Deciphering the Immunostimulatory Effects of β-Glucan on a Rainbow Trout (Oncorhynchus mykiss) Macrophage-like Cell Line (RTS11) by Whole Transcriptome Analysis
Abstract
1. Introduction
2. Methodology
2.1. In Vitro Cell Culture
2.2. Stimulation with β-Glucans
2.3. RNA Extraction
2.4. Transcriptional Analysis: RNA-seq
3. Results
3.1. Identification of Gene Expression Patterns in RTS11
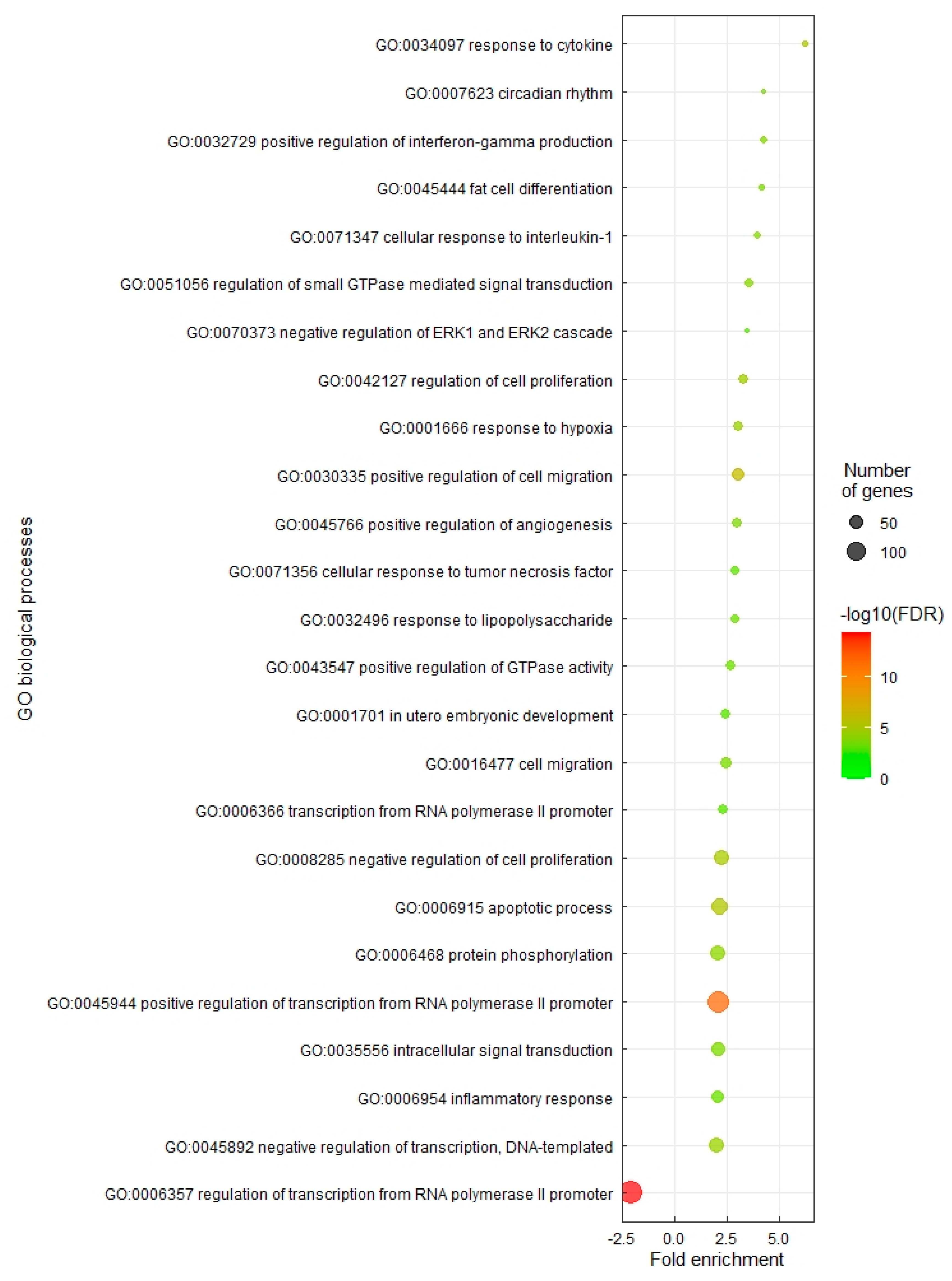
3.2. Gene Set Enrichment Using Gene Ontology
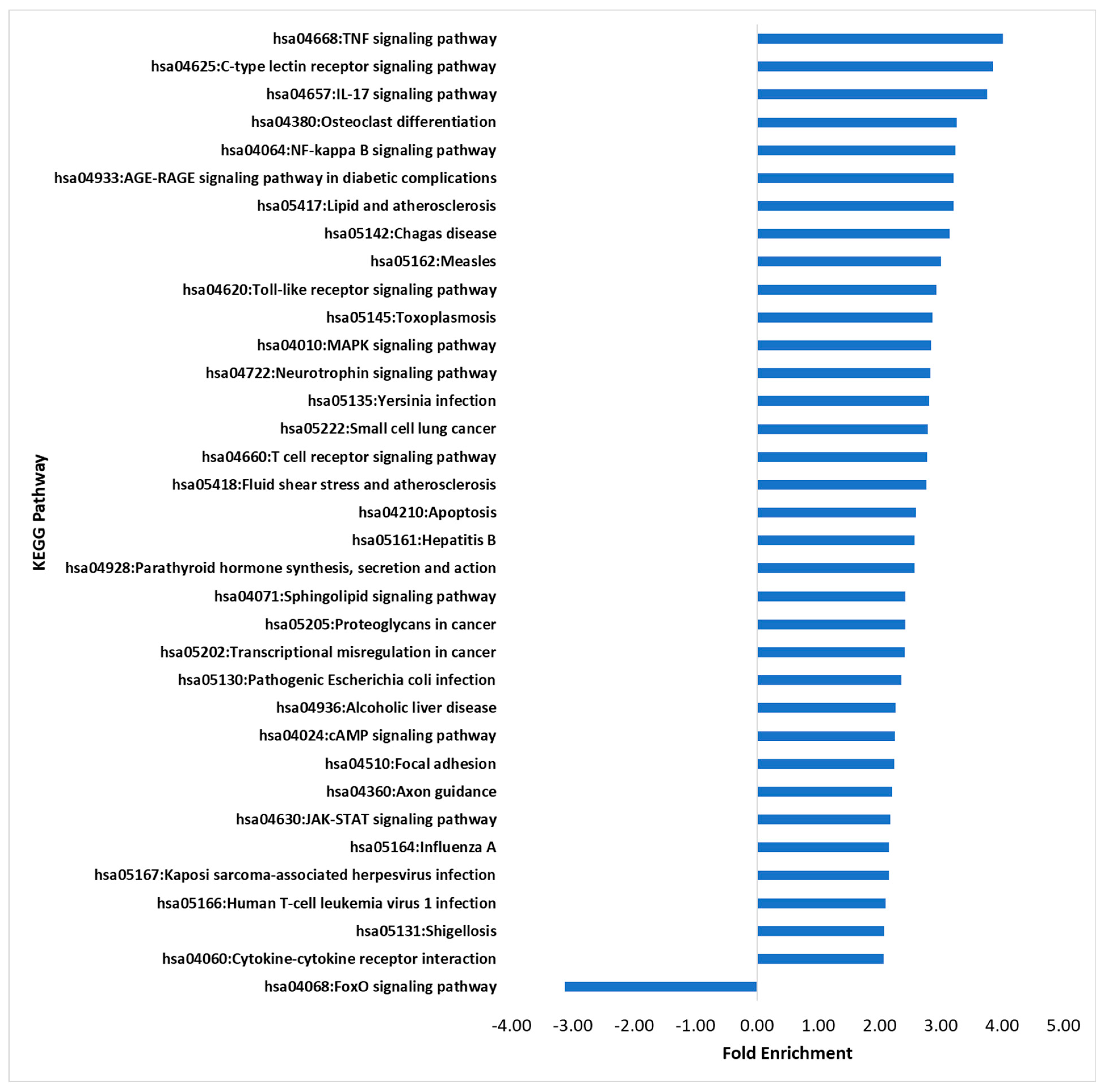
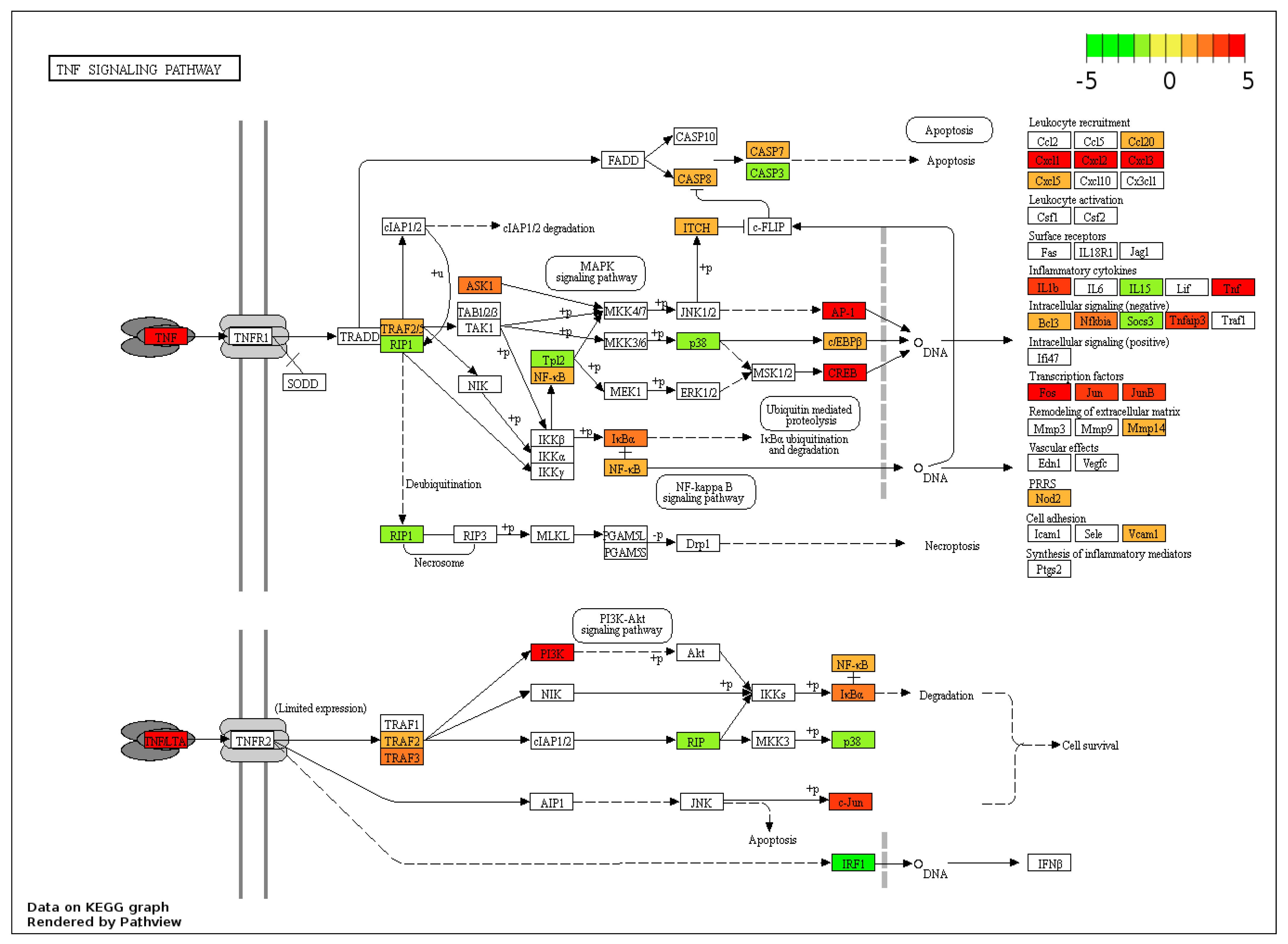
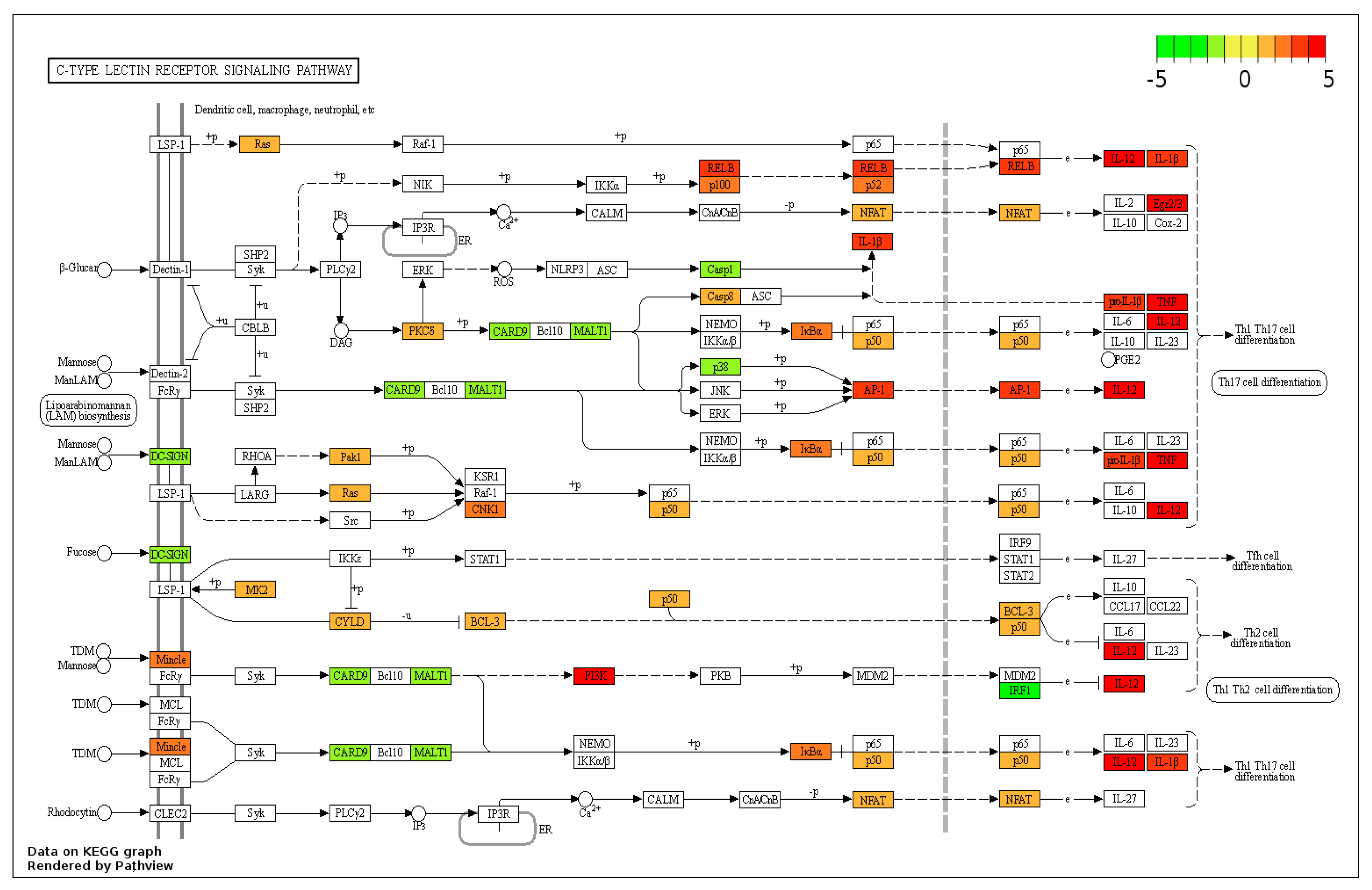
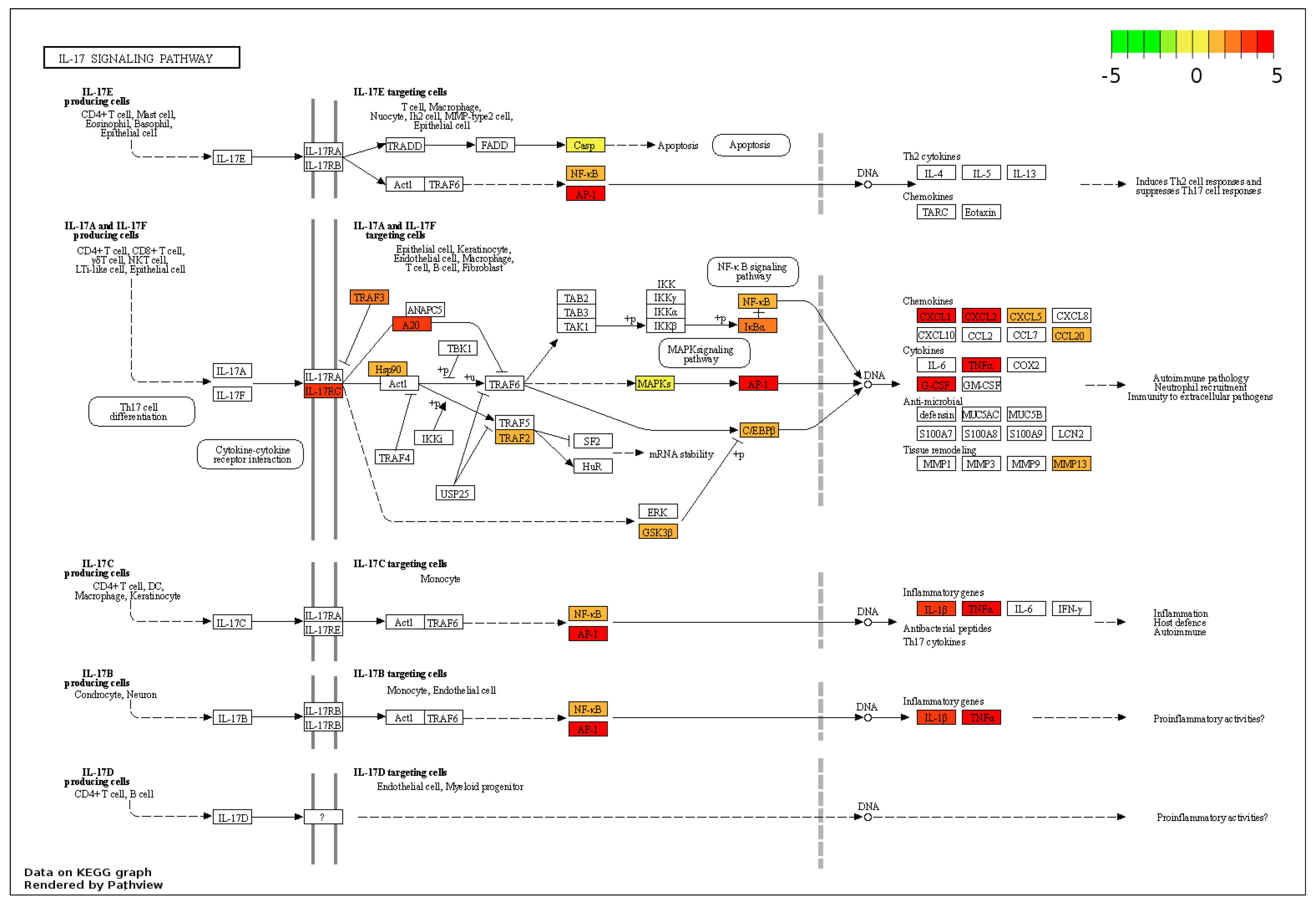
3.3. KEGG Pathway Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hunter, M.C.; Smith, R.G.; Schipanski, M.E.; Atwood, L.W.; Mortensen, D.A. Agriculture in 2050: Recalibrating Targets for Sustainable Intensification. BioScience 2017, 67, 386–391. [Google Scholar] [CrossRef]
- Jones, A.R.; Alleway, H.K.; McAfee, D.; Reis-Santos, P.; Theuerkauf, S.J.; Jones, R.C. Climate-Friendly Seafood: The Potential for Emissions Reduction and Carbon Capture in Marine Aquaculture. BioScience 2022, 72, 123–143. [Google Scholar] [CrossRef] [PubMed]
- Rodger, H.D. Fish Disease Causing Economic Impact in Global Aquaculture. In Fish Vaccines; Birkhäuser Advances in Infectious Diseases; Adams, A., Ed.; Springer: Basel, Switzerland, 2016; pp. 1–34. ISBN 978-3-0348-0980-1. [Google Scholar]
- Reverter, M.; Sarter, S.; Caruso, D.; Avarre, J.-C.; Combe, M.; Pepey, E.; Pouyaud, L.; Vega-Heredía, S.; de Verdal, H.; Gozlan, R.E. Aquaculture at the Crossroads of Global Warming and Antimicrobial Resistance. Nat. Commun. 2020, 11, 1870. [Google Scholar] [CrossRef] [PubMed]
- Leal, M.C.; Calado, R. The Key Role of Functional Aquafeeds to Achieve a More Sustainable Aquaculture. J. World Aquac. Soc. 2019, 50, 1044–1047. [Google Scholar] [CrossRef]
- Cornet, V.; Khuyen, T.D.; Mandiki, S.N.M.; Betoulle, S.; Bossier, P.; Reyes-López, F.E.; Tort, L.; Kestemont, P. GAS1: A New β-Glucan Immunostimulant Candidate to Increase Rainbow Trout (Oncorhynchus Mykiss) Resistance to Bacterial Infections With Aeromonas Salmonicida Achromogenes. Front. Immunol. 2021, 12, 693613. [Google Scholar] [CrossRef]
