Multi-Omics Pipeline and Omics-Integration Approach to Decipher Plant’s Abiotic Stress Tolerance Responses
Abstract
:1. Introduction
2. Genomics
2.1. Functional Genomics
2.1.1. Sequencing-Based Approaches
2.1.2. Hybridization-Based Approaches
2.1.3. Expansions to Functional Genomics Approaches
2.2. Structural Genomics
2.2.1. Genomic Selection (GS)
2.2.2. Genome Sequencing and Mapping
| Plants | QTLs/Markers | Chr. Location | Methods Used | Abiotic Stresses | References |
|---|---|---|---|---|---|
| Rice | OsHKT1;1 | Chr 1 | GWAS | Salinity | [39] |
| qWUE.STI6 | Chr 6 | Linkage mapping | Drought | [80] | |
| Saltol | Chr 1 | Linkage mapping | Salinity | [87] | |
| qCTBB2 qCTBB3 | Chr 2 Chr 3 | Linkage mapping | Cold | [89] | |
| qSTS4 | Chr 4 | QTL-seq | Salinity | [90] | |
| Soybean | AX-93897192 | Chr 19 | GWAS | Phosphorus efficiency | [40] |
| qGI10-1 | Chr 10 | GWAS | Drought | [79] | |
| qSFT_3-38, qSFT_7-3 | Chr 3 Chr 7 | Linkage mapping | Flooding | [91] | |
| qST6 qST10 | Chr 6 Chr 10 | Genotype-based sequencing (GBS) | Salinity | [92] | |
| Wheat | MQTL1D.4 MQTL2D.5 MQTL3A.1 | Chr 1D Chr 2D Chr 3A | MetaQTL | Drought stress Heat, Salinity Waterlogging | [41] |
| QNa.asl-2A | Chr 2A | Genotype-based sequencing (GBS) | Salinity | [81] | |
| YIELD_MQTL4B.2_D | Chr 4B | MetaQTL | Heat, Drought | [85] | |
| qWMs108_7-1 | Chr 7-3A | Linkage mapping | Drought | [93] | |
| QSpad3.ua-1D.5 | Chr 1D | GWAS | Waterlogging | [94] | |
| QMrl3B(T2|T1) | Chr 3B | Linkage mapping | Salinity | [95] | |
| Maize | Zm00001eb013650 | Chr 1-10 | GWAS + RNAseq | Salinity | [42] |
| qPOD2b | Chr 2 | Genome-Wide Association Study (GWAS) | Cold | [96] | |
| Rapeseed | SA07_23415428 | Chr SA07 | GWAS | Freezing | [45] |
| qDSI_SL-11-3 qDSI_RL-11-1 qDSI_RL-11-4 qDSI_SL11-3 | Chr C01 | Linkage mapping | Drought, Freezing | [97] | |
| qRRL.A3b | Chr A03 | Linkage mapping | Waterlogging | [98] | |
| Barley | QcRWC.3H_2.1 QcWC.3H_1 3H | Chr 3H | Linkage mapping | Drought | [76] |
| HORVU2Hr1G111780.3 | Chr 2H | Linkage mapping | Salinity | [82] | |
| qSLS-4 | Chr 4H | Linkage mapping | Salinity | [88] | |
| QBIO.2H | Chr 2H | GWAS | Waterlogging | [99] | |
| Cotton | qtlCSI01 | Chr 3 | Composite interval mapping | Drought | [47] |
| qGR-Chr4-3, qFER-Chr12-3, qFER-Chr15-1 | Chr 4 Chr 12 Chr 15 | Linkage mapping | Salinity | [48] | |
| qEC_A02_ck qFW_A06_150.1 | Chr 2 Chr 6 | Genotyping by Sequencing (GBS) | Salinity | [49] | |
| qFSHa1 | Chr 15 | Composite interval mapping | Heat | [86] | |
| Sorghum | qPH-6 qMC2-9 | Chr 6 Chr 9 | Genotype-based sequencing (GBS) | Excess soil nitrogen | [50] |
| qTB45_4.S | Chr 4 | Linkage mapping | Salinity | [51] |
2.2.3. Molecular Marker Resources
2.3. Comparative Genomics
3. Transcriptomics
4. Proteomics
5. Bioinformatics
6. Epigenetics-Aided Epigenomics
7. Metabolomics
8. Proteogenomics, Lipidomics, Ionomics and Interactomics
9. Phenomics
10. Integration of Multi-Omics Data and Interpretation for Abiotic Stress Response in Plants
11. Conclusions and Perspectives
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abobatta, W.F. The influence of climate change on interactions between environmental stresses and plants. In Plant Stress Mitigators; Ghorbanpour, M., Shahid, M.A., Eds.; Academic Press: Cambridge, MA, USA, 2023; pp. 425–434. [Google Scholar] [CrossRef]
- Li, Y.; Roychowdhury, R.; Govta, L.; Jaiwar, S.; Wei, Z.Z.; Shams, I.; Fahima, T. Intracellular reactive oxygen species (intraROS)-aided localized cell death contributing to immune responses against wheat powdery mildew pathogen. Phytopathology 2023. [Google Scholar] [CrossRef]
- Del Buono, D.; Regni, L.; Proietti, P. Abiotic stresses, biostimulants and plant activity. Agriculture 2023, 13, 191. [Google Scholar] [CrossRef]
- Roychowdhury, R. Crop Improvement in the Era of Climate Change; IK International Publisher: New Delhi, India, 2014; p. 496. ISBN 978-939-045-590-4. [Google Scholar]
- Roychowdhury, R.; Choudhury, S.; Hasanuzzaman, M.; Srivastava, S. Sustainable Agriculture in the Era of Climate Change; Springer-Nature: Basel, Switzerland, 2020; p. 690. ISBN 978-303-045-669-6. [Google Scholar]
- Hasanuzzaman, M.; Roychowdhury, R.; Karmakar, J.; Dey, N.; Nahar, K.; Fujita, M. Recent advances in biotechnology and genomic approaches for abiotic stress tolerance in crop plants. In Genomics and Proteomics: Concepts, Technologies and Applications, 1st ed.; Thangadurai, D., Sangeetha, J., Eds.; Apple Academic Press: Burlington, ON, Canada, 2015; pp. 333–366. ISBN 978-177-463-537-7. [Google Scholar]
- Rai, K.K.; Kumar, A.; Rai, A.; Rai, V.P.; Rai, A.C. Conventional breeding approaches for abiotic stress management in horticultural crops. In Stress Tolerance in Horticultural Crops; Rai, A.C., Rai, A., Rai, K.K., Rai, V.P., Kumar, A., Eds.; Woodhead Publishing: Cambridge, UK; Elsevier: Alpharetta, GA, USA, 2021; pp. 21–32. [Google Scholar] [CrossRef]
- Rai, A.C.; Rai, A.; Rai, K.K.; Rai, V.P.; Kumar, A. Stress Tolerance in Horticultural Crops; Woodhead Publishing: Cambridge, UK; Elsevier: Alpharetta, GA, USA, 2021. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Tah, J. Mutagenesis—A potential approach for crop improvement. In Crop Improvement—New Approaches and Modern Techniques, 1st ed.; Hakeem, K.R., Ahmad, P., Ozturk, M., Eds.; Springer: Boston, MA, USA, 2013; pp. 149–187. ISBN 978-148-997-357-3. [Google Scholar]
- Roychowdhury, R.; Karmakar, J.; Adak, M.K.; Dey, N. Physio-biochemical and microsatellite-based profiling of lowland rice (Oryza sativa L.) landraces for osmotic stress tolerance. Am. J. Plant Sci. 2013, 4, 52–63. [Google Scholar] [CrossRef] [Green Version]
- Roychowdhury, R.; Taoutaou, A.; Hakeem, K.R.; Gawwad, M.R.A.; Tah, J. Molecular marker-assisted technologies for crop improvement. In Crop Improvement in the Era of Climate Change, 1st ed.; Roychowdhury, R., Ed.; IK International Publishing House: New Delhi, India, 2014; pp. 241–258. ISBN 978-939-045-590-4. [Google Scholar]
- Deshmukh, R.; Sonah, H.; Patil, G.; Chen, W.; Prince, S.; Mutava, R.; Vuong, T.; Valliyodan, B.; Nguyen, H.T. Integrating omic approaches for abiotic stress tolerance in soybean. Front. Plant Sci. 2014, 5, 244. [Google Scholar] [CrossRef] [PubMed]
- Henry, V.J.; Bandrowski, A.E.; Pepin, A.S.; Gonzalez, B.J.; Desfeux, A. OMICtools: An informative directory for multi-omic data analysis. Database 2014, 2014, bau069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sharma, T.; Gupta, A. Integrated OMICS approaches to ameliorate the abiotic stress in Brassica napus. In Plant Stress: Challenges and Management in the New Decade; Roy, S., Mathur, P., Chakraborty, A.P., Saha, S.P., Eds.; Springer-Nature: Basel, Switzerland, 2022; pp. 361–373. ISBN 978-303-095-365-2. [Google Scholar]
- Khandagale, K.; Krishna, R.; Roylawar, P.; Ade, A.B.; Benke, A.; Shinde, B.; Singh, M.; Gawande, S.J.; Rai, A. Omics approaches in Allium research: Progress and way ahead. PeerJ 2020, 8, e9824. [Google Scholar] [CrossRef]
- Muthuramalingam, P.; Jeyasri, R.; Rakkammal, K.; Satish, L.; Shamili, S.; Karthikeyan, A.; Valliammai, A.; Priya, A.; Selvaraj, A.; Gowri, P.; et al. Multi-Omics and integrative approach towards understanding salinity tolerance in rice: A review. Biology 2022, 11, 1022. [Google Scholar] [CrossRef]
- Raza, A. Plant biotechnological tools: Solutions for raising climate-resilient crop plants. Mod. Phytomorphol. 2022, 15, 132–133. [Google Scholar]
- Ashraf, U.; Mahmood, S.; Shahid, N.; Imran, M.; Siddique, M.; Abrar, M. Multi-omics approaches for strategic improvements of crops under changing climatic conditions. In Principles and Practices of Omics and Genome Editing for Crop Improvement; Prakash, C.S., Fiaz, S., Fahad, S., Eds.; Springer-Nature: Basel, Switzerland, 2022; pp. 57–92. ISBN 978-303-096-925-7. [Google Scholar]
- Zhou, R.; Jiang, F.; Niu, L.; Song, X.; Yu, L.; Yang, Y.; Wu, Z. Increase crop resilience to heat stress using omic strategies. Front. Plant. Sci. 2022, 13, 891861. [Google Scholar] [CrossRef]
- Weckwerth, W.; Ghatak, A.; Bellaire, A.; Chaturvedi, P.; Varshney, R.K. PANOMICS meets germplasm. Plant Biotechnol. J. 2020, 18, 1507–1525. [Google Scholar] [CrossRef] [Green Version]
- Subramanian, I.; Verma, S.; Kumar, S.; Jere, A.; Anamika, K. Multi-omics data integration, interpretation, and its application. Bioinform. Biol. Insights 2020, 14, 1177932219899051. [Google Scholar] [CrossRef] [Green Version]
- Derbyshire, M.C.; Batley, J.; Edwards, D. Use of multiple ‘omics techniques to accelerate the breeding of abiotic stress tolerant crops. Curr. Plant Biol. 2022, 32, 100262. [Google Scholar] [CrossRef]
- Khan, M.K.R.; Ditta, A.; Wang, B.; Fang, L.; Anwar, Z.; Ijaz, A.; Ahmed, S.R.; Khan, S.M. The intervention of multi-omics approaches for developing abiotic stress resistance in cotton crop under climate change. In Sustainable Agriculture in the Era of the OMICs Revolution; Prakash, C.S., Fiaz, S., Nadeem, M.A., Baloch, F.S., Qayyum, A., Eds.; Springer: Cham, Switzerland, 2023; pp. 37–82. [Google Scholar] [CrossRef]
- Vennapusa, A.R.; Nimmakayala, P.; Zaman-Allah, M.A.; Ratnakumar, P. Physiological, molecular, and genetic perspectives of environmental stress response in plants. Front. Plant Sci. 2023, 14, 1213762. [Google Scholar] [CrossRef]
- Parida, S.K.; Mondal, N.; Yadav, R.; Vishwakarma, H.; Rana, J.C. Mining legume germplasm for genetic gains: An Indian perspective. Front. Genet. 2023, 14, 996828. [Google Scholar] [CrossRef]
- Parray, J.A.; Yaseen Mir, M.; Shameem, N.; Parray, J.A.; Yaseen Mir, M.; Shameem, N. Advancement in sustainable agriculture: Computational and bioinformatics tools. In Sustainable Agriculture: Biotechniques in Plant Biology; Parray, J.A., Yaseen Mir, M., Shameem, N., Eds.; Springer: Singapore, 2019; pp. 465–547. [Google Scholar] [CrossRef]
- Thakkar, S.; Banerjee, A.; Goel, S.; Roy, S.; Bansal, K.C. Genomics-based approaches to improve abiotic stress tolerance in plants: Present status and future prospects. In Plant Perspectives to Global Climate Changes—Developing Climate-Resilient Plants; Aftab, T., Roychoudhury, A., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 195–219. [Google Scholar] [CrossRef]
