–170. [Google Scholar] [CrossRef]
- Fernandes, T.J.; Costa, J.; Oliveira, M.B.P.; Mafra, I. DNA barcoding coupled to HRM analysis as a new and simple tool for the authentication of Gadidae fish species. Food Chem. 2017, 230, 49–57. [Google Scholar] [CrossRef]
- Teletchea, F.; Maudet, C.; Hänni, C. Food and forensic molecular identification: Update and challenges. Trends Biotechnol. 2005, 23, 359–366. [Google Scholar] [CrossRef]
- Barcaccia, G.; Lucchin, M.; Cassandro, M. DNA Barcoding as a Molecular Tool to Track Down Mislabeling and Food Piracy. Diversity 2015, 8, 2. [Google Scholar] [CrossRef] [Green Version]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database resources of the national center for biotechnology information. Nucleic Acids Res. 2021, 50, D20–D26. [Google Scholar] [CrossRef]
- Handy, S.M.; Deeds, J.R.; Ivanova, N.V.; Hebert, P.D.N.; Hanner, R.H.; Ormos, A.; Weigt, L.A.; Moore, M.M.; Yancy, H.F. A single-laboratory validated method for the generation of DNA barcodes for the identification of fish for regulatory compliance. J. AOAC Int. 2011, 94, 201–211. [Google Scholar] [CrossRef] [Green Version]
- Silva, A.J.; Hellberg, R.S. DNA-Based Techniques for Seafood Species Authentication. In Advances in Food and Nutrition Research; Toldrá, F., Ed.; Elsevier Inc.: Cambridge, MA, USA, 2021; Volume 95, pp. 207–255. ISBN 9780128215203. [Google Scholar]
- Fernandes, T.J.R.; Amaral, J.S.; Mafra, I. DNA Barcode Markers Applied to Seafood Authentication: An Updated Review. Crit. Rev. Food Sci. Nutr. 2021, 61, 3904–3935. [Google Scholar] [CrossRef]
- Wolf, C.; Burgener, M.; Hübner, P.; Lüthy, J. PCR-RFLP Analysis of Mitochondrial DNA: Differentiation of Fish Species. LWT-Food Sci. Technol. 2000, 33, 144–150. [Google Scholar] [CrossRef]
- Hellberg, R.S.R.; Morrissey, M.T. Advances in DNA-Based Techniques for the Detection of Seafood Species Substitution on the Commercial Market. JALA J. Assoc. Lab. Autom. 2011, 16, 308–321. [Google Scholar] [CrossRef]
- Wittwer, C.T. High-resolution DNA melting analysis: Advancements and limitations. Hum. Mutat. 2009, 30, 857–859. [Google Scholar] [CrossRef]
- Bio-Rad Laboratories, Inc. Precision Melt Analysis TM Software Instruction Manual; Bio-Rad Laboratories, Inc.: Hercules, CA, USA, 2019. [Google Scholar]
- Fernandes, T.J.; Costa, J.; Oliveira, M.B.P.; Mafra, I. Exploiting 16S rRNA gene for the detection and quantification of fish as a potential allergenic food: A comparison of two real-time PCR approaches. Food Chem. 2018, 245, 1034–1041. [Google Scholar] [CrossRef]
- Shi, R.; Xiong, X.; Huang, M.; Xu, W.; Li, Y.; Cao, M.; Xiong, X. High resolution melting (HRM) analysis of a 12S rRNA mini barcode as a novel approach for codfish species authentication in processed fish products. Eur. Food Res. Technol. 2020, 246, 891–899. [Google Scholar] [CrossRef]
- Bariche, M. Field Identification Guide to the Living Marine Resources of the Eastern and Southern Mediterranean. FAO Species Identification Guide for Fishery Purposes. 2012. Available online: http://www.fao.org/3/i1276b/i1276b00.htm (accessed on 19 April 2023).