- Douxfils, J.; Fierro-Castro, C.; Mandiki, S.N.M.; Emile, W.; Tort, L.; Kestemont, P. Dietary β-Glucans Differentially Modulate Immune and Stress-Related Gene Expression in Lymphoid Organs from Healthy and Aeromonas Hydrophila-Infected Rainbow Trout (Oncorhynchus Mykiss). Fish Shellfish Immunol. 2017, 63, 285–296. [Google Scholar] [CrossRef]
- Ji, L.; Fu, S.; Sun, G.; Li, X.; Liu, Y. Dietary β-Glucan Modulate Haematological Parameters, Cytokines and Gene Expression in TLR and ERK Pathways of Rainbow Trout (Oncorhynchus Mykiss) during Infection by Aeromonas Salmonicida. Aquac. Res. 2020, 51, 906–917. [Google Scholar] [CrossRef]
- Ji, L.; Sun, G.; Li, X.; Liu, Y. Comparative Transcriptome Analysis Reveals the Mechanism of β-Glucan in Protecting Rainbow Trout (Oncorhynchus Mykiss) from Aeromonas Salmonicida Infection. Fish Shellfish Immunol. 2020, 98, 87–99. [Google Scholar] [CrossRef]
- Ordás, M.C.; González-Torres, L.; Arense, P.; Heavyside, R.; Zarza, C.; Tafalla, C. Analysis of Immunostimulatory Responses and Immune Tolerance to β-Glucans in Rainbow Trout Cell Lines. Aquaculture 2021, 541, 736805. [Google Scholar] [CrossRef]
- Løvoll, M.; Fischer, U.; Mathisen, G.S.; Bøgwald, J.; Ototake, M.; Dalmo, R.A. The C3 Subtypes Are Differentially Regulated after Immunostimulation in Rainbow Trout, but Head Kidney Macrophages Do Not Contribute to C3 Transcription. Vet. Immunol. Immunopathol. 2007, 117, 284–295. [Google Scholar] [CrossRef]
- Porter, D.; Peggs, D.; McGurk, C.; Martin, S.A.M. Gut Associated Lymphoid Tissue (GALT) Primary Cells and Stable Cell Lines as Predictive Models for Intestinal Health in Rainbow Trout (Oncorhynchus Mykiss). Front. Immunol. 2022, 13, 1023235. [Google Scholar] [CrossRef] [PubMed]
- Hadiuzzaman, M.; Moniruzzaman, M.; Shahjahan, M.; Bai, S.C.; Min, T.; Hossain, Z. β-Glucan: Mode of Action and Its Uses in Fish Immunomodulation. Front. Mar. Sci. 2022, 9, 905986. [Google Scholar] [CrossRef]
- Porter, D.; Peggs, D.; McGurk, C.; Martin, S.A.M. Immune Responses to Prebiotics in Farmed Salmonid Fish: How Transcriptomic Approaches Help Interpret Responses. Fish Shellfish Immunol. 2022, 127, 35–47. [Google Scholar] [CrossRef]
- Mu, D.; Yang, J.; Jiang, Y.; Wang, Z.; Chen, W.; Huang, J.; Zhang, Y.; Liu, Q.; Yang, D. Single-Cell Transcriptomic Analysis Reveals Neutrophil as Orchestrator during β-Glucan–Induced Trained Immunity in a Teleost Fish. J. Immunol. 2022, 209, 783–795. [Google Scholar] [CrossRef]