- Xu, W.; Zhang, H.; Zhang, Y.; Shen, P.; Li, X.; Li, R.; Yang, L. A paired-end whole-genome sequencing approach enables comprehensive characterization of transgene integration in rice. Commun. Biol. 2022, 5, 667. [Google Scholar] [CrossRef]
- Zanini, S.F.; Bayer, P.E.; Wells, R.; Snowdon, R.J.; Batley, J.; Varshney, R.K.; Nguyen, H.T.; Edwards, D.; Golicz, A.A. Pangenomics in crop improvement-from coding structural variations to finding regulatory variants with pangenome graphs. Plant Genome 2022, 15, e20177. [Google Scholar] [CrossRef]
- Sharma, N.; Siddappa, S.; Malhotra, N.; Thakur, K.; Salaria, N.; Sood, S.; Bhardwaj, V. Advances in potato functional genomics: Implications for crop improvement. Plant Cell Tissue Organ Cult. 2022, 148, 447–464. [Google Scholar] [CrossRef]
- Singh, R.; Kumar, K.; Bharadwaj, C.; Verma, P.K. Broadening the horizon of crop research: A decade of advancements in plant molecular genetics to divulge phenotype governing genes. Planta 2022, 255, 46. [Google Scholar] [CrossRef]
- Jain, D.; Ashraf, N.; Khurana, J.P.; Kameshwari, S. The ‘omics’ approach for crop improvement against drought stress. In Genetic Enhancement of Crops for Tolerance to Abiotic Stress: Mechanisms and Approaches; Rajpal, V.R., Sehgal, D., Kumar, A., Raina, S.N., Eds.; Springer-Nature: Basel, Switzerland, 2019; Volume I, pp. 183–204. ISBN 978-331-991-956-0. [Google Scholar]
- Deokar, A.A.; Kondawar, V.; Jain, P.K.; Karuppayil, S.M.; Raju, N.L.; Vadez, V.; Varshney, R.K.; Srinivasan, R. Comparative analysis of expressed sequence tags (ESTs) between drought-tolerant and -susceptible genotypes of chickpea under terminal drought stress. BMC Plant Biol. 2011, 11, 70. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Scholtens, D.; Datta, S. Bioinformatics Methods: From Omics to Next Generation Sequencing; Chapman and Hall/CRC Press: London, UK, 2022; p. 336. ISBN 978-149-876-515-2. [Google Scholar]
- Girma, G.; Natsume, S.; Carluccio, A.V.; Takagi, H.; Matsumura, H.; Uemura, A.; Muranaka, S.; Takagi, H.; Stavolone, L.; Gedil, M.; et al. Identification of candidate flowering and sex genes in white Guinea yam (D. rotundata Poir.) by SuperSAGE transcriptome profiling. PLoS ONE 2019, 14, e0216912. [Google Scholar] [CrossRef] [Green Version]
- El-Sappah, A.H.; Rather, S.A. Genomics approaches to study abiotic stress tolerance in plants. In Plant Abiotic Stress Physiology; Aftab, T., Hakeem, R., Eds.; Apple Academic Press: Palm Bay, FL, USA, 2022; pp. 25–46. ISBN 978-100-318-057-9. [Google Scholar]
- Kulwal, P.L.; Mir, R.R.; Varshney, R.K. Efficient breeding of crop plants. In Fundamentals of Field Crop Breeding; Yadava, D.K., Dikshit, H.K., Mishra, G.P., Tripathi, S., Eds.; Springer-Nature: Singapore, 2022; pp. 745–777. ISBN 978-981-169-257-4. [Google Scholar]
- Zhu, F.; Ahchige, M.W.; Brotman, Y.; Alseekh, S.; Zsögön, A.; Fernie, A.R. Bringing more players into play: Leveraging stress in genome wide association studies. J. Plant Physiol. 2022, 271, 153657. [Google Scholar] [CrossRef] [PubMed]
- Lv, Y.; Ma, J.; Wei, H.; Xiao, F.; Wang, Y.; Jahan, N.; Hazman, M.; Qian, Q.; Shang, L.; Guo, L. Combining GWAS, genome-wide domestication and a transcriptomic analysis reveals the loci and natural alleles of salt tolerance in rice (Oryza sativa L.). Front. Plant Sci. 2022, 13, 912637. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Ning, L.; Yu, W.; Zhao, W.; Huang, F.; Yu, D.; Wang, H.; Cheng, H. Detection of candidate loci and genes related to phosphorus efficiency at maturity through a genome-wide association study in Soybean. Agronomy 2022, 12, 2031. [Google Scholar] [CrossRef]
- Tanin, M.J.; Saini, D.K.; Sandhu, K.S.; Pal, N.; Gudi, S.; Chaudhary, J.; Sharma, A. Consensus genomic regions associated with multiple abiotic stress tolerance in wheat and implications for wheat breeding. Sci. Rep. 2022, 12, 13680. [Google Scholar] [CrossRef]
- Liu, P.; Zhu, Y.; Liu, H.; Liang, Z.; Zhang, M.; Zou, C.; Yuan, G.; Gao, S.; Pan, G.; Shen, Y.; et al. A Combination of a genome-wide association study and a transcriptome analysis reveals circRNAs as new regulators involved in the response to salt stress in maize. Int. J. Mol. Sci. 2022, 23, 9755. [Google Scholar] [CrossRef]
- Zhang, X.; You, J.; Miao, H.; Zhang, H. Genomic designing for sesame resistance to abiotic stresses. In Genomic Designing for Abiotic Stress Resistant Oilseed Crops; Kole, C., Ed.; Springer-Nature: Basel, Switzerland, 2022; pp. 219–234. ISBN 978-303-090-044-1. [Google Scholar]
- Fatemi, F.; Kianersi, F.; Pour-Aboughadareh, A.; Poczai, P.; Jadidi, O. Overview of identified genomic regions associated with various agronomic and physiological traits in barley under abiotic stresses. Appl. Sci. 2022, 12, 5189. [Google Scholar] [CrossRef]
- Samineni, S.; Mahendrakar, M.D.; Hotti, A.; Chand, U.; Rathore, A.; Gaur, P.M. Impact of heat and drought stresses on grain nutrient content in chickpea: Genome-wide marker-trait associations for protein, Fe and Zn. Environ. Exp. Bot. 2022, 194, 104688. [Google Scholar] [CrossRef]
- Chao, W.S.; Li, X.; Horvath, D.P.; Anderson, J.V. Genetic loci associated with freezing tolerance in a European rapeseed (Brassica napus L.) diversity panel identified by genome-wide association mapping. Plant Direct 2022, 6, e405. [Google Scholar] [CrossRef]
- Shukla, R.P.; Tiwari, G.J.; Joshi, B.; Song-Beng, K.; Tamta, S.; Boopathi, N.M.; Jena, S.N. GBS-SNP and SSR based genetic mapping and QTL analysis for drought tolerance in upland cotton. Physiol. Mol. Biol. Plants 2021, 27, 1731–1745. [Google Scholar] [CrossRef]
- Guo, A.; Su, Y.; Nie, H.; Li, B.; Ma, X.; Hua, J. Identification of candidate genes involved in salt stress response at germination and seedling stages by QTL mapping in upland cotton. G3 2022, 12, jkac099. [Google Scholar] [CrossRef]
- Diouf, L.; Pan, Z.; He, S.P.; Gong, W.F.; Jia, Y.H.; Magwanga, R.O.; Romy, K.R.E.; Or Rashid, H.; Kirungu, J.N.; Du, X. High-density linkage map construction and mapping of salt-tolerant QTLs at seedling stage in upland cotton using genotyping by sequencing (GBS). Int. J. Mol. Sci. 2017, 18, 2622. [Google Scholar] [CrossRef] [Green Version]
- Gelli, M.; Konda, A.R.; Liu, K.; Zhang, C.; Clemente, T.E.; Holding, D.R.; Dweikat, I.M. Validation of QTL mapping and transcriptome profiling for identification of candidate genes associated with nitrogen stress tolerance in sorghum. BMC Plant Biol. 2017, 17, 123. [Google Scholar] [CrossRef] [Green Version]
- Hostetler, A.N.; Govindarajulu, R.; Hawkins, J.S. QTL mapping in an interspecific sorghum population uncovers candidate regulators of salinity tolerance. Plant Stress 2021, 2, 100024. [Google Scholar] [CrossRef]
- Parihar, A. Molecular breeding and marker-assisted selection for crop improvement. In Plant Genomics for Sustainable Agriculture; Singh, R.L., Mondal, S., Parihar, A., Singh, P.K., Eds.; Springer-Nature: Singapore, 2022; pp. 129–164. ISBN 978-981-166-974-3. [Google Scholar]
- Salvi, S.; Tuberosa, R. The crop QTLome comes of age. Curr. Opin. Biotechnol. 2015, 32, 179–185. [Google Scholar] [CrossRef]
- Ha, B.K.; Vuong, T.D.; Velusamy, V.; Nguyen, H.T.; Shannon, J.G.; Lee, J.D. Genetic mapping of quantitative trait loci conditioning salt tolerance in wild soybean (Glycine soja) PI 483463. Euphytica 2013, 193, 79–88. [Google Scholar] [CrossRef]
- Sheoran, S.; Gupta, M.; Kumari, S.; Kumar, S.; Rakshit, S. Meta-QTL analysis and candidate genes identification for various abiotic stresses in maize (Zea mays L.) and their implications in breeding programs. Mol. Breed. 2022, 42, 26. [Google Scholar] [CrossRef]
- Selamat, N.; Nadarajah, K.K. Meta-analysis of quantitative traits loci (QTL) identified in drought response in rice (Oryza sativa L.). Plants 2021, 10, 716. [Google Scholar] [CrossRef]
- Prakash, N.R.; Lokeshkumar, B.M.; Rathor, S.; Warraich, A.S.; Yadav, S.; Vinaykumar, N.M.; Dushynthkumar, B.M.; Krishnamurthy, S.L.; Sharma, P.C. Meta-analysis and validation of genomic loci governing seedling and reproductive stage salinity tolerance in rice. Physiol. Plant. 2022, 174, e13629. [Google Scholar] [CrossRef]
- Khan, I.; Zhang, Y.; Akbar, F.; Khan, J. Abiotic stress tolerance in cereals through genome editing. In Omics Approach to Manage Abiotic Stress in Cereals; Roychoudhury, A., Aftab, T., Acharya, K., Eds.; Springer-Nature: Singapore, 2022; pp. 295–319. ISBN 978-981-190-140-9. [Google Scholar]
- Singh, A.; Roychowdhury, R.; Singh, T.; Wang, W.; Yadav, D.; Kumar, A.; Modi, A.; Rai, A.C.; Ghughe, S.; Kumar, A.; et al. Improvement of crop’s stress tolerance by gene editing CRISPR/CAS9 system. In Sustainable Agriculture in the Era of Climate Change; Roychowdhury, R., Choudhury, S., Hasanuzzaman, M., Srivastava, S., Eds.; Springer-Nature: Basel, Switzerland, 2020; pp. 557–587. ISBN 978-303-045-669-6. [Google Scholar]
- Lou, D.; Wang, H.; Liang, G.; Yu, D. OsSAPK2 confers abscisic acid sensitivity and tolerance to drought stress in rice. Front. Plant Sci. 2017, 8, 993. [Google Scholar] [CrossRef] [Green Version]
- Jain, A.; Bhar, A.; Das, S. Improving biotic and abiotic stress tolerance in plants: A CRISPR-Cas approach. In Genome Engineering for Crop Improvement; Sarmah, B.K., Borah, B.K., Eds.; Springer-Nature: Basel, Switzerland, 2021; pp. 217–237. ISBN 978-303-063-372-1. [Google Scholar]
- Zhang, A.; Liu, Y.; Wang, F.; Li, T.; Chen, Z.; Kong, D.; Bi, J.; Zhang, F.; Luo, X.; Wang, J.; et al. Enhanced rice salinity tolerance via CRISPR/Cas9-targeted mutagenesis of the OsRR22 gene. Mol. Breed. 2019, 39, 47. [Google Scholar] [CrossRef] [Green Version]
- Bouzroud, S.; Gasparini, K.; Hu, G.; Barbosa, M.A.M.; Rosa, B.L.; Fahr, M.; Bendaou, N.; Bouzayen, M.; Zsögön, A.; Smouni, A.; et al. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes 2020, 11, 272. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Debbarma, J.; Sarki, Y.N.; Saikia, B.; Boruah, H.P.D.; Singha, D.L.; Chikkaputtaiah, C. Ethylene Response Factor (ERF) family proteins in abiotic stresses and CRISPR-Cas9 genome editing of ERFs for multiple abiotic stress tolerance in crop plants: A review. Mol. Biotechnol. 2019, 61, 153–172. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Que, Z.; Xia, Y.; Tang, N.; Li, D.; He, R.; Cao, M. Knock out of the annexin gene OsAnn3 via CRISPR/Cas9-mediated genome editing decreased cold tolerance in rice. J. Plant Biol. 2017, 60, 539–547. [Google Scholar] [CrossRef]