- Chen, Y.; Liu, J.; Wu, R. A new record of blue-spotted seabream Pagrus caeruleostictus from Chinese coastal waters documented from morphology and DNA barcoding. Chin. J. Oceanol. Limnol. 2014, 33, 500–505. [Google Scholar] [CrossRef]
- Carpenter, K.E.; De Angelis, N. The Living Marine Resources of the Eastern Central Atlantic. In Bony Fishes Part 2 (Perciformes to Tetradontiformes) and Sea Turtles; Carpenter, K.E., De Angelis, N., Eds.; Food and Agriculture Organisation: Rome, Italy, 2016; Volume 4, pp. 1511–2350. ISBN 9789251064771. [Google Scholar]
- Geneious Prime 2023.0.3. Available online: https://www.geneious.com (accessed on 7 May 2023).
- Ivanova, N.V.; Zemlak, T.S.; Hanner, R.H.; Hebert, P.D.N. Universal primer cocktails for fish DNA barcoding. Mol. Ecol. Notes 2007, 7, 544–548. [Google Scholar] [CrossRef]
- Ceruso, M.; Mascolo, C.; De Luca, P.; Venuti, I.; Biffali, E.; Ambrosio, R.L.; Smaldone, G.; Sordino, P.; Pepe, T. Dentex dentex Frauds: Establishment of a New DNA Barcoding Marker. Foods 2021, 10, 580. [Google Scholar] [CrossRef]
- McGinnis, S.; Madden, T.L. BLAST: At the core of a powerful and diverse set of sequence analysis tools. Nucleic Acids Res. 2004, 32, 20–25. [Google Scholar] [CrossRef]
- Sievers, F.; Wilm, A.; Dineen, D.; Gibson, T.J.; Karplus, K.; Li, W.; Lopez, R.; McWilliam, H.; Remmert, M.; Söding, J.; et al. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 2011, 7, 539. [Google Scholar] [CrossRef]
- Keskin, E.; Atar, H.H. DNA barcoding commercially important fish species of Turkey. Mol. Ecol. Resour. 2013, 13, 788–797. [Google Scholar] [CrossRef] [PubMed]
- Armani, A.; Guardone, L.; Castigliego, L.; D’Amico, P.; Messina, A.; Malandra, R.; Gianfaldoni, D.; Guidi, A. DNA and Mini-DNA barcoding for the identification of Porgies species (family Sparidae) of commercial interest on the international market. Food Control 2015, 50, 589–596. [Google Scholar] [CrossRef]
- Landi, M.; Dimech, M.; Arculeo, M.; Biondo, G.; Martins, R.; Carneiro, M.; Carvalho, G.R.; Brutto, S.L.; Costa, F. DNA Barcoding for Species Assignment: The Case of Mediterranean Marine Fishes. PLoS ONE 2014, 9, e106135. [Google Scholar] [CrossRef] [PubMed]