- Petit, J.; Bailey, E.C.; Wheeler, R.T.; de Oliveira, C.A.F.; Forlenza, M.; Wiegertjes, G.F. Studies Into β-Glucan Recognition in Fish Suggests a Key Role for the C-Type Lectin Pathway. Front. Immunol. 2019, 10, 280. [Google Scholar] [CrossRef]
- Furukawa, A.; Kamishikiryo, J.; Mori, D.; Toyonaga, K.; Okabe, Y.; Toji, A.; Kanda, R.; Miyake, Y.; Ose, T.; Yamasaki, S.; et al. Structural Analysis for Glycolipid Recognition by the C-Type Lectins Mincle and MCL. Proc. Natl. Acad. Sci. USA 2013, 110, 17438–17443. [Google Scholar] [CrossRef]
- Patin, E.C.; Orr, S.J.; Schaible, U.E. Macrophage Inducible C-Type Lectin As a Multifunctional Player in Immunity. Front. Immunol. 2017, 8, 861. [Google Scholar] [CrossRef]
- Johansson, P.; Wang, T.; Collet, B.; Corripio-Miyar, Y.; Monte, M.M.; Secombes, C.J.; Zou, J. Identification and Expression Modulation of a C-Type Lectin Domain Family 4 Homologue That Is Highly Expressed in Monocytes/Macrophages in Rainbow Trout (Oncorhynchus Mykiss). Dev. Comp. Immunol. 2016, 54, 55–65. [Google Scholar] [CrossRef]
- Wiegertjes, G.F.; Wentzel, A.S.; Spaink, H.P.; Elks, P.M.; Fink, I.R. Polarization of Immune Responses in Fish: The ‘Macrophages First’ Point of View. Mol. Immunol. 2016, 69, 146–156. [Google Scholar] [CrossRef]
- Salinas, I. The Mucosal Immune System of Teleost Fish. Biology 2015, 4, 525–539. [Google Scholar] [CrossRef] [PubMed]
- Goswami, M.; Yashwanth, B.S.; Trudeau, V.; Lakra, W.S. Role and Relevance of Fish Cell Lines in Advanced in Vitro Research. Mol. Biol. Rep. 2022, 49, 2393–2411. [Google Scholar] [CrossRef]
- Ganassin, R.C.; Bols, N.C. Development of a Monocyte/Macrophage-like Cell Line, RTS11, from Rainbow Trout Spleen. Fish Shellfish Immunol. 1998, 8, 457–476. [Google Scholar] [CrossRef]
- Martin, S.A.; Zou, J.; Houlihan, D.F.; Secombes, C.J. Directional responses following recombinant cytokine stimulation of rainbow trout (Oncorhynchus mykiss) RTS-11 macrophage cells as revealed by transcriptome profiling. BMC Genom. 2007, 8, 15025. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, I.; Belmonte, R.; Anderson, V.L.; Saraiva, M.; Wang, T.; van West, P.; Secombes, C.J. Immune Gene Expression in Trout Cell Lines Infected with the Fish Pathogenic Oomycete Saprolegnia Parasitica. Dev. Comp. Immunol. 2012, 38, 44–54. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Secombes, C.J. The Function of Fish Cytokines. Biology 2016, 5, 23. [Google Scholar] [CrossRef] [PubMed]