- Zhou, J. Sequence-based modeling of three-dimensional genome architecture from kilobase to chromosome scale. Nat. Genet. 2022, 54, 725–734. [Google Scholar] [CrossRef] [PubMed]
- Mérot, C.; Oomen, R.A.; Tigano, A.; Wellenreuther, M. A roadmap for understanding the evolutionary significance of structural genomic variation. Trends Ecol. Evol. 2020, 35, 561–572. [Google Scholar] [CrossRef] [PubMed]
- DoVale, J.C.; Carvalho, H.F.; Sabadin, F.; Fritsche-Neto, R. Reduction of genotyping marker density for genomic selection is not an affordable approach to long-term breeding in cross-pollinated crops. bioRxiv 2021. [Google Scholar] [CrossRef]
- Merrick, L.F.; Herr, A.W.; Sandhu, K.S.; Lozada, D.N.; Carter, A.H. Optimizing plant breeding programs for genomic selection. Agronomy 2022, 12, 714. [Google Scholar] [CrossRef]
- Rio, S.; Gallego-Sánchez, L.; Montilla-Bascón, G.; Canales, F.J.; Sánchez, J.I.Y.; Prats, E. Genomic prediction and training set optimization in a structured Mediterranean oat population. Theor. Appl. Genet. 2021, 134, 3595–3609. [Google Scholar] [CrossRef]
- Rogers, A.R.; Holland, J.B. Environment-specific genomic prediction ability in maize using environmental covariates depends on environmental similarity to training data. G3 2022, 12, jkab440. [Google Scholar] [CrossRef]
- Rio, S.; Akdemir, D.; Carvalho, T.; Sánchez, J.I.Y. Assessment of genomic prediction reliability and optimization of experimental designs in multi-environment trials. Theor. Appl. Genet. 2022, 135, 405–419. [Google Scholar] [CrossRef]
- Beyene, Y.; Gowda, M.; Pérez-Rodríguez, P.; Olsen, M.; Robbins, K.R.; Burgueño, J.; Prasanna, B.M.; Crossa, J. Application of genomic selection at the early stage of breeding pipeline in tropical maize. Front. Plant Sci. 2021, 12, 685488. [Google Scholar] [CrossRef]
- Udriște, A.A.; Iordachescu, M.; Ciceoi, R.; Bădulescu, L. Next-generation sequencing of local Romanian tomato varieties and bioinformatics analysis of the Ve locus. Int. J. Mol. Sci. 2022, 23, 9750. [Google Scholar] [CrossRef]
- Kress, W.J.; Soltis, D.E.; Kersey, P.J.; Wegrzyn, J.L.; Leebens-Mack, J.H.; Gostel, M.R.; Liu, X.; Soltis, P.S. Green plant genomes: What we know in an era of rapidly expanding opportunities. Proc. Natl. Acad. Sci. USA 2022, 119, e2115640118. [Google Scholar] [CrossRef]
- Giacopuzzi, E.; Popitsch, N.; Taylor, J.C. GREEN-DB: A framework for the annotation and prioritization of non-coding regulatory variants from whole-genome sequencing data. Nucleic Acids Res. 2022, 50, 2522–2535. [Google Scholar] [CrossRef]
- Pérez-Wohlfeil, E.; Diaz-Del-Pino, S.; Trelles, O. Ultra-fast genome comparison for large-scale genomic experiments. Sci. Rep. 2019, 9, 10274. [Google Scholar] [CrossRef] [Green Version]
- Ramkumar, T.R.; Arya, S.S.; Kumari, D.D.; Lenka, S.K. Brassica juncea genome sequencing: Structural and functional insights. In The Brassica juncea Genome; Kole, C., Mohapatra, T., Eds.; Springer-Nature: Basel, Switzerland, 2022; pp. 221–240. ISBN 978-303-091-507-0. [Google Scholar]
- Song, J.M.; Zhang, Y.; Zhou, Z.W.; Lu, S.; Ma, W.; Lu, C.; Chen, L.L.; Guo, L. Oil plant genomes: Current state of the science. J. Exp. Bot. 2022, 73, 2859–2874. [Google Scholar] [CrossRef]
- Song, J.; Xu, D.; Dong, Y.; Li, F.; Bian, Y.; Li, L.; Luo, X.; Fei, S.; Li, L.; Zhao, C.; et al. Fine mapping and characterization of a major QTL for grain weight on wheat chromosome arm 5DL. Theor. Appl. Genet. 2022, 135, 3237–3246. [Google Scholar] [CrossRef]
- Satrio, R.D.; Fendiyanto, M.H.; Supena, E.D.J.; Suharsono, S.; Miftahudin, M. Genome-wide SNP discovery, linkage mapping, and analysis of QTL for morpho-physiological traits in rice during vegetative stage under drought stress. Physiol. Mol. Biol. Plants 2021, 27, 2635–2650. [Google Scholar] [CrossRef]
- Sun, M.; Li, Y.; Zheng, J.; Wu, D.; Li, C.; Li, Z.; Zang, Z.; Zhang, Y.; Fang, Q.; Li, W.; et al. A nuclear factor Y-B transcription factor, GmNFYB17, regulates resistance to drought stress in soybean. Int. J. Mol. Sci. 2022, 23, 7242. [Google Scholar] [CrossRef]
- Mwando, E.; Han, Y.; Angessa, T.; Zhang, X.Q.; Li, C. Fine-mapping and characterisation of genes on barley (Hordeum vulgare) chromosome 2H for salinity stress tolerance during germination. Crop. J. 2022, 10, 754–766. [Google Scholar] [CrossRef]
- Makhtoum, S.; Sabouri, H.; Gholizadeh, A.; Ahangar, L.; Katouzi, M. QTLs controlling physiological and morphological traits of barley (Hordeum vulgare L.) seedlings under salinity, drought, and normal conditions. BioTech 2022, 11, 26. [Google Scholar] [CrossRef] [PubMed]
- Singh, R.K.; Gregorio, G.B.; Jain, R.K. QTL mapping for salinity tolerance in rice. Physiol. Mol. Biol. Plant. 2007, 13, 87–99. [Google Scholar]
- Asif, M.A.; Garcia, M.; Tilbrook, J.; Brien, C.; Dowling, K.; Berger, B.; Schilling, R.K.; Short, L.; Trittermann, C.; Gilliham, M.; et al. Identification of salt tolerance QTL in a wheat RIL mapping population using destructive and non-destructive phenotyping. Funct. Plant Biol. 2022, 49, 672. [Google Scholar] [CrossRef] [PubMed]
- Touzy, G.; Lafarge, S.; Redondo, E.; Lievin, V.; Decoopman, X.; Le Gouis, J.; Praud, S. Identification of QTLs affecting post-anthesis heat stress responses in European bread wheat. Theor. Appl. Genet. 2022, 135, 947–964. [Google Scholar] [CrossRef]
- Rani, S.; Baber, M.; Naqqash, T.; Malik, S.A. Identification and genetic mapping of potential QTLs conferring heat tolerance in cotton (Gossypium hirsutum L.) by using micro satellite marker’s approach. Agronomy 2022, 12, 1381. [Google Scholar] [CrossRef]
- Yang, L.; Lei, L.; Li, P.; Wang, J.; Wang, C.; Yang, F.; Chen, J.; Liu, H.; Zheng, H.; Xin, W.; et al. Identification of candidate genes conferring cold tolerance to rice (Oryza sativa L.) at the bud-bursting stage using bulk segregant analysis sequencing and linkage mapping. Front. Plant Sci. 2021, 12, 647239. [Google Scholar] [CrossRef]
- Lei, L.; Cao, L.; Ding, G.; Zhou, J.; Luo, Y.; Bai, L.; Xia, T.; Chen, L.; Wang, J.; Liu, K.; et al. OsBBX11 on qSTS4 links to salt tolerance at the seeding stage in Oryza sativa L. ssp. Japonica. Front. Plant Sci. 2023, 14, 1139961. [Google Scholar] [CrossRef]
- Dhungana, S.K.; Kim, H.S.; Kang, B.K.; Seo, J.H.; Kim, H.T.; Shin, S.O.; Oh, J.H.; Baek, I.Y. Identification of QTL for tolerance to flooding stress at seedling stage of soybean (Glycine max L. Merr.). Agronomy 2021, 11, 908. [Google Scholar] [CrossRef]
- Cho, K.H.; Kim, M.Y.; Kwon, H.; Yang, X.; Lee, S.H. Novel QTL identification and candidate gene analysis for enhancing salt tolerance in soybean (Glycine max (L.) Merr.). Plant Sci. 2021, 313, 111085. [Google Scholar] [CrossRef]
- Khaled, K.A.M.; Habiba, R.M.M.; Bashasha, J.A.; El-Aziz, M.H.A. Identification and mapping of QTL associated with some traits related for drought tolerance in wheat using SSR markers. Beni-Suef Univ. J. Basic Appl. Sci. 2022, 11, 38. [Google Scholar] [CrossRef]
- Xu, L.; Zhao, C.; Pang, J.; Niu, Y.; Liu, H.; Zhang, W.; Zhou, M. Genome-wide association study reveals quantitative trait loci for waterlogging-triggered adventitious roots and aerenchyma formation in common wheat. Front. Plant Sci. 2022, 13, 1066752. [Google Scholar] [CrossRef]
- Guo, X.; Wu, C.; Wang, D.; Wang, G.; Jin, K.; Zhao, Y.; Tian, J.; Deng, Z. Conditional QTL mapping for seed germination and seedling traits under salt stress and candidate gene prediction in wheat. Sci. Rep. 2022, 12, 21010. [Google Scholar] [CrossRef]
- Jin, Y.; Zhang, Z.; Xi, Y.; Yang, Z.; Xiao, Z.; Guan, S.; Qu, J.; Wang, P.; Zhao, R. Identification and functional verification of cold tolerance genes in spring maize seedlings based on a genome-wide association study and quantitative trait locus mapping. Front. Plant Sci. 2021, 12, 776972. [Google Scholar] [CrossRef]
- Gad, M.; Chao, H.; Li, H.; Zhao, W.; Lu, G.; Li, M. QTL Mapping for seed germination response to drought stress in Brassica napus. Front. Plant Sci. 2020, 11, 629970. [Google Scholar] [CrossRef]
- Ding, X.Y.; Xu, J.S.; Huang, H.; Xing, Q.I.A.O.; Shen, M.Z.; Cheng, Y.; Zhang, X. Unraveling waterlogging tolerance-related traits with QTL analysis in reciprocal intervarietal introgression lines using genotyping by sequencing in rapeseed (Brassica napus L.). J. Integr. Agric. 2020, 19, 1974–1983. [Google Scholar] [CrossRef]
- Borrego-Benjumea, A.; Carter, A.; Zhu, M.; Tucker, J.R.; Zhou, M.; Badea, A. Genome-wide association study of waterlogging tolerance in barley (Hordeum vulgare L.) under controlled field conditions. Front. Plant Sci. 2021, 12, 711654. [Google Scholar] [CrossRef]
- Sarkar, B.; Varalaxmi, Y.; Vanaja, M.; Kumar, N.R.; Prabhakar, M.; Jyothilakshmi, N.; Yadav, S.K.; Maheswari, M.; Singh, V.K. Genome-wide SNP discovery, identification of QTLs and candidate genes associated with morpho-physiological and yield related traits for drought tolerance in maize. ResearchSquare 2022. [Google Scholar] [CrossRef]
- Wijerathna-Yapa, A.; Ramtekey, V.; Ranawaka, B.; Basnet, B.R. Applications of in vitro tissue culture technologies in breeding and genetic improvement of wheat. Plants 2022, 11, 2273. [Google Scholar] [CrossRef]
- Maldonado-Alconada, A.M.; Castillejo, M.Á.; Rey, M.D.; Labella-Ortega, M.; Tienda-Parrilla, M.; Hernández-Lao, T.; Honrubia-Gómez, I.; Ramírez-García, J.; Guerrero-Sanchez, V.M.; López-Hidalgo, C.; et al. Multiomics molecular research into the recalcitrant and orphan Quercus ilex tree species: Why, what for, and how. Int. J. Mol. Sci. 2022, 23, 9980. [Google Scholar] [CrossRef]
- Brake, M.; Al-Qadumii, L.; Hamasha, H.; Migdadi, H.; Awad, A.; Haddad, N.; Sadder, M.T. Development of SSR markers linked to stress responsive genes along tomato chromosome 3 (Solanum lycopersicum L.). BioTech 2022, 11, 34. [Google Scholar] [CrossRef]
- Shamim, M.; Kumar, M.; Srivastava, D. Molecular markers mediated heat stress tolerance in crop plants. In Thermotolerance in Crop Plants; Kumar, R.R., Praveen, S., Rai, G.K., Eds.; Springer-Nature: Singapore, 2022; pp. 23–44. ISBN 978-981-193-800-9. [Google Scholar]