- Abbas, E.M.; Soliman, T.; El-Magd, M.A.; Farrag, M.M.; Ismail, R.F.; Kato, M. Phylogeny and DNA Barcoding of the Family Sparidae Inferred from Mitochondrial DNA of the Egyptian Waters. J. Fish. Aquat. Sci. 2017, 12, 73–81. [Google Scholar] [CrossRef] [Green Version]
- Ball, A.O.; Beal, M.G.; Chapman, R.W.; Sedberry, G.R. Population structure of red porgy, Pagrus pagrus, in the Atlantic Ocean. Mar. Biol. 2007, 150, 1321–1332. [Google Scholar] [CrossRef]
- Viret, A.; Tsaparis, D.; Tsigenopoulos, C.S.; Berrebi, P.; Sabatini, A.; Arculeo, M.; Fassatoui, C.; Magoulas, A.; Marengo, M.; Morales-Nin, B.; et al. Absence of spatial genetic structure in common dentex (Dentex dentex Linnaeus, 1758) in the Mediterranean Sea as evidenced by nuclear and mitochondrial molecular markers. PLoS ONE 2018, 13, e0203866. [Google Scholar] [CrossRef]
- Ceruso, M.; Mascolo, C.; Palma, G.; Anastasio, A.; Pepe, T.; Sordino, P. The complete mitochondrial genome of the common dentex, Dentex dentex (perciformes: Sparidae). Mitochondrial DNA Part B 2018, 3, 391–392. [Google Scholar] [CrossRef] [Green Version]
- Bargelloni, L.; Alarcon, J.A.; Alvarez, M.C.; Penzo, E.; Magoulas, A.; Reis, C.A.F.; Patarnello, T. Discord in the family Sparidae (Teleostei): Divergent phylogeographical patterns across the Atlantic-Mediterranean divide. J. Evol. Biol. 2003, 16, 1149–1158. [Google Scholar] [CrossRef] [Green Version]
- Dulčić, J.; Kraljevic, M. On the record of Red seabream Pagrus major (Temminck and Schlegel, 1843) (Osteichthyes: Sparidae) in the Adriatic Sea. Sci. Mar. 2007, 71, 15–17. [Google Scholar] [CrossRef] [Green Version]
- Saad, A.; Sabour, W.; Masri, M.; Barakat, I.; Capapé, C. On the Occurrence of Red Seabream Pagrus major (Osteichthyes: Sparidae) in the Eastern Mediterranean Sea, First Record from the Syrian Coast. Cah. Biol. Mar. 2022, 63, 89–92. [Google Scholar] [CrossRef]
- Rodriguez-Ezpeleta, N.; Mendibil, I.; Álvarez, P.; Cotano, U. Effect of Fish Sampling and Tissue Storage Conditions in DNA Quality. Rev. Investig. Mar. AZTI-Tecnalia 2013, 20, 77–87. [Google Scholar]
- Oosting, T.; Hilario, E.; Wellenreuther, M.; Ritchie, P.A. DNA degradation in fish: Practical solutions and guidelines to improve DNA preservation for genomic research. Ecol. Evol. 2020, 10, 8643–8651. [Google Scholar] [CrossRef] [PubMed]