- Di Tommaso, P.; Chatzou, M.; Floden, E.W.; Barja, P.P.; Palumbo, E.; Notredame, C. Nextflow Enables Reproducible Computational Workflows. Nat. Biotechnol. 2017, 35, 316–319. [Google Scholar] [CrossRef] [PubMed]
- Ewels, P.A.; Peltzer, A.; Fillinger, S.; Patel, H.; Alneberg, J.; Wilm, A.; Garcia, M.U.; Di Tommaso, P.; Nahnsen, S. The Nf-Core Framework for Community-Curated Bioinformatics Pipelines. Nat. Biotechnol. 2020, 38, 276–278. [Google Scholar] [CrossRef]
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast Universal RNA-Seq Aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Li, B.; Dewey, C.N. RSEM: Accurate Transcript Quantification from RNA-Seq Data with or without a Reference Genome. BMC Bioinformatics 2011, 12, 323. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An Efficient General Purpose Program for Assigning Sequence Reads to Genomic Features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef]
- Ewels, P.; Magnusson, M.; Lundin, S.; Käller, M. MultiQC: Summarize Analysis Results for Multiple Tools and Samples in a Single Report. Bioinformatics 2016, 32, 3047–3048. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated Estimation of Fold Change and Dispersion for RNA-Seq Data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Huang, D.W.; Sherman, B.T.; Lempicki, R.A. Systematic and Integrative Analysis of Large Gene Lists Using DAVID Bioinformatics Resources. Nat. Protoc. 2009, 4, 44–57. [Google Scholar] [CrossRef]
- Sherman, B.T.; Hao, M.; Qiu, J.; Jiao, X.; Baseler, M.W.; Lane, H.C.; Imamichi, T.; Chang, W. DAVID: A Web Server for Functional Enrichment Analysis and Functional Annotation of Gene Lists (2021 Update). Nucleic Acids Res. 2022, 50, W216–W221. [Google Scholar] [CrossRef]
- Luo, W.; Pant, G.; Bhavnasi, Y.K.; Blanchard, S.G., Jr.; Brouwer, C. Pathview Web: User Friendly Pathway Visualization and Data Integration. Nucleic Acids Res. 2017, 45, W501–W508. [Google Scholar] [CrossRef] [PubMed]