- Kumar, P.; Patni, B.; Singh, M. Wheat genome sequence opens new opportunities to understand the genetic basis of frost tolerance (FT) and marker-assisted breeding in wheat (Triticum aestivum L.). J. Stress Physiol. Biochem. 2022, 18, 17–27. [Google Scholar]
- Chugh, V.; Kaur, D.; Purwar, S.; Kaushik, P.; Sharma, V.; Kumar, H.; Rai, A.; Singh, C.M.; Kamaluddin; Dubey, R.B. Applications of molecular markers for developing abiotic-stress-resilient oilseed crops. Life 2023, 13, 88. [Google Scholar] [CrossRef] [PubMed]
- Bhatia, D.; Bajwa, G.S. Molecular marker techniques and recent advancements. In Genotyping by Sequencing for Crop Improvement; Sonah, H., Goyal, V., Shivaraj, S.M., Deshmukh, R.K., Eds.; John Wiley & Sons Ltd.: Chichester, UK, 2022; pp. 1–21. ISBN 978-111-974-568-6. [Google Scholar]
- Das, A.; Singh, S.; Islam, Z.; Munshi, A.D.; Behera, T.K.; Dutta, S.; Weng, Y.; Dey, S.S. Current progress in genetic and genomics-aided breeding for stress resistance in cucumber (Cucumis sativus L.). Sci. Hortic. 2022, 300, 111059. [Google Scholar] [CrossRef]
- Khan, N.; You, F.M.; Cloutier, S. Designing genomic solutions to enhance abiotic Stress resistance in flax. In Genomic Designing for Abiotic Stress Resistant Oilseed Crops; Kole, C., Ed.; Springer-Nature: Basel, Switzerland, 2022; pp. 251–283. ISBN 978-303-090-044-1. [Google Scholar]
- Avni, R.; Lux, T.; Minz-Dub, A.; Millet, E.; Sela, H.; Distelfeld, A.; Deek, J.; Yu, G.; Steuernagel, B.; Pozniak, C.; et al. Genome sequences of three Aegilops species of the section Sitopsis reveal phylogenetic relationships and provide resources for wheat improvement. Plant J. 2022, 110, 179–192. [Google Scholar] [CrossRef] [PubMed]
- Kalyana Babu, B.; Sood, S.; Gaur, V.S.; Kumar, A. Comparative genomics of finger millet. In The Finger Millet Genome; Kumar, A., Sood, S., Kalyana Babu, B., Gupta, S.M., Dayakar Rao, B., Eds.; Springer-Nature: Basel, Switzerland, 2022; pp. 113–121. ISBN 978-303-100-868-9. [Google Scholar]
- Fang, Y.; Qin, X.; Liao, Q.; Du, R.; Luo, X.; Zhou, Q.; Li, Z.; Chen, H.; Jin, W.; Yuan, Y.; et al. The genome of homosporous maidenhair fern sheds light on the euphyllophyte evolution and defences. Nat. Plants 2021, 8, 1024–1037. [Google Scholar] [CrossRef] [PubMed]
- Hill, M.J.; Penning, B.W.; McCann, M.C.; Carpita, N.C. COMPILE: A GWAS computational pipeline for gene discovery in complex genomes. BMC Plant Biol. 2022, 22, 315. [Google Scholar] [CrossRef]
- Kaur, B.; Sandhu, K.S.; Kamal, R.; Kaur, K.; Singh, J.; Röder, M.S.; Muqaddasi, Q.H. Omics for the improvement of abiotic, biotic, and agronomic traits in major cereal crops: Applications, challenges, and prospects. Plants 2021, 10, 1989. [Google Scholar] [CrossRef]
- McGettigan, P.A. Transcriptomics in the RNA-seq era. Curr. Opin. Chem. Biol. 2013, 17, 4–11. [Google Scholar] [CrossRef]
- Kwasniewski, M.; Daszkowska-Golec, A.; Janiak, A.; Chwialkowska, K.; Nowakowska, U.; Sablok, G.; Szarejko, I. Transcriptome analysis reveals the role of the root hairs as environmental sensors to maintain plant functions under water-deficiency conditions. J. Exp. Bot. 2016, 67, 1079–1094. [Google Scholar] [CrossRef]
- Lowe, R.; Shirley, N.; Bleackley, M.; Dolan, S.; Shafee, T. Transcriptomics technologies. PLoS Comput. Biol. 2017, 13, e1005457. [Google Scholar] [CrossRef] [Green Version]
- Karkute, S.G.; Gujjar, R.S.; Rai, A.; Akhtar, M.; Singh, M.; Singh, B. Genome wide expression analysis of WRKY genes in tomato (Solanum lycopersicum) under drought stress. Plant Gene 2018, 13, 8–17. [Google Scholar] [CrossRef]
- Rai, A.C.; Rai, A.; Shah, K.; Singh, M. Engineered BcZAT12 gene mitigates salt stress in tomato seedlings. Physiol. Mol. Biol. Plants 2021, 27, 535–541. [Google Scholar] [CrossRef]
- Mishra, P.; Singh, P.; Rai, A.; Abhishek, K.; Shanmugam, V.; Aamir, M.; Kumar, A.; Malik, M.Z.; Singh, S.K. Abiotic stress-mediated transcription regulation, chromatin dynamics, and gene expression in plants: Arabidopsis as a role model. In Mitigation of Plant Abiotic Stress by Microorganisms; Santoyo, G., Kumar, A., Aamir, M., Uthandi, S., Eds.; Academic Press: Cambridge, MA, USA, 2022; pp. 321–345. [Google Scholar] [CrossRef]
- Tyagi, S.; Kabade, P.G.; Gnanapragasam, N.; Singh, U.M.; Gurjar, A.K.S.; Rai, A.; Sinha, P.; Kumar, A.; Singh, V.K. Codon usage provide insights into the adaptation of rice genes under stress condition. Int. J. Mol. Sci. 2023, 24, 1098. [Google Scholar] [CrossRef]
- Shah, T.; Xu, J.; Zou, X.; Cheng, Y.; Nasir, M.; Zhang, X. Omics approaches for engineering wheat production under abiotic stresses. Int. J. Mol. Sci. 2018, 19, 2390. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Liu, J.; Huang, Y.; Wu, H.; Hu, X.; Cheng, B.; Ma, Q.; Zhao, Y. Comparative transcriptomics reveals the molecular mechanism of the parental lines of Maize hybrid An’nong876 in response to salt stress. Int. J. Mol. Sci. 2022, 23, 5231. [Google Scholar] [CrossRef]
- Ramkumar, M.K.; Mulani, E.; Jadon, V.; Sureshkumar, V.; Krishnan, S.G.; Senthil Kumar, S.; Raveendran, M.; Singh, A.K.; Solanke, A.U.; Singh, N.K.; et al. Identification of major candidate genes for multiple abiotic stress tolerance at seedling stage by network analysis and their validation by expression profiling in rice (Oryza sativa L.). 3 Biotech 2022, 12, 127. [Google Scholar] [CrossRef]
- Zinati, Z.; Sazegari, S. Identification of important genes involved in priming induced drought tolerance in barley through transcriptomic data mining. Crop Pasture Sci. 2022, 73, 1011–1025. [Google Scholar] [CrossRef]
- Chen, C.; Shang, X.; Sun, M.; Tang, S.; Khan, A.; Zhang, D.; Yan, H.; Jiang, Y.; Yu, F.; Wu, Y.; et al. Comparative transcriptome analysis of two sweet Sorghum genotypes with different salt tolerance abilities to reveal the mechanism of salt tolerance. Int. J. Mol. Sci. 2022, 23, 2272. [Google Scholar] [CrossRef]
- Han, B.; Wang, F.; Liu, Z.; Chen, L.; Yue, D.; Sun, W.; Lin, Z.; Zhang, X.; Zhou, X.; Yang, X. Transcriptome and metabolome profiling of interspecific CSSLs reveals general and specific mechanisms of drought resistance in cotton. Theor. Appl. Genet. 2022, 135, 3375–3391. [Google Scholar] [CrossRef]
- Wang, X.; Song, S.; Wang, X.; Liu, J.; Dong, S. Transcriptomic and metabolomic analysis of seedling-stage soybean responses to PEG-simulated drought stress. Int. J. Mol. Sci. 2022, 23, 6869. [Google Scholar] [CrossRef]
- Guo, H.; Mao, M.; Deng, Y.; Sun, L.; Chen, R.; Cao, P.; Lai, J.; Zhang, Y.; Wang, C.; Li, C.; et al. Multi-omics analysis reveals that SlERF.D6 synergistically regulates SGAs and fruit development. Front. Plant Sci. 2022, 13, 860577. [Google Scholar] [CrossRef] [PubMed]
- Smita, S.; Katiyar, A.; Lenka, S.K.; Dalal, M.; Kumar, A.; Mahtha, S.K.; Yadav, G.; Chinnusamy, V.; Pandey, D.M.; Bansal, K.C. Gene network modules associated with abiotic stress response in tolerant rice genotypes identified by transcriptome meta-analysis. Funct. Integr. Genom. 2020, 20, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Azzouz-Olden, F.; Hunt, A.G.; Dinkins, R. Transcriptome analysis of drought-tolerant Sorghum genotype SC56 in response to water stress reveals an oxidative stress defense strategy. Mol. Biol. Rep. 2020, 47, 3291–3303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Liu, X.; Zhang, D.; Tang, H.; Sun, B.; Li, C.; Hao, L.; Liu, C.; Li, Y.; Shi, Y.; et al. Genome-wide identification of gene expression in contrasting maize inbred lines under field drought conditions reveals the significance of transcription factors in drought tolerance. PLoS ONE 2017, 12, e0179477. [Google Scholar] [CrossRef] [Green Version]
- Satya, P.; Bhattacharjee, S.; Sarkar, D.; Roy, S.; Sharma, L.; Mandal, N.A. Transcriptomics in plant. In Plant Genomics for Sustainable Agriculture; Singh, R.L., Mondal, S., Parihar, A., Singh, P.K., Eds.; Springer-Nature: Singapore, 2022; pp. 99–127. ISBN 978-981-166-974-3. [Google Scholar]
- Tiwari, J.K.; Buckseth, T.; Zinta, R.; Saraswati, A.; Singh, R.K.; Rawat, S.; Dua, V.K.; Chakrabarti, S.K. Transcriptome analysis of potato shoots, roots and stolons under nitrogen stress. Sci. Rep. 2020, 10, 1152. [Google Scholar] [CrossRef] [Green Version]
- Song, Y.; Lv, J.; Ma, Z.; Dong, W. The mechanism of alfalfa (Medicago sativa L.) response to abiotic stress. Plant Growth Regul. 2019, 89, 239–249. [Google Scholar] [CrossRef]
- Samtani, H.; Sharma, A.; Khurana, P. Overexpression of HVA1 enhances drought and heat stress tolerance in Triticum aestivum doubled haploid plants. Cells 2022, 11, 912. [Google Scholar] [CrossRef]
- El-Esawi, M.A.; Al-Ghamdi, A.A.; Ali, H.M.; Ahmad, M. Overexpression of AtWRKY30 transcription factor enhances heat and drought stress tolerance in wheat (Triticum aestivum L.). Genes 2019, 10, 163. [Google Scholar] [CrossRef] [Green Version]
- Sun, J.Q.; Jiang, H.L.; Xu, Y.X.; Li, H.M.; Wu, X.Y.; Xie, Q.; Li, C.Y. The CCCH-type zinc finger proteins AtSZF1 and AtSZF2 regulate salt stress responses in Arabidopsis. Plant Cell Physiol. 2007, 48, 1148–1158. [Google Scholar] [CrossRef] [Green Version]
- Yun, S.D.; Kim, M.H.; Oh, S.A.; Soh, M.S.; Park, S.K. Overexpression of C-Repeat Binding Factor1 (CBF1) gene enhances heat stress tolerance in Arabidopsis. J. Plant Biol. 2022, 65, 253–260. [Google Scholar] [CrossRef]
- Yang, R.; Hong, Y.; Ren, Z.; Tang, K.; Zhang, H.; Zhu, J.K.; Zhao, C. A role for PICKLE in the regulation of cold and salt stress tolerance in Arabidopsis. Front. Plant Sci. 2019, 10, 900. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Wang, F.; Ma, Y.; Dang, H.; Hu, X. Transcription factor SlAREB1 is involved in the antioxidant regulation under saline–alkaline stress in tomato. Antioxidants 2022, 11, 1673. [Google Scholar] [CrossRef]
- Klay, I.; Gouia, S.; Liu, M.; Mila, I.; Khoudi, H.; Bernadac, A.; Bouzayen, M.; Pirrello, J. Ethylene Response Factors (ERF) are differentially regulated by different abiotic stress types in tomato plants. Plant Sci. 2018, 274, 137–145. [Google Scholar] [CrossRef]
- Liu, X.; Zhang, Q.; Yang, G.; Zhang, C.; Dong, H.; Liu, Y.; Yin, R.; Lin, L. Pivotal roles of tomato photoreceptor SlUVR8 in seedling development and UV-B stress tolerance. Biochem. Biophys. Res. Commun. 2020, 522, 177–183. [Google Scholar] [CrossRef]