- Fernandes, T.J.; Costa, J.; Oliveira, M.B.P.; Mafra, I. COI barcode-HRM as a novel approach for the discrimination of hake species. Fish. Res. 2018, 197, 50–59. [Google Scholar] [CrossRef]
- Morgan, J.A.T.; Welch, D.J.; Harry, A.V.; Street, R. A mitochondrial species identification assay for Australian blacktip sharks (Carcharhinus tilstoni, C. limbatus and C. amblyrhynchoides) using real-time PCR and high-resolution melt analysis. Mol. Ecol. Resour. 2011, 11, 813–819. [Google Scholar] [CrossRef]
- Buddhachat, K.; Attakitbancha, C.; Ritbamrung, O.; Chanthap, K.; Suwannapoom, C.; Nganvongpanit, K. Using mini-barcodes coupled with high resolution melting (minibar-HRM) method for species discrimination across Pangasianodon gigas, Pangasianodon hypophthalmus and Pangasius larnaudii. Aquaculture 2020, 530, 735773. [Google Scholar] [CrossRef]
- Zhao, Y.; Song, W.; Li, R.; Wang, C.; Mu, C. A high-resolution melting analysis method for the effective discrimination of three Sepia species. J. Consum. Prot. Food Saf. 2020, 15, 383–385. [Google Scholar] [CrossRef]
- Quintrel, M.; Jilberto, F.; Sepúlveda, M.; Marín, M.E.; Véliz, D.; Araneda, C.; Larraín, M.A. Development and Validation of a Multi-Locus PCR-HRM Method for Species Identification in Mytilus Genus with Food Authenticity Purposes. Foods 2021, 10, 1684. [Google Scholar] [CrossRef]
- Monteiro, C.S.; Deconinck, D.; Eljasik, P.; Sobczak, M.; Derycke, S.; Panicz, R.; Kane, N.; Mazloomrezaei, M.; Devlin, R.H.; Faria, M.A. A fast HRMA tool to authenticate eight salmonid species in commercial food products. Food Chem. Toxicol. 2021, 156, 112440. [Google Scholar] [CrossRef]
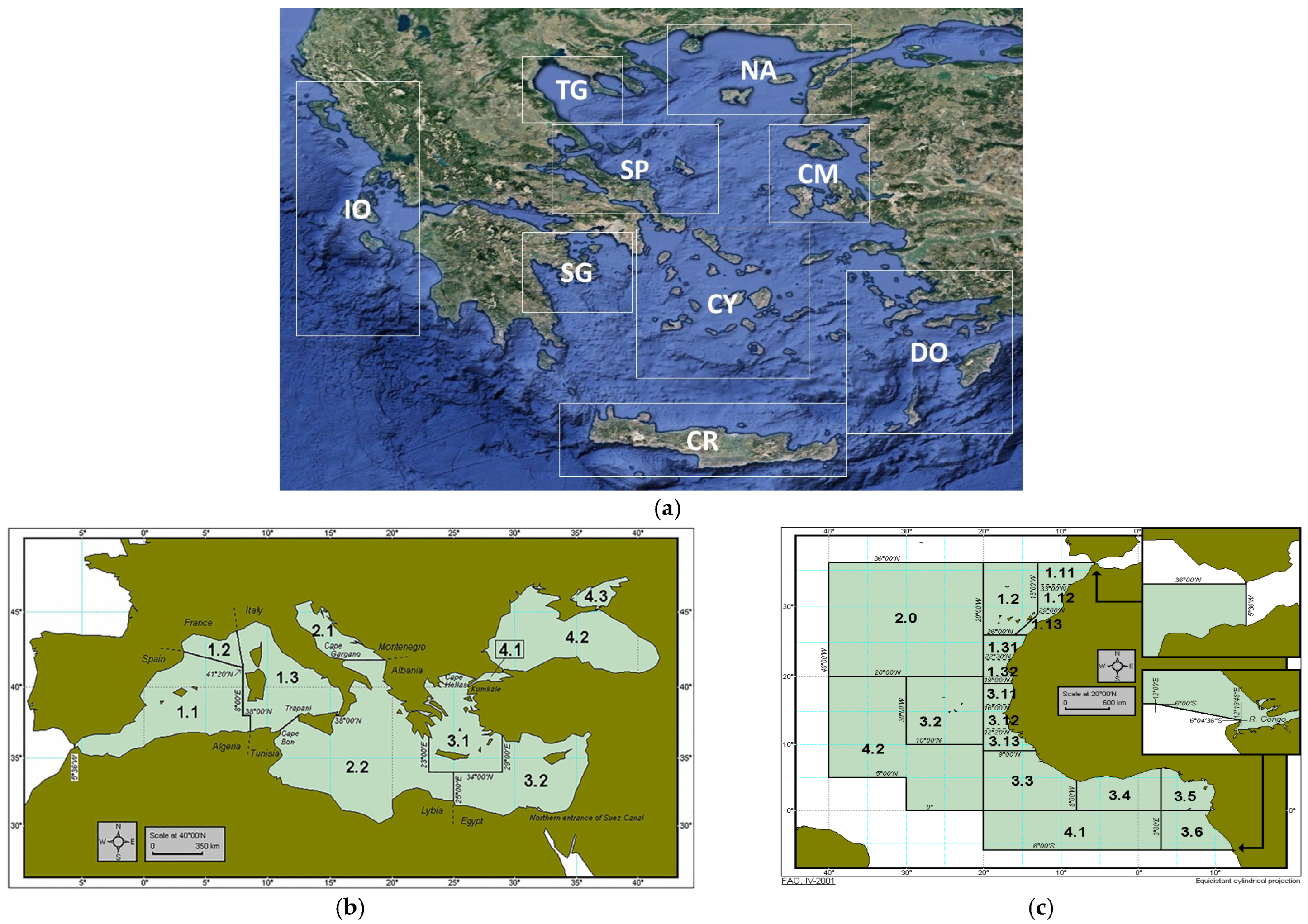




| Species | Sample Id | Greek Seas/Other Region | FAO Region | Total Number of Samples | FC Out of Total | FM/SM Out of Total | RE Out of Total | Frozen/ Filleted Out of Total | Cooked/ Out of Total |
|---|---|---|---|---|---|---|---|---|---|
| P. pagrus | PpG | CY | 37.3.1 | 12 | 12 | 2 | |||
| (Pp) | PpG | SP | 37.3.1 | 25 | 24 | 1 | 1 | 3 | |
| PpG | SG | 37.3.1 | 11 | 10 | 1 | 2 | |||
| PpG | DO | 37.3.1 | 10 | 8 | 2 | 2 | |||
| PpG | CR | 37.3.1 | 3 | 2 | 1 | 2 | |||
| PpG | TG | 37.3.1 | 1 | 1 | 1 | 1 | |||
| PpG | CM | 37.3.1 | 1 | 1 | 1 | ||||