- Kiron, V.; Kulkarni, A.; Dahle, D.; Vasanth, G.; Lokesh, J.; Elvebo, O. Recognition of Purified β 1,3/1,6 Glucan and Molecular Signalling in the Intestine of Atlantic Salmon. Dev. Comp. Immunol. 2016, 56, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Vanaporn, M.; Titball, R.W. Trehalose and Bacterial Virulence. Virulence 2020, 11, 1192–1202. [Google Scholar] [CrossRef]
- Kawano, A.; Haiduk, C.; Schirmer, K.; Hanner, R.; Lee, L.E.J.; Dixon, B.; Bols, N.C. Development of a Rainbow Trout Intestinal Epithelial Cell Line and Its Response to Lipopolysaccharide. Aquac. Nutr. 2011, 17, e241–e252. [Google Scholar] [CrossRef]
- Pasquariello, R.; Verdile, N.; Pavlovic, R.; Panseri, S.; Schirmer, K.; Brevini, T.A.L.; Gandolfi, F. New Stable Cell Lines Derived from the Proximal and Distal Intestine of Rainbow Trout (Oncorhynchus Mykiss) Retain Several Properties Observed In Vivo. Cells 2021, 10, 1555. [Google Scholar] [CrossRef] [PubMed]
- Petit, J.; de Bruijn, I.; Goldman, M.R.G.; van den Brink, E.; Pellikaan, W.F.; Forlenza, M.; Wiegertjes, G.F. β-Glucan-Induced Immuno-Modulation: A Role for the Intestinal Microbiota and Short-Chain Fatty Acids in Common Carp. Front. Immunol. 2022, 12, 761820. [Google Scholar] [CrossRef] [PubMed]

| HGNC ID | Gene Description | ENSEMBL Gene ID | log2FoldChange | Padj |
|---|---|---|---|---|
| NR4A3 | nuclear receptor subfamily 4, group A, member 3 | ENSOMYG00000016957 | 10.84 | 2.47 × 10 −24 |
| NR4A3 | nuclear receptor subfamily 4, group A, member 3 | ENSOMYG00000056982 | 9.50 | 2.48 × 10 −19 |
| EGR3 | early growth response 3 | ENSOMYG00000003397 | 8.01 | 1.31 × 10 −12 |
| DUSP2 | dual specificity phosphatase 2 | ENSOMYG00000015118 | 7.97 | 1.81 × 10 −13 |
| SYT11 | synaptotagmin 11 | ENSOMYG00000021392 | 7.95 | 1.85 × 10 −12 |
| EGR1 | early growth response 1 | ENSOMYG00000015637 | 7.93 | 1.08 × 10 −73 |
| OCSTAMP | osteoclast stimulatory transmembrane protein | ENSOMYG00000033187 | 7.90 | 6.59 × 10 −25 |
| INHBA | inhibin subunit β A | ENSOMYG00000053580 | 7.84 | 7.24 × 10 −43 |
| CNTF | ciliary neurotrophic factor | ENSOMYG00000033868 | 7.71 | 1.45 × 10−138 |
| CCL4L2 | C-C motif chemokine ligand 4 like 2 | ENSOMYG00000026069 | 7.55 | 2.01 × 10−44 |
| TRIO | trio Rho guanine nucleotide exchange factor | ENSOMYG00000037143 | 7.03 | 5.13 × 10−292 |