- Munir, S.; Liu, H.; Xing, Y.; Hussain, S.; Ouyang, B.; Zhang, Y.; Li, H.; Ye, Z. Overexpression of calmodulin-like (ShCML44) stress-responsive gene from Solanum habrochaites enhances tolerance to multiple abiotic stresses. Sci. Rep. 2016, 6, 31772. [Google Scholar] [CrossRef]
- Lee, S.S.; Jung, W.Y.; Park, H.J.; Lee, A.; Kwon, S.Y.; Kim, H.S.; Cho, H.S. genome-wide analysis of alternative splicing in an inbred cabbage (Brassica oleracea L.) line ’HO’ in response to heat stress. Curr. Genom. 2018, 19, 12–20. [Google Scholar] [CrossRef]
- Meena, R.P.; Ghosh, G.; Vishwakarma, H.; Padaria, J.C. Expression of a Pennisetum glaucum gene DREB2A confers enhanced heat, drought and salinity tolerance in transgenic Arabidopsis. Mol. Biol. Rep. 2022, 49, 7347–7358. [Google Scholar] [CrossRef]
- Hu, J.; Zhou, J.; Peng, X.; Xu, H.; Liu, C.; Du, B.; Yuan, H.; Zhu, L.; He, G. The Bphi008a gene interacts with the ethylene pathway and transcriptionally regulates MAPK genes in the response of rice to brown planthopper feeding. Plant Physiol. 2011, 156, 856–872. [Google Scholar] [CrossRef] [Green Version]
- Yan, L.; Baoxiang, W.; Jingfang, L.; Zhiguang, S.; Ming, C.; Yungao, X.; Bo, X.; Bo, Y.; Jian, L.; Jinbo, L.; et al. A novel SAPK10-WRKY87-ABF1 biological pathway synergistically enhance abiotic stress tolerance in transgenic rice (Oryza sativa). Plant Physiol. Biochem. 2021, 168, 252–262. [Google Scholar] [CrossRef]
- Gupta, A.; Shaw, B.P. Biochemical and molecular characterisations of salt tolerance components in rice varieties tolerant and sensitive to NaCl: The relevance of Na+ exclusion in salt tolerance in the species. Funct. Plant Biol. 2020, 48, 72–87. [Google Scholar] [CrossRef]
- Zhao, H.; Li, Z.; Wang, Y.; Wang, J.; Xiao, M.; Liu, H.; Quan, R.; Zhang, H.; Huang, R.; Zhu, L.; et al. Cellulose synthase-like protein OsCSLD4 plays an important role in the response of rice to salt stress by mediating abscisic acid biosynthesis to regulate osmotic stress tolerance. Plant Biotechnol. J. 2022, 20, 468–484. [Google Scholar] [CrossRef] [PubMed]
- Zhong, R.; Wang, Y.; Gai, R.; Xi, D.; Mao, C.; Ming, F. Rice SnRK protein kinase OsSAPK8 acts as a positive regulator in abiotic stress responses. Plant Sci. 2020, 292, 110373. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Han, X.; Lu, Z.; Qiu, W.; Yu, M.; Li, H.; He, Z.; Zhuo, R. MAPK cascades and transcriptional factors: Regulation of heavy metal tolerance in plants. Int. J. Mol. Sci. 2022, 23, 4463. [Google Scholar] [CrossRef] [PubMed]
- Kokkanti, R.R.; Vemuri, H.; Gaddameedi, A.; Rayalacheruvu, U. Variability in drought stress-induced physiological, biochemical responses and expression of DREB2A, NAC4 and HSP70 genes in groundnut (Arachis hypogaea L.). S. Afr. J. Bot. 2022, 144, 448–457. [Google Scholar] [CrossRef]
- Chen, Y.; Li, C.; Zhang, B.; Yi, J.; Yang, Y.; Kong, C.; Lei, C.; Gong, M. The role of the late embryogenesis-abundant (LEA) protein family in development and the abiotic stress response: A comprehensive expression analysis of potato (Solanum Tuberosum). Genes 2019, 10, 148. [Google Scholar] [CrossRef] [Green Version]
- Conde, D.; Kirst, M. Decoding exceptional plant traits by comparative single-cell genomics. Trends Plant Sci. 2022, 27, 1095–1098. [Google Scholar] [CrossRef]
- Barkla, B.J.; Vera-Estrella, R.; Raymond, C. Single-cell-type quantitative proteomic and ionomic analysis of epidermal bladder cells from the halophyte model plant Mesembryanthemum crystallinum to identify salt-responsive proteins. BMC Plant Biol. 2016, 16, 110. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, P.; Abdel Latef, A.A.; Rasool, S.; Akram, N.A.; Ashraf, M.; Gucel, S. Role of proteomics in crop stress tolerance. Front. Plant Sci. 2016, 7, 1336. [Google Scholar] [CrossRef] [Green Version]
- Jan, N.; Rather, A.M.; John, R.; Chaturvedi, P.; Ghatak, A.; Weckwerth, W.; Zargar, S.M.; Mir, R.A.; Khan, M.A.; Mir, R.R. Proteomics for abiotic stresses in legumes: Present status and future directions. Crit. Rev. Biotechnol. 2022, 43, 171–190. [Google Scholar] [CrossRef]
- Li, J.; Sohail, H.; Nawaz, M.A.; Liu, C.; Yang, P. Physiological and proteomic analyses reveals that Brassinosteroids application improves the chilling stress tolerance of pepper seedlings. Plant Growth Regul. 2022, 96, 315–329. [Google Scholar] [CrossRef]
- Lohani, N.; Singh, M.B.; Bhalla, P.L. Biological parts for engineering abiotic stress tolerance in plants. BioDesign Res. 2022, 2022, 9819314. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Cereal crop proteomics: Systemic analysis of crop drought stress responses towards marker-assisted selection breeding. Front. Plant Sci. 2017, 8, 757. [Google Scholar] [CrossRef] [Green Version]
- Kausar, R.; Wang, X.; Komatsu, S. Crop proteomics under abiotic stress: From data to insights. Plants 2022, 11, 2877. [Google Scholar] [CrossRef]
- Yan, S.; Bhawal, R.; Yin, Z.; Thannhauser, T.W.; Zhang, S. Recent advances in proteomics and metabolomics in plants. Mol. Hortic. 2022, 2, 17. [Google Scholar] [CrossRef]
- Ahmad, J.; Ali, A.A.; Iqbal, M.; Ahmad, A.; Qureshi, M.I. Proteomics of mercury-induced responses and resilience in plants: A review. Environ. Chem. Lett. 2022, 20, 3335–3355. [Google Scholar] [CrossRef]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [Green Version]
- Vats, S.; Bansal, R.; Rana, N.; Kumawat, S.; Bhatt, V.; Jadhav, P.; Kale, V.; Sathe, A.; Sonah, H.; Jugdaohsingh, R.; et al. Unexplored nutritive potential of tomato to combat global malnutrition. Crit. Rev. Food Sci. Nutr. 2022, 62, 1003–1034. [Google Scholar] [CrossRef]
- Singh, P.K.; Indoliya, Y.; Agrawal, L.; Awasthi, S.; Deeba, F.; Dwivedi, S.; Chakrabarty, D.; Shirke, P.A.; Pandey, V.; Singh, N.; et al. Genomic and proteomic responses to drought stress and biotechnological interventions for enhanced drought tolerance in plants. Curr. Plant Biol. 2022, 29, 100239. [Google Scholar] [CrossRef]
- Mancini, I.; Domingo, G.; Bracale, M.; Loreto, F.; Pollastri, S. Isoprene emission influences the proteomic profile of Arabidopsis plants under well-watered and drought-stress conditions. Int. J. Mol. Sci. 2022, 23, 3836. [Google Scholar] [CrossRef]
- Camp, E.F.; Kahlke, T.; Signal, B.; Oakley, C.A.; Lutz, A.; Davy, S.K.; Suggett, D.J.; Leggat, W.P. Proteome metabolome and transcriptome data for three Symbiodiniaceae under ambient and heat stress conditions. Sci. Data 2022, 9, 153. [Google Scholar] [CrossRef]
- Zhang, X.; Feng, Y.; Khan, A.; Ullah, N.; Li, Z.; Zaheer, S.; Zhou, R.; Zhang, Z. Quantitative proteomics-based analysis reveals molecular mechanisms of chilling tolerance in grafted cotton seedlings. Agronomy 2022, 12, 1152. [Google Scholar] [CrossRef]
- Zhao, Y.; Zhang, F.; Mickan, B.; Wang, D.; Wang, W. Physiological, proteomic, and metabolomic analysis provide insights into Bacillus sp.-mediated salt tolerance in wheat. Plant Cell Rep. 2022, 41, 95–118. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.N.; Ahmed, I.; Ud Din, I.; Noureldeen, A.; Darwish, H.; Khan, M. Proteomic insight into soybean response to flooding stress reveals changes in energy metabolism and cell wall modifications. PLoS ONE 2022, 17, e0264453. [Google Scholar] [CrossRef] [PubMed]
- Komatsu, S.; Yamaguchi, H.; Hitachi, K.; Tsuchida, K.; Rehman, S.U.; Ohno, T. Morphological, biochemical, and proteomic analyses to understand the promotive effects of plant-derived smoke solution on wheat growth under flooding stress. Plants 2022, 11, 1508. [Google Scholar] [CrossRef] [PubMed]
- Long, R.; Li, M.; Zhang, T.; Kang, J.; Sun, Y.; Cong, L.; Gao, Y.; Liu, F.; Yang, Q. Comparative proteomic analysis reveals differential root proteins in Medicago sativa and Medicago truncatula in response to salt stress. Front. Plant Sci. 2016, 7, 424. [Google Scholar] [CrossRef] [Green Version]
- Zhu, Y.; Jia, X.; Wu, Y.; Hu, Y.; Cheng, L.; Zhao, T.; Huang, Z.; Wang, Y. Quantitative proteomic analysis of Malus halliana exposed to salt-alkali mixed stress reveals alterations in energy metabolism and stress regulation. Plant Growth Regul. 2020, 90, 205–222. [Google Scholar] [CrossRef]
- Liu, Y.L.; Shen, Z.J.; Simon, M.; Li, H.; Ma, D.N.; Zhu, X.Y.; Zheng, H.L. Comparative proteomic analysis reveals the regulatory effects of H2S on salt tolerance of mangrove plant Kandelia obovata. Int. J. Mol. Sci. 2019, 21, 118. [Google Scholar] [CrossRef] [Green Version]
- Laha, A.; Chakraborty, P.; Banerjee, C.; Panja, A.S.; Bandopadhyay, R. Application of bioinformatics for crop stress response and mitigation. In Sustainable Agriculture in the Era of Climate Change; Roychowdhury, R., Choudhury, S., Hasanuzzaman, M., Srivastava, S., Eds.; Springer-Nature: Basel, Switzerland, 2020; pp. 589–614. ISBN 978-303-045-669-6. [Google Scholar] [CrossRef]
- Ambrosino, L.; Colantuono, C.; Diretto, G.; Fiore, A.; Chiusano, M.L. Bioinformatics resources for plant abiotic stress responses: State of the art and opportunities in the fast evolving—Omics era. Plants 2020, 9, 591. [Google Scholar] [CrossRef]
- Orozco, A.; Morera, J.; Jiménez, S.; Boza, R. A review of bioinformatics training applied to research in molecular medicine, agriculture and biodiversity in Costa Rica and Central America. Brief Bioinform. 2013, 14, 661–670. [Google Scholar] [CrossRef] [Green Version]
- Noor, W.; Sadia, B.; Baloch, I.A.; Ahmad, A.J.; Safia, B.; Farida. Identification and characterization of abiotic stress responsive genes in Ricinus communis L. using bioinformatics tools. Int. J. Biosci. 2020, 16, 23–34. [Google Scholar]
- Raza, A.; Tabassum, J.; Fakhar, A.Z.; Sharif, R.; Chen, H.; Zhang, C.; Ju, L.; Fotopoulos, V.; Siddique, K.H.M.; Singh, R.K.; et al. Smart reprograming of plants against salinity stress using modern biotechnological tools. Crit. Rev. Biotechnol. 2022, 1–28. [Google Scholar] [CrossRef]
- Larmande, P.; Todorov, K. Revealing genotype-phenotype interactions: The AgroLD experience and challenges. In Integrative Bioinformatics—History and Future; Chen, M., Hofestädt, R., Eds.; Springer-Nature: Singapore, 2022; pp. 321–342. ISBN 978-981-166-795-4. [Google Scholar]
- Hussain, B.; Akpınar, B.A.; Alaux, M.; Algharib, A.M.; Sehgal, D.; Ali, Z.; Aradottir, G.I.; Batley, J.; Bellec, A.; Bentley, A.R.; et al. Capturing wheat phenotypes at the genome level. Front. Plant Sci. 2022, 13, 851079. [Google Scholar] [CrossRef]