| PpG | IO | 37.2.2 | 8 | 8 | 1 | 2 | |||
| PpI | IMP | 34.1 | 10 | 10 | 9 | 3 | |||
| PpM | ME | 37.1.1 | 3 | 3 | 3 | ||||
| P. major | Pm | AQ | 16 | 14 | 2 | 2 | 2 | ||
| (Pm) | Pm | LFT | 3 | 3 | 2 | ||||
| Pm | CR | 37.1.1 | 1 | 1 | 1 | 1 | |||
| Pm | SP | 37.1.1 | 1 | 1 | 1 | 1 | |||
| P. caeruleostictus (Pc) | Pc | IMP | 1 | 1 | 1 | 1 | |||
| P. erythrinus (Pe) | Pe | SP | 37.1.1 | 2 | 1 | 1 | 2 | ||
| Pe | DO | 37.1.1 | 8 | 4 | 3 | 3 | |||
| PeI | IMP | 34.1 | 1 | 1 | 1 | 1 | |||
| D. dentex | DdG | CY | 37.3.1 | 6 | 4 | 1 | 1 | 2 | |
| (Dd) | DdG | SP | 37.3.1 | 3 | 2 | 1 | 1 | ||
| DdG | SG | 37.3.1 | 7 | 5 | 1 | 1 | 2 | ||
| DdG | DO | 37.3.1 | 2 | 2 | 2 | ||||
| DdG | CR | 37.3.1 | 6 | 6 | 4 | ||||
| DdG | ΝA | 37.3.1 | 4 | 4 | 2 | ||||
| DdG | IO | 37.2.2 | 29 | 29 | 6 | ||||
| DdM | ME | 37.1.1 | 3 | 3 | 3 | ||||
| D. gibbosus | Dg | SP | 2 | 1 | 1 | 1 | 1 | ||
| (Dg) | Dg | CY | 3 | 1 | 2 | 1 | |||
| Dg | IO | 2 | 1 | 1 | 2 | 1 | |||
| Dg | SG | 2 | 2 | 1 | 1 | ||||
| DgI | IMP | 34.1 | 3 | 3 | 3 | 2 | |||
| D. angolensis (Da) | Da | IMP | 34.1 | 2 | 2 | 1 |
| Name | Primer | Tm °C | |
|---|---|---|---|
| COIpp | TCAACCAACCATAAAGACATCGGCAC | 63.2 | primers from [37] modified in this study |
| COIUnR | TAGACTTCTGGGTGGCCRAARAAYCA | 64.0 | primers from [50] |
| cytbF1 | CATGCTAACGGAGCATCCTTCT | 60.3 | This study |
| cytbUR | GCAAATAGGAARTATCAYTCRGG | 58.0 | This study |
| 16sF2 | AGTATGRGCGACAGAAAAGGA | 56.9 | This study |
| 16SR2 | GATTCGGTGGTTGGTCCGTTC | 61.8 | This study |
| cytbF2 | CATATTAAACCCGAATGATATTT | 51.7 | This study |
| CRR | GGGAAGAAACAGCATATTATG | 54 | This study |
| COI4FU | CTAGGCGACGACCAGATTTATAATGT | 60.8 | This study |
| COIR3 | ACTCCTGAGGAGGCAAGTAGG | 61.8 | This study |
| COIR4U | GTTAGGTCTACTGATGCTCCTGC | 62.4 | This study |
| COIR5 | GACTGGCAGGGACAGAAGG | 61.0 | This study |
| COIIF | CGCCTAAACCAAACAGCATTC | 58.4 | This study |
| ATP6R | GTAAAGGTRTAAGGGAGGAG | 55.5 | This study |
| ND2Fd | CACCCTAGCTATCCTCCCCCTCATAGC | 66.9 | primers from [51] |
| ND2Rd | AATAACTTCGGGGAGTCACGAGTGTAGG | 65.7 | primers from [51] |
| HRMCOIF1 | AATGTTATTGTTACAGCACACGC | 57.1 | This study |
| HRMCOIR1 | GCTATGTCAGGGGCACCAA | 58.8 | This study |
| HRMcytbF2 | GCAGGCGTAATTATTCTATTAAC | 55.3 | This study |