| NAB2 | NGFI-A binding protein 2 (EGR1 binding protein 2) | ENSOMYG00000012721 | 6.72 | 5.32 × 10−272 |
| EGR3 | early growth response 3 | ENSOMYG00000045806 | 6.64 | 3.34 × 10−122 |
| CXCL2 | C-X-C motif chemokine ligand 6 | ENSOMYG00000022444 | 6.60 | 6.8 × 10−213 |
| SPRY4 | sprouty RTK signalling antagonist 4 | ENSOMYG00000031862 | 6.35 | 1.84 × 10−30 |
| TNF | tumour necrosis factor a | ENSOMYG00000058346 | 6.04 | 4.29 × 10−254 |
| AMER2 | APC membrane recruitment protein 2 | ENSOMYG00000061958 | 6.01 | 0.00000239 |
| PRDM1 | PR/SET domain 1 | ENSOMYG00000011583 | 5.92 | 2.88 × 10−10 |
| RNF208 | ring finger protein 208 | ENSOMYG00000009419 | 5.89 | 0.00000372 |
| TMCC3 | transmembrane and coiled-coil domain family 3 | ENSOMYG00000005847 | 5.84 | 0.00000231 |
| TIMM13 | translocase of inner mitochondrial membrane 13 | ENSOMYG00000009641 | −3.48 | 0.030884 |
| TESK2 | Testis-associated actin remodelling kinase 2 | ENSOMYG00000066315 | −3.49 | 5.72 × 10−11 |
| HHLA1 | HERV-H LTR-associating 1 | ENSOMYG00000045198 | −3.50 | 0.019347 |
| GPX2 | glutathione peroxidase 2 | ENSOMYG00000015731 | −3.63 | 0.015558 |
| CCNI | cyclin I | ENSOMYG00000006476 | −3.75 | 4.85 × 10−59 |
| ARMH4 | Armadillo-like helical domain containing 4 | ENSOMYG00000000520 | −3.82 | 0.007779 |
| C1orf21 | chromosome 1 open reading frame 21 | ENSOMYG00000003998 | −3.85 | 0.010249 |
| CD93 | CD93 molecule | ENSOMYG00000014356 | −3.86 | 1.01 × 10−6 |
| ADORA2A | adenosine A2a receptor | ENSOMYG00000041298 | −3.96 | 0.0024 |
| IGKC | immunoglobulin kappa constant | ENSOMYG00000062277 | −4.09 | 0.002041 |
| TIRAP | TIR domain containing adaptor protein | ENSOMYG00000047353 | −4.19 | 5.59 × 10−9 |
| HES6 | hes family bHLH transcription factor 6 | ENSOMYG00000047424 | −4.20 | 3.67 × 10−10 |
| TMEM132D | transmembrane protein 132D | ENSOMYG00000046943 | −4.28 | 0.004874 |
| ZBTB45 | zinc finger and BTB domain containing 45 | ENSOMYG00000011021 | −4.31 | 1.29 × 10−21 |
| MYC | MYC proto-oncogene, bHLH transcription factor | ENSOMYG00000039293 | −4.38 | 7.09 × 10−13 |
| LHX4 | LIM homeobox 4 | ENSOMYG00000031792 | −4.40 | 0.006305 |
| PDXP | pyridoxal phosphatase | ENSOMYG00000015314 | −4.45 | 0.001192 |
| MFSD11 | major facilitator superfamily domain containing 11 | ENSOMYG00000003467 | −4.46 | 9.54 × 10−7 |