- Li, C.; Chu, W.; Gill, R.A.; Sang, S.; Shi, Y.; Hu, X.; Yang, Y.; Zaman, Q.U.; Zhang, B. Computational tools and resources for CRISPR/Cas genome editing. Genom. Proteom. Bioinform. 2022, in press. [CrossRef]
- Bhati, J.; Avashthi, H.; Kumar, A.; Majumdar, S.G.; Budhlakoti, N.; Mishra, D.C. Protocol for identification and annotation of differentially expressed genes using reference-based transcriptomic approach. In Genomics of Cereal Crops; Wani, S.H., Anuj Kumar, A., Eds.; Springer-Nature: Humana, NY, USA, 2022; pp. 175–193. ISBN 978-107-162-533-0. [Google Scholar]
- Waddington, C.H. The epigenotype. 1942. Int. J. Epidemiol. 2012, 41, 10–13. [Google Scholar] [CrossRef] [Green Version]
- Manning, K.; Tör, M.; Poole, M.; Hong, Y.; Thompson, A.J.; King, G.J.; Giovannoni, J.J.; Seymour, G.B. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006, 38, 948–952. [Google Scholar] [CrossRef]
- Schmitz, R.J.; Schultz, M.D.; Lewsey, M.G.; O’Malley, R.C.; Urich, M.A.; Libiger, O.; Schork, N.J.; Ecker, J.R. Transgenerational epigenetic instability is a source of novel methylation variants. Science 2011, 334, 369–373. [Google Scholar] [CrossRef] [Green Version]
- Lloyd, J.P.B.; Lister, R. Epigenome plasticity in plants. Nat. Rev. Genet. 2022, 23, 55–68. [Google Scholar] [CrossRef]
- Zentner, G.E.; Henikoff, S. Regulation of nucleosome dynamics by histone modifications. Nat. Struct. Mol. Biol. 2013, 20, 259–266. [Google Scholar] [CrossRef]
- Fitz-James, M.H.; Cavalli, G. Molecular mechanisms of transgenerational epigenetic inheritance. Nat. Rev. Genet. 2022, 23, 325–341. [Google Scholar] [CrossRef]
- Cong, W.; Miao, Y.; Xu, L.; Zhang, Y.; Yuan, C.; Wang, J.; Zhuang, T.; Lin, X.; Jiang, L.; Wang, N.; et al. Transgenerational memory of gene expression changes induced by heavy metal stress in rice (Oryza sativa L.). BMC Plant Biol. 2019, 19, 282. [Google Scholar] [CrossRef] [Green Version]
- Yung, W.S.; Huang, C.; Li, M.W.; Lam, H.M. Changes in epigenetic features in legumes under abiotic stresses. Plant Genome 2022, e20237. [Google Scholar] [CrossRef] [PubMed]
- Dar, F.A.; Mushtaq, N.U.; Saleem, S.; Rehman, R.U.; Dar, T.U.H.; Hakeem, K.R. Role of epigenetics in modulating phenotypic plasticity against abiotic stresses in plants. Int. J. Genom. 2022, 2022, 1092894. [Google Scholar] [CrossRef] [PubMed]
- Tiwari, A.; Pandey-Rai, S.; Rai, K.K.; Tiwari, A.; Pandey, N. Molecular and epigenetic basis of heat stress responses and acclimatization in plants. Nucleus 2023, 66, 69–79. [Google Scholar] [CrossRef]
- Ali, S.; Khan, N.; Tang, Y. Epigenetic marks for mitigating abiotic stresses in plants. J. Plant Physiol. 2022, 275, 153740. [Google Scholar] [CrossRef] [PubMed]
- De Kort, H.; Toivainen, T.; Van Nieuwerburgh, F.; Andrés, J.; Hytönen, T.P.; Honnay, O. Signatures of polygenic adaptation align with genome-wide methylation patterns in wild strawberry plants. New Phytol. 2022, 235, 1501–1514. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Zhao, F.; Zhou, B. The Characters of Non-Coding RNAs and Their Biological Roles in Plant Development and Abiotic Stress Response. Int. J. Mol. Sci. 2022, 23, 4124. [Google Scholar] [CrossRef]
- Lu, X.; Hyun, T.K. The role of epigenetic modifications in plant responses to stress. Bot. Serbica 2021, 45, 3–12. [Google Scholar] [CrossRef]
- Choi, Y.; Gehring, M.; Johnson, L.; Hannon, M.; Harada, J.J.; Goldberg, R.B.; Jacobsen, S.E.; Fischer, R.L. DEMETER, a DNA glycosylase domain protein, is required for endosperm gene imprinting and seed viability in Arabidopsis. Cell 2002, 110, 33–42. [Google Scholar] [CrossRef] [Green Version]
- Van Oosten, M.J.; Bressan, R.A.; Zhu, J.K.; Bohnert, H.J.; Chinnusamy, V. The role of the epigenome in gene expression control and the epimark changes in response to the environment. Critic. Rev. Plant Sci. 2014, 33, 64–87. [Google Scholar] [CrossRef]
- Luo, M.; Cheng, K.; Xu, Y.; Yang, S.; Wu, K. Plant responses to abiotic stress regulated by histone deacetylases. Front. Plant Sci. 2017, 8, 2147. [Google Scholar] [CrossRef] [Green Version]
- Tang, K.; Lang, Z.; Zhang, H.; Zhu, J.K. The DNA demethylase ROS1 targets genomic regions with distinct chromatin modifications. Nat. Plants 2016, 2, 16169. [Google Scholar] [CrossRef] [Green Version]
- He, L.; Huang, H.; Bradai, M.; Zhao, C.; You, Y.; Ma, J.; Zhao, L.; Lozano-Durán, R.; Zhu, J.K. DNA methylation-free Arabidopsis reveals crucial roles of DNA methylation in regulating gene expression and development. Nat. Commun. 2022, 13, 1335. [Google Scholar] [CrossRef]
- Pandey, R.; Müller, A.; Napoli, C.A.; Selinger, D.A.; Pikaard, C.S.; Richards, E.J.; Bender, J.; Mount, D.W.; Jorgensen, R.A. Analysis of histone acetyltransferase and histone deacetylase families of Arabidopsis thaliana suggests functional diversification of chromatin modification among multicellular eukaryotes. Nucleic Acids Res. 2002, 30, 5036–5055. [Google Scholar] [CrossRef] [Green Version]
- Earley, K.W.; Shook, M.S.; Brower-Toland, B.; Hicks, L.; Pikaard, C.S. In vitro specificities of Arabidopsis co-activator histone acetyltransferases: Implications for histone hyperacetylation in gene activation. Plant J. 2007, 52, 615–626. [Google Scholar] [CrossRef]
- Song, Y.; Ji, D.; Li, S.; Wang, P.; Li, Q.; Xiang, F. The dynamic changes of DNA methylation and histone modifications of salt responsive transcription factor genes in soybean. PLoS ONE 2012, 7, e41274. [Google Scholar] [CrossRef]
- Zheng, M.; Liu, X.; Lin, J.; Liu, X.; Wang, Z.; Xin, M.; Yao, Y.; Peng, H.; Zhou, D.X.; Ni, Z.; et al. Histone acetyltransferase GCN5 contributes to cell wall integrity and salt stress tolerance by altering the expression of cellulose synthesis genes. Plant J. 2019, 97, 587–602. [Google Scholar] [CrossRef]
- Hu, Z.; Song, N.; Zheng, M.; Liu, X.; Liu, Z.; Xing, J.; Ma, J.; Guo, W.; Yao, Y.; Peng, H.; et al. Histone acetyltransferase GCN5 is essential for heat stress-responsive gene activation and thermotolerance in Arabidopsis. Plant J. 2015, 84, 1178–1191. [Google Scholar] [CrossRef] [Green Version]
- Hwarari, D.; Guan, Y.; Ahmad, B.; Movahedi, A.; Min, T.; Hao, Z.; Lu, Y.; Chen, J.; Yang, L. ICE-CBF-COR signaling cascade and its regulation in plants responding to cold stress. Int. J. Mol. Sci. 2022, 23, 1549. [Google Scholar] [CrossRef]
- Singh, R.K.; Prasad, M. Delineating the epigenetic regulation of heat and drought response in plants. Crit. Rev. Biotechnol. 2022, 42, 548–561. [Google Scholar] [CrossRef]
- Sharma, M.; Kumar, P.; Verma, V.; Sharma, R.; Bhargava, B.; Irfan, M. Understanding plant stress memory response for abiotic stress resilience: Molecular insights and prospects. Plant Physiol. Biochem. 2022, 179, 10–24. [Google Scholar] [CrossRef]
- Boden, S.A.; Kavanová, M.; Finnegan, E.J.; Wigge, P.A. Thermal stress effects on grain yield in Brachypodium distachyon occur via H2A.Z-nucleosomes. Genome Biol. 2013, 14, R65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Obata, T.; Fernie, A.R. The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol. Life Sci. 2012, 69, 3225–3243. [Google Scholar] [CrossRef] [Green Version]
- Hong, J.; Yang, L.; Zhang, D.; Shi, J. Plant metabolomics: An indispensable system biology tool for plant science. Int. J. Mol. Sci. 2016, 17, 767. [Google Scholar] [CrossRef] [PubMed]
- Chugh, V.; Kaur, N.; Gupta, A.K.; Rai, A. The seed biochemical signature as a potent marker for water logging tolerance in maize. Plant Stress 2022, 4, 100085. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Ivanisevic, J.; Giera, M.; Siuzdak, G. Identification of bioactive metabolites using activity metabolomics. Nat. Rev. Mol. Cell Biol. 2019, 20, 353–367. [Google Scholar] [CrossRef]
- Fiehn, O.; Kopka, J.; Dörmann, P.; Altmann, T.; Trethewey, R.N.; Willmitzer, L. Metabolite profiling for plant functional genomics. Nat. Biotechnol. 2000, 18, 1157–1161. [Google Scholar] [CrossRef]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Verslues, P.E.; Bailey-Serres, J.; Brodersen, C.; Buckley, T.N.; Conti, L.; Christmann, A.; Dinneny, J.R.; Grill, E.; Hayes, S.; Heckman, R.W.; et al. Burning questions for a warming and changing world: 15 unknowns in plant abiotic stress. Plant Cell 2023, 35, 67–105. [Google Scholar] [CrossRef]
- Mashabela, M.D.; Masamba, P.; Kappo, A.P. Metabolomics and chemoinformatics in agricultural biotechnology research: Complementary probes in unravelling new metabolites for crop improvement. Biology 2022, 11, 1156. [Google Scholar] [CrossRef]
- Hasanuzzaman, M.; Nahar, K.; Alam, M.M.; Roychowdhury, R.; Fujita, M. Physiological, biochemical, and molecular mechanisms of heat stress tolerance in plants. Int. J. Mol. Sci. 2013, 14, 9643–9684. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Khan, M.H.; Choudhury, S. Arsenic in rice: An overview on stress implications, tolerance and mitigation strategies. In Plants under Metal and Metalloid Stress: Responses, Tolerance and Remediation; Hasanuzzaman, M., Nahar, K., Fujita, M., Eds.; Springer-Nature: Singapore, 2018; pp. 401–416. ISBN 978-981-132-242-6. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Khan, M.H.; Choudhury, S. Physiological and molecular responses for metalloid stress in rice—A Comprehensive Overview. In Advances in Rice Research for Abiotic Stress Tolerance; Hasanuzzaman, M., Fujita, M., Nahar, K., Biswas, J., Eds.; Woodhead Publishing: Cambridge, UK; Elsevier: Amsterdam, The Netherlands, 2019; pp. 341–369. ISBN 978-012-814-333-9. [Google Scholar] [CrossRef]
- Gujjar, R.S.; Karkute, S.G.; Rai, A.; Singh, M.; Singh, B. Proline-rich proteins may regulate free cellular proline levels during drought stress in tomato. Curr. Sci. 2018, 114, 915–920. [Google Scholar] [CrossRef]
- Patel, J.; Khandwal, D.; Choudhary, B.; Ardeshana, D.; Jha, R.K.; Tanna, B.; Yadav, S.; Mishra, A.; Varshney, R.K.; Siddique, K.H.M. Differential physio-biochemical and metabolic responses of peanut (Arachis hypogaea L.) under multiple abiotic stress conditions. Int. J. Mol. Sci. 2022, 23, 660. [Google Scholar] [CrossRef]
- Xu, Y.; Fu, X. Reprogramming of Plant Central Metabolism in Response to Abiotic Stresses: A metabolomics view. Int. J. Mol. Sci. 2022, 23, 5716. [Google Scholar] [CrossRef]