| HRMcytbR1 | GGTTTAATATGAGGGGGTGTAAC | 58.9 | This study |
| HRM16sF1 | GTAAAGATCATAATTAACCCAAG | 53.5 | This study |
| HRM16sR1 | GTTTGCAGCGGATAGTCTGATAT | 58.9 | This study |
| GENE | Forward Primer | Reverse Primer | Amplicon Length | Annealing T/°C | Restriction Enzyme |
|---|---|---|---|---|---|
| COI | COIpp | COIUnR | 706 | 54 | HindIII or Sau3AI |
| cytb | cytbF1 | cytbUR | 583 | 54 | Sau3AI |
| CR | cytbF2 | CRR | 1292 | 53 | XbaI |
| Species | Forward Primer | Reverse Primer | Length (bp) | Annealing T °C |
|---|---|---|---|---|
| P. pagrus vs. other Sparidae species | COI4FU | COIR5 | 461 | 57 |
| COI4FU | COI4RU | 295 | ||
| COI4FU | COIR3 | 211 | ||
| D. dentex vs. other Sparidae species | COIIF | ATP6R | 673 | 52 |
| cytbF1 | cytbUR | 583 | ||
| COI4FU | COI4RU | 295 | ||
| ND2Fd | ND2Rd | 249 |
| GENE | Forward Primer | Reverse Primer | Fragment Length (bp) | Annealing T °C |
|---|---|---|---|---|
| COI | HRMCOIF1 | HRMCOIR1 | 116 | 53 |
| cytb | HRMcytbF2 | HRMcytbR1 | 113 | 52 |
| 16s | HRM16sF1 | HRM16sR1 | 156 | 53 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatzoglou, E.; Tsaousi, N.; Apostolidis, A.P.; Exadactylos, A.; Sandaltzopoulos, R.; Giantsis, I.A.; Gkafas, G.A.; Malandrakis, E.E.; Sarantopoulou, J.; Tokamani, M.; et al. High-Resolution Melting (HRM) Analysis for Rapid Molecular Identification of Sparidae Species in the Greek Fish Market. Genes 2023, 14, 1255. https://doi.org/10.3390/genes14061255
Chatzoglou E, Tsaousi N, Apostolidis AP, Exadactylos A, Sandaltzopoulos R, Giantsis IA, Gkafas GA, Malandrakis EE, Sarantopoulou J, Tokamani M, et al. High-Resolution Melting (HRM) Analysis for Rapid Molecular Identification of Sparidae Species in the Greek Fish Market. Genes. 2023; 14(6):1255. https://doi.org/10.3390/genes14061255
Chicago/Turabian StyleChatzoglou, Evanthia, Nefeli Tsaousi, Apostolos P. Apostolidis, Athanasios Exadactylos, Raphael Sandaltzopoulos, Ioannis A. Giantsis, Georgios A. Gkafas, Emmanouil E. Malandrakis, Joanne Sarantopoulou, Maria Tokamani, and et al. 2023. "High-Resolution Melting (HRM) Analysis for Rapid Molecular Identification of Sparidae Species in the Greek Fish Market" Genes 14, no. 6: 1255. https://doi.org/10.3390/genes14061255