| RPE65 | retinoid isomerohydrolase RPE65 | ENSOMYG00000015503 | −4.91 | 0.000398 |
| ZBED4 | zinc finger BED-type containing 4 | ENSOMYG00000069563 | −4.93 | 7.16 × 10−5 |
| HGNC | Gene Descprition | ENSEMBL_ID | log2foldchange | padj |
|---|---|---|---|---|
| TIMP3 | TIMP Metallopeptidase Inhibitor 3 | ENSOMYG00000011029 | 3.20 | 0 |
| JUN | Jun Proto-Oncogene | ENSOMYG00000036861 | 4.06 | 9.9 × 10−223 |
| RELB | RELB Proto-Oncogene | ENSOMYG00000021944 | 3.07 | 1.3 × 10−195 |
| FOSL1 | FOS ligand 1 | ENSOMYG00000025915 | 5.07 | 6.6 × 10−126 |
| NFKB2 | Nuclear Factor Kappa B Subunit 2 | ENSOMYG00000002188 | 2.42 | 2.5 × 10−124 |
| FOS | Fos Proto-Oncogene | ENSOMYG00000029885 | 4.65 | 5.4 × 10−103 |
| ALDH1A2 | Aldehyde Dehydrogenase 1 Family Member A2 | ENSOMYG00000046040 | 4.65 | 7.6 × 10−99 |
| PML | PML Nuclear Body Scaffold | ENSOMYG00000027472 | 2.45 | 3.08 × 10−42 |
| CD274 | Programmed death-ligand 1 | ENSOMYG00000032862 | 1.28 | 6.31 × 10−42 |
| MAPKAPK2 | MAPK Activated Protein Kinase 2 | ENSOMYG00000032252 | 1.28 | 7.44 × 10−36 |
| BCL2L1 | BCL2 ligand 1 | ENSOMYG00000043310 | 2.37 | 6.37 × 10−32 |
| ITIH4 | Inter-α-Trypsin Inhibitor Heavy Chain 4 | ENSOMYG00000017068 | 2.40 | 8.07 × 10−30 |
| SKIL | SKI-Like Proto-Oncogene | ENSOMYG00000042548 | 2.15 | 5.68 × 10−21 |
| RARA | Retinoic Acid Receptor α | ENSOMYG00000041123 | 1.83 | 9.96 × 10−15 |
| MAPKAPK3 | MAPK Activated Protein Kinase 3 | ENSOMYG00000042013 | 1.06 | 5.17 × 10−8 |
| BCL2 | BCL2 Apoptosis Regulator | ENSOMYG00000038826 | 1.48 | 1.71 × 10−6 |
| SRF | Serum Response Factor | ENSOMYG00000038445 | 1.38 | 2.84 × 10−5 |
| HGNC | Gene Description | ENSEMBL_ID | log2foldchange | padj |
|---|---|---|---|---|
| NFKB1 | Nuclear Factor Kappa B Subunit 1 | ENSOMYG00000040602 | 1.86 | 1.75 × 10−152 |
| ZC3H12A | Zinc Finger CCCH-Type Containig 12A | ENSOMYG00000038385 | 3.32 | 2.34 × 10−105 |
| CCL4 | C-C Motif Chemokine Ligand 4 | ENSOMYG00000008241 | 3.31 | 7.05 × 10−86 |
| CCL3L1 | C-C Motif Chemokine Ligand 3 Like 1 | ENSOMYG00000026069 | 7.55 | 2.01 × 10−44 |
| SOX9 | SRY-Box Transcription Factor 9 | ENSOMYG00000071421 | 3.48 | 6.63 × 10−39 |
| CCL8 | C-C Motif Chemokine Ligand 8 | ENSOMYG00000002731 | 2.65 | 2.68 × 10−32 |
| HIF1A | Hypoxia Inducible Factor 1 Subunit α | ENSOMYG00000040205 | 1.19 | 2.42 × 10−31 |