- Huchzermeyer, B.; Menghani, E.; Khardia, P.; Shilu, A. Metabolic pathway of natural antioxidants, antioxidant enzymes and ROS providence. Antioxidants 2022, 11, 761. [Google Scholar] [CrossRef]
- Ren, G.; Mo, H.; Xu, R. Arginine decarboxylase gene ADC2 regulates fiber elongation in cotton. Genes 2022, 13, 784. [Google Scholar] [CrossRef]
- Quan, N.T.; Anh, L.H.; Khang, D.T.; Tuyen, P.T.; Toan, N.P.; Minh, T.N.; Minh, L.T.; Bach, D.T.; Ha, P.T.T.; Elzaawely, A.A.; et al. Involvement of secondary metabolites in response to drought stress of rice (Oryza sativa L.). Agriculture 2016, 6, 23. [Google Scholar] [CrossRef] [Green Version]
- Piasecka, A.; Sawikowska, A.; Kuczyńska, A.; Ogrodowicz, P.; Mikołajczak, K.; Krystkowiak, K.; Gudyś, K.; Guzy-Wróbelska, J.; Krajewski, P.; Kachlicki, P. Drought-related secondary metabolites of barley (Hordeum vulgare L.) leaves and their metabolomic quantitative trait loci. Plant J. 2017, 89, 898–913. [Google Scholar] [CrossRef] [Green Version]
- Radwan, A.; Kleinwächter, M.; Selmar, D. Impact of drought stress on specialised metabolism: Biosynthesis and the expression of monoterpene synthases in sage (Salvia officinalis). Phytochemistry 2017, 141, 20–26. [Google Scholar] [CrossRef]
- Nawaz, M.; Hassan, M.U.; Chattha, M.U.; Mahmood, A.; Shah, A.N.; Hashem, M.; Alamri, S.; Batool, M.; Rasheed, A.; Thabit, M.A.; et al. Trehalose: A promising osmo-protectant against salinity stress-physiological and molecular mechanisms and future prospective. Mol. Biol. Rep. 2022, 49, 11255–11271. [Google Scholar] [CrossRef]
- Biondi, S.; Antognoni, F.; Marincich, L.; Lianza, M.; Tejos, R.; Ruiz, K.B. The polyamine “multiverse” and stress mitigation in crops: A case study with seed priming in quinoa. Sci. Hort. 2022, 304, 111292. [Google Scholar] [CrossRef]
- Masouleh, S.S.S.; Sassine, Y.N. Molecular and biochemical responses of horticultural plants and crops to heat stress. Ornam. Hort. 2020, 26, 148–158. [Google Scholar] [CrossRef]
- Paupière, M.J.; Tikunov, Y.; Schleiff, E.; Bovy, A.; Fragkostefanakis, S. Reprogramming of tomato leaf metabolome by the activity of heat stress transcription factor HsfB1. Front. Plant Sci. 2020, 11, 610599. [Google Scholar] [CrossRef] [PubMed]
- Rouphael, Y.; Raimondi, G.; Lucini, L.; Carillo, P.; Kyriacou, M.C.; Colla, G.; Cirillo, V.; Pannico, A.; El-Nakhel, C.; De Pascale, S. Physiological and metabolic responses triggered by omeprazole improve tomato plant tolerance to NaCl stress. Front. Plant Sci. 2018, 9, 249. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sabreena; Hassan, S. Plant life under changing environment: An exertion of environmental factors in oxidative stress modulation. In Antioxidant Defense in Plants-Molecular Basis of Regulation; Aftab, T., Hakeem, K.R., Eds.; Springer-Nature: Singapore, 2022; pp. 421–433. ISBN 978-981-167-981-0. [Google Scholar]
- Rai, G.K.; Mushtaq, M.; Bhat, B.A.; Kumar, R.R.; Singh, M.; Rai, P.K. Reactive Oxygen Species: Friend or Foe. In Thermotolerance in Crop Plants; Kumar, R.R., Praveen, S., Rai, G.K., Eds.; Springer-Nature: Singapore, 2022; pp. 129–162. ISBN 978-981-193-800-9. [Google Scholar]
- Parida, A.K.; Kumari, A.; Rangani, J.; Patel, M. Halophytes: Potential resources of coastal ecosystems and their economic, ecological and bioprospecting significance. In Halophytes and Climate Change: Adaptive Mechanisms and Potential Uses; Hasanuzzaman, M., Shabala, S., Fujita, M., Eds.; CABI: Oxfordshire, UK, 2019; pp. 287–323. ISBN 978-178-639-433-0. [Google Scholar]
- Sudhakar, C.; Veeranagamallaiah, G.; Nareshkumar, A.; Sudhakarbabu, O.; Sivakumar, M.; Pandurangaiah, M.; Kiranmai, K.; Lokesh, U. Polyamine metabolism influences antioxidant defense mechanism in foxtail millet (Setaria italica L.) cultivars with different salinity tolerance. Plant Cell Rep. 2015, 34, 141–156. [Google Scholar] [CrossRef] [PubMed]
- Hossain, M.A.; Hoque, T.S.; Zaid, A.; Wani, S.H.; Mostofa, M.G.; Henry, R. Targeting the ascorbate-glutathione pathway and the glyoxalase pathway for genetic engineering of abiotic stress-tolerance in rice. In Molecular Breeding for Rice Abiotic Stress Tolerance and Nutritional Quality; Hossain, M.A., Hassan, L., Ifterkharuddaula, K.M., Kumar, A., Henry, R., Eds.; John Wiley & Sons, Ltd.: Chichester, UK, 2021; pp. 398–427. ISBN 978-111-963-317-4. [Google Scholar]
- Sharma, P.; Dubey, R.S. Protein synthesis by plants under stressful conditions. In Handbook of Plant and Crop Stress, 4th ed.; Pessarakli, M., Ed.; CRC Press: Boca Raton, UK, 2019; pp. 405–449. ISBN 978-135-110-460-9. [Google Scholar]
- Chaturvedi, S.; Khan, S.; Bhunia, R.K.; Kaur, K.; Tiwari, S. Metabolic engineering in food crops to enhance ascorbic acid production: Crop biofortification perspectives for human health. Physiol. Mol. Biol. Plants 2022, 28, 871–884. [Google Scholar] [CrossRef]
- Putri, S.P.; Yamamoto, S.; Tsugawa, H.; Fukusaki, E. Current metabolomics: Technological advances. J. Biosci. Bioeng. 2013, 116, 9–16. [Google Scholar] [CrossRef]
- Ghatak, A.; Chaturvedi, P.; Weckwerth, W. Metabolomics in plant stress physiology. In Plant Genetics and Molecular Biology (Advances in Biochemical Engineering/Biotechnology, Vol 164); Varshney, R., Pandey, M., Chitikineni, A., Eds.; Springer: Cham, Switzerland, 2018; pp. 187–236. [Google Scholar] [CrossRef]
- Jurowski, K.; Kochan, K.; Walczak, J.; Barańska, M.; Piekoszewski, W.; Buszewski, B. Analytical techniques in lipidomics: State of the art. Crit. Rev. Anal. Chem. 2017, 47, 418–437. [Google Scholar] [CrossRef]
- Alseekh, S.; Aharoni, A.; Brotman, Y.; Contrepois, K.; D’auria, J.; Ewald, J.; Ewald, J.C.; Fraser, P.D.; Giavalisco, P.; Hall, R.D.; et al. Mass spectrometry-based metabolomics: A guide for annotation, quantification and best reporting practices. Nat. Methods 2021, 18, 747–756. [Google Scholar] [CrossRef]
- Pua, A.; Goh, R.M.V.; Huang, Y.; Tang, V.C.Y.; Ee, K.H.; Cornuz, M.; Liu, S.Q.; Lassabliere, B.; Yu, B. Recent advances in analytical strategies for coffee volatile studies: Opportunities and challenges. Food Chem. 2022, 388, 132971. [Google Scholar] [CrossRef]
- Brunetti, A.E.; Carnevale Neto, F.; Vera, M.C.; Taboada, C.; Pavarini, D.P.; Bauermeister, A.; Lopes, N.P. An integrative omics perspective for the analysis of chemical signals in ecological interactions. Chem. Soc. Rev. 2018, 47, 1574–1591. [Google Scholar] [CrossRef]
- Zhu, F.Y.; Chen, M.X.; Ye, N.H.; Shi, L.; Ma, K.L.; Yang, J.F.; Cao, Y.Y.; Zhang, Y.; Yoshida, T.; Fernie, A.R.; et al. Proteogenomic analysis reveals alternative splicing and translation as part of the abscisic acid response in Arabidopsis seedlings. Plant J. 2017, 91, 518–533. [Google Scholar] [CrossRef] [Green Version]
- Ruggles, K.V.; Krug, K.; Wang, X.; Clauser, K.R.; Wang, J.; Payne, S.H.; Fenyö, D.; Zhang, B.; Mani, D.R. Methods, tools and current perspectives in proteogenomics. Mol. Cell. Proteom. 2017, 16, 959–981. [Google Scholar] [CrossRef] [Green Version]
- Ullah, M.A.; Abdullah-zawawi, M.R.; Zainal-abidin, R.A.; Sukiran, N.L.; Uddin, M.I.; Zainal, Z. A review of integrative omic approaches for understanding rice salt response mechanisms. Plants 2022, 11, 1430. [Google Scholar] [CrossRef]
- Low, T.Y.; Mohtar, M.A.; Ang, M.Y.; Jamal, R. Connecting proteomics to next-generation sequencing: Proteogenomics and its current applications in biology. Proteomics 2019, 19, e1800235. [Google Scholar] [CrossRef]
- Gupta, A.; Shaw, B.P.; Sahu, B.B. Post-translational regulation of the membrane transporters contributing to salt tolerance in plants. Funct. Plant Biol. 2021, 48, 1199–1212. [Google Scholar] [CrossRef]
- Pranneshraj, V.; Sangha, M.K.; Djalovic, I.; Miladinovic, J.; Djanaguiraman, M. Lipidomics-assisted GWAS (LGWAS) approach for improving high-temperature stress tolerance of crops. Int. J. Mol. Sci. 2022, 23, 9389. [Google Scholar] [CrossRef]
- Buffagni, V.; Zhang, L.; Senizza, B.; Rocchetti, G.; Ferrarini, A.; Miras-Moreno, B.; Lucini, L. Metabolomics and lipidomics insight into the effect of different polyamines on tomato plants under non-stress and salinity conditions. Plant Sci. 2022, 322, 111346. [Google Scholar] [CrossRef]
- Zhang, D.; Li, J.; Li, M.; Cheng, Z.; Xu, Q.; Song, X.; Shang, X.; Guo, W. Overexpression of a cotton nonspecific lipid transfer protein gene, GhLTP4, enhances drought tolerance by remodeling lipid profiles, regulating abscisic acid homeostasis and improving tricarboxylic acid cycle in cotton. Environ. Exp. Bot. 2022, 201, 104991. [Google Scholar] [CrossRef]
- Moradi, P.; Mahdavi, A.; Khoshkam, M.; Iriti, M. Lipidomics unravels the role of leaf lipids in thyme plant response to drought stress. Int. J. Mol. Sci. 2017, 18, 2067. [Google Scholar] [CrossRef] [Green Version]
- Tarazona, P.; Feussner, K.; Feussner, I. An enhanced plant lipidomics method based on multiplexed liquid chromatography-mass spectrometry reveals additional insights into cold- and drought-induced membrane remodeling. Plant J. 2015, 84, 621–633. [Google Scholar] [CrossRef]
- Liu, B.; Wang, X.; Li, K.; Cai, Z. Spatially resolved metabolomics and lipidomics reveal salinity and drought-tolerant mechanisms of cottonseeds. J. Agric. Food Chem. 2021, 69, 8028–8037. [Google Scholar] [CrossRef] [PubMed]
- Engel, K.M.; Prabutzki, P.; Leopold, J.; Nimptsch, A.; Lemmnitzer, K.; Vos, D.R.N.; Hopf, C.; Schiller, J. A new update of MALDI-TOF mass spectrometry in lipid research. Prog. Lipid Res. 2022, 86, 101145. [Google Scholar] [CrossRef] [PubMed]
- Sun, A.Z.; Chen, L.S.; Tang, M.; Chen, J.H.; Li, H.; Jin, X.Q.; Yi, Y.; Guo, F.Q. Lipidomic remodeling in Begonia grandis under heat stress. Front. Plant Sci. 2022, 13, 843942. [Google Scholar] [CrossRef] [PubMed]
- Baxter, I. Ionomics: Studying the social network of mineral nutrients. Curr. Opin. Plant Biol. 2009, 12, 381–386. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salt, D.E.; Baxter, I.; Lahner, B. Ionomics and the study of the plant ionome. Annu. Rev. Plant Biol. 2008, 59, 709–733. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Du, P.; Ji, J.; He, X.; Zhang, J.; Shang, Y.; Liu, H.; Xu, J.; Liang, B. Ionomic combined with transcriptomic and metabolomic analyses to explore the mechanism underlying the effect of melatonin in relieving nutrient stress in apple. Int. J. Mol. Sci. 2022, 23, 9855. [Google Scholar] [CrossRef]