| CEBPB | CCAAT Enhancer Binding Protein β | ENSOMYG00000019888 | 1.28 | 5.05 × 10−28 |
| CD40 | CD40 Molecule | ENSOMYG00000029243 | 1.34 | 1.92 × 10−27 |
| MYC | MYC Proto-Oncogene | ENSOMYG00000041344 | 2.20 | 3.08 × 10−23 |
| RC3H1 | Ring Finger And CCCH-Type Domains 1 | ENSOMYG00000029982 | 1.52 | 1.1 × 10−21 |
| KLF2 | KLF Transcription Factor 2 | ENSOMYG00000039184 | 1.37 | 4.29 × 10−17 |
| TANK | TRAF Family Member Associated NFKB Activator | ENSOMYG00000047138 | 1.18 | 1.83 × 10−13 |
| GBP1 | Guanylate Binding Protein 1 | ENSOMYG00000036400 | 1.75 | 2.18 × 10−11 |
| CCL20 | C-C Motif Chemokine Ligand 20 | ENSOMYG00000026966 | 1.70 | 0.0000010 |
| ACOD1 | Aconitate Decarboxylase 1 | ENSOMYG00000028719 | 5.62 | 0.0000023 |
| ADAMTS7 | ADAM Metallopeptidase with Thrombospondin Type 1 Motif 7 | ENSOMYG00000063119 | 1.17 | 0.00034 |
| PTGIS | Prostaglandin I2 Synthase | ENSOMYG00000027171 | 2.85 | 0.02 |
| NR1D1 | Nuclear Receptor Subfamily 1 Group D Member 1 | ENSOMYG00000024434 | 2.23 | 0.046 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Porter, D.; Naseer, S.; Peggs, D.; McGurk, C.; Martin, S.A.M. Deciphering the Immunostimulatory Effects of β-Glucan on a Rainbow Trout (Oncorhynchus mykiss) Macrophage-like Cell Line (RTS11) by Whole Transcriptome Analysis. Genes 2023, 14, 1261. https://doi.org/10.3390/genes14061261
Porter D, Naseer S, Peggs D, McGurk C, Martin SAM. Deciphering the Immunostimulatory Effects of β-Glucan on a Rainbow Trout (Oncorhynchus mykiss) Macrophage-like Cell Line (RTS11) by Whole Transcriptome Analysis. Genes. 2023; 14(6):1261. https://doi.org/10.3390/genes14061261
Chicago/Turabian StylePorter, Dean, Shahmir Naseer, David Peggs, Charles McGurk, and Samuel Allen Moore Martin. 2023. "Deciphering the Immunostimulatory Effects of β-Glucan on a Rainbow Trout (Oncorhynchus mykiss) Macrophage-like Cell Line (RTS11) by Whole Transcriptome Analysis" Genes 14, no. 6: 1261. https://doi.org/10.3390/genes14061261
APA StylePorter, D., Naseer, S., Peggs, D., McGurk, C., & Martin, S. A. M. (2023). Deciphering the Immunostimulatory Effects of β-Glucan on a Rainbow Trout (Oncorhynchus mykiss) Macrophage-like Cell Line (RTS11) by Whole Transcriptome Analysis. Genes, 14(6), 1261. https://doi.org/10.3390/genes14061261