- Guo, J.; Lu, X.; Tao, Y.; Guo, H.; Min, W. Comparative ionomics and metabolic responses and adaptive strategies of cotton to salt and alkali stress. Front. Plant Sci. 2022, 13, 871387. [Google Scholar] [CrossRef]
- Hua, Y.P.; Wang, Y.; Zhou, T.; Huang, J.Y.; Yue, C.P. Combined morpho-physiological, ionomic and transcriptomic analyses reveal adaptive responses of allohexaploid wheat (Triticum aestivum L.) to iron deficiency. BMC Plant Biol. 2022, 22, 234. [Google Scholar] [CrossRef]
- Muszyńska, E.; Labudda, M. Dual role of metallic trace elements in stress biology—From negative to beneficial impact on plants. Int. J. Mol. Sci. 2019, 20, 3117. [Google Scholar] [CrossRef] [Green Version]
- Kirkby, E. Introduction, definition and classification of nutrients. In Marschner’s Mineral Nutrition of Higher Plants; Marschner, P., Ed.; Academic Press: Cambridge, MA, USA, 2012; pp. 3–5. ISBN 978-012-384-905-2. [Google Scholar] [CrossRef]
- Zhang, R.; Wang, Y.; Hussain, S.; Yang, S.; Li, R.; Liu, S.; Chen, Y.; Wei, H.; Dai, Q.; Hou, H. Study on the effect of salt stress on yield and grain quality among different rice varieties. Front. Plant Sci. 2022, 13, 918460. [Google Scholar] [CrossRef]
- Gupta, A.; Shaw, B.P. Augmenting salt tolerance in rice by regulating uptake and tissue specific accumulation of Na+- through Ca2+-induced alteration of biochemical events. Plant Biol. 2021, 23, 122–130. [Google Scholar] [CrossRef]
- Balemi, T.; Negisho, K. Management of soil phosphorus and plant adaptation mechanisms to phosphorus stress for sustainable crop production: A review. J. Soil Sci. Plant Nutr. 2012, 12, 547–561. [Google Scholar] [CrossRef] [Green Version]
- Yugandhar, P.; Veronica, N.; Subrahmanyam, D.; Brajendra, P.; Nagalakshmi, S.; Srivastava, A.; Voleti, S.R.; Sarla, N.; Sundaram, R.M.; Sevanthi, A.M.; et al. Revealing the effect of seed phosphorus concentration on seedling vigour and growth of rice using mutagenesis approach. Sci. Rep. 2022, 12, 1203. [Google Scholar] [CrossRef]
- Shafi, A.; Zahoor, I.; Habib, H. Omics technologies to unravel plant-microbe interactions. In Plant-Microbe Dynamics: Recent Advances for Sustainable Agriculture; Pirzadah, T.B., Malik, B., Hakeem, K.R., Eds.; CRC Press: Boca Raton, UK, 2021; pp. 201–220. ISBN 978-036-761-838-4. [Google Scholar]
- Gupta, O.P.; Deshmukh, R.; Kumar, A.; Singh, S.K.; Sharma, P.; Ram, S.; Singh, G.P. From gene to biomolecular networks: A review of evidences for understanding complex biological function in plants. Curr. Opin. Biotechnol. 2022, 74, 66–74. [Google Scholar] [CrossRef]
- Hajheidari, M.; Huang, S.C. Elucidating the biology of transcription factor–DNA interaction for accurate identification of Cis-regulatory elements. Curr. Opin. Plant Biol. 2022, 68, 102232. [Google Scholar] [CrossRef]
- Di Silvestre, D.; Bergamaschi, A.; Bellini, E.; Mauri, P.L. Large scale proteomic data and network-based systems biology approaches to explore the plant world. Proteomes 2018, 6, 27. [Google Scholar] [CrossRef] [Green Version]
- Kapoor, B.; Kumar, P.; Gill, N.S.; Sharma, R.; Thakur, N.; Irfan, M. Molecular mechanisms underpinning the silicon-selenium (Si-Se) interactome and cross-talk in stress-induced plant responses. Plant Soil 2023, 486, 45–68. [Google Scholar] [CrossRef]
- Yadav, R.; Santal, A.R.; Singh, N.P. Root protein interactomics of salt stress-induced proteins of wheat genotypes KH-65 (salt-tolerant) and PBW-373 (salt-susceptible). J. Appl. Biotechnol. Rep. 2022, 9, 632–639. [Google Scholar] [CrossRef]
- Perez-Sanz, F.; Navarro, P.J.; Egea-Cortines, M. Plant phenomics: An overview of image acquisition technologies and image data analysis algorithms. Gigascience 2017, 6, gix092. [Google Scholar] [CrossRef] [Green Version]
- Kumar, J.; Kumar, S.; Pratap, A. Phenomics in Crop Plants: Trends, Options and Limitations; Springer: New Delhi, India, 2015. [Google Scholar] [CrossRef]
- Plomin, R.; DeFries, J.C.; Loehlin, J.C. Genotype-environment interaction and correlation in the analysis of human behavior. Psychol. Bull. 1977, 84, 309–322. [Google Scholar] [CrossRef]
- Roychowdhury, R.; Zilberman, O.; Chandrasekhar, K.; Curzon, A.Y.; Nashef, K.; Abbo, S.; Slafer, G.A.; Bonfil, D.J.; Ben-David, R. Pre-anthesis spike growth dynamics and its association to yield components among elite bread wheat cultivars (Triticum aestivum L. spp.) under Mediterranean climate. Field Crops Res. 2023, 298, 108948. [Google Scholar] [CrossRef]
- Van Bezouw, R.F.H.M.; Keurentjes, J.J.B.; Harbinson, J.; Aarts, M.G.M. Converging phenomics and genomics to study natural variation in plant photosynthetic efficiency. Plant J. 2019, 97, 112–133. [Google Scholar] [CrossRef] [PubMed]
- Pasala, R.; Pandey, B.B. Plant phenomics: High-throughput technology for accelerating genomics. J. Biosci. 2020, 45, 111. [Google Scholar] [CrossRef]
- Furbank, R.T.; Tester, M. Phenomics—Technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef]
- Arya, S.; Sandhu, K.S.; Singh, J.; Kumar, S. Deep learning: As the new frontier in high-throughput plant phenotyping. Euphytica 2022, 218, 47. [Google Scholar] [CrossRef]
- Ninomiya, S. High-throughput field crop phenotyping: Current status and challenges. Breed. Sci. 2022, 72, 3–18. [Google Scholar] [CrossRef]
- Laha, S.D.; Naskar, A.J.; Sarkar, T.; Guha, S.; Mondal, H.A.; Das, M. Field phenotyping for salt tolerance and imaging techniques for crop stress biology. In Intelligent Image Analysis for Plant Phenotyping; Samal, A., Choudhury, S.D., Eds.; CRC Press: Boca Raton, UK, 2020; pp. 287–304. ISBN 978-131-517-730-4. [Google Scholar]
- Tayade, R.; Yoon, J.; Lay, L.; Khan, A.L.; Yoon, Y.; Kim, Y. Utilization of spectral indices for high-throughput phenotyping. Plants 2022, 11, 1712. [Google Scholar] [CrossRef]
- Al-Tamimi, N.; Langan, P.; Bernád, V.; Walsh, J.; Mangina, E.; Negrão, S. Capturing crop adaptation to abiotic stress using image-based technologies. Open Biol. 2022, 12, 210353. [Google Scholar] [CrossRef]
- Langan, P.; Bernád, V.; Walsh, J.; Henchy, J.; Khodaeiaminjan, M.; Mangina, E.; Negrão, S. Phenotyping for waterlogging tolerance in crops: Current trends and future prospects. J. Exp. Bot. 2022, 73, 5149–5169. [Google Scholar] [CrossRef]
- Vines, P.L.; Zhang, J. High-throughput plant phenotyping for improved turfgrass breeding applications. Grass Res. 2022, 2, 1. [Google Scholar] [CrossRef]
- Burchard-Levine, V.; Arribas, D.R.; del Hoyo, L.V.; Isabel, M.D.P.M.; Corral, J.B. A review of in-situ sampling protocols to support the remote sensing of vegetation. GeoFocus—Int. Rev. Geo. Info. Sci. Technol. 2022, 29, 59–87. [Google Scholar]
- Maphosa, L.; Emily Thoday-Kennedy, E.; Vakani, J.; Phelan, A.; Badenhorst, P.; Slater, A.; Spangenberg, G.; Kant, S. Phenotyping wheat under salt stress conditions using a 3D laser scanner. Israel J. Plant Sci. 2017, 64, 55–62. [Google Scholar] [CrossRef]
- Solovchenko, A.; Lukyanov, A.; Vasilieva, S.; Lobakova, E. Chlorophyll fluorescence as a valuable multitool for microalgal biotechnology. Biophys. Rev. 2022, 14, 973–983. [Google Scholar] [CrossRef]
- Berger, K.; Machwitz, M.; Kycko, M.; Kefauver, S.C.; Van Wittenberghe, S.; Gerhards, M.; Verrelst, J.; Atzberger, C.; van der Tol, C.; Damm, A.; et al. Multi-sensor spectral synergies for crop stress detection and monitoring in the optical domain: A review. Remote Sens. Environ. 2022, 280, 113198. [Google Scholar] [CrossRef]
- Xiao, Q.; Bai, X.; Zhang, C.; He, Y. Advanced high-throughput plant phenotyping techniques for genome-wide association studies: A review. J. Adv. Res. 2022, 35, 215–230. [Google Scholar] [CrossRef]
- Mishra, A.; Mishra, K.B.; Höermiller, I.I.; Heyer, A.G.; Nedbal, L. Chlorophyll fluorescence emission as a reporter on cold tolerance in Arabidopsis thaliana accessions. Plant Signal. Behav. 2011, 6, 301–310. [Google Scholar] [CrossRef] [Green Version]
- Zuo, G.; Aiken, R.M.; Feng, N.; Zheng, D.; Zhao, H.; Avenson, T.J.; Lin, X. Fresh perspectives on an established technique: Pulsed amplitude modulation chlorophyll a fluorescence. Plant-Environ. Interact 2022, 3, 41–59. [Google Scholar] [CrossRef]
- Calzadilla, P.I.; Carvalho, F.E.L.; Gomez, R.; Lima Neto, M.C.; Signorelli, S. Assessing photosynthesis in plant systems: A cornerstone to aid in the selection of resistant and productive crops. Environ. Exp. Bot. 2022, 201, 104950. [Google Scholar] [CrossRef]
- Fu, P.; Montes, C.M.; Siebers, M.H.; Gomez-Casanovas, N.; McGrath, J.M.; Ainsworth, E.A.; Bernacchi, C.J. Advances in field-based high-throughput photosynthetic phenotyping. J. Exp. Bot. 2022, 73, 3157–3172. [Google Scholar] [CrossRef]
- Osmolovskaya, N.; Shumilina, J.; Kim, A.; Didio, A.; Grishina, T.; Bilova, T.; Keltsieva, O.A.; Zhukov, V.; Tikhonovich, I.; Tarakhovskaya, E.; et al. Methodology of drought stress research: Experimental setup and physiological characterization. Int. J. Mol. Sci. 2018, 19, 4089. [Google Scholar] [CrossRef] [Green Version]
- Singh, B.; Mishra, S.; Bohra, A.; Joshi, R.; Siddique, K.H. Crop phenomics for abiotic stress tolerance in crop plants. In Biochemical, Physiological and Molecular Avenues for Combating Abiotic Stress Tolerance in Plants; Wani, S.H., Ed.; Academic Press: Cambridge, MA, USA, 2018; pp. 277–296. ISBN 978-012-813-066-7. [Google Scholar]
- Jansen, M.; Gilmer, F.; Biskup, B.; Nagel, K.A.; Rascher, U.; Fischbach, A.; Briem, S.; Dreissen, G.; Tittmann, S.; Braun, S.; et al. Simultaneous phenotyping of leaf growth and chlorophyll fluorescence via growscreen fluoro allows detection of stress tolerance in Arabidopsis thaliana and other rosette plants. Funct. Plant Biol. 2009, 36, 902–914. [Google Scholar] [CrossRef] [PubMed]
- Dornbusch, T.; Lorrain, Ś.; Kuznetsov, D.; Fortier, A.; Liechti, R.; Xenarios, I.; Fankhauser, C. Measuring the diurnal pattern of leaf hyponasty and growth in Arabidopsis a novel phenotyping approach using laser scanning. Funct. Plant Biol. 2012, 39, 860–869. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acosta-Gamboa, L.M.; Suxing, L.; Jarrod, W.C.; Zachary, C.C.; Raquel, T.; Jessica, P.Y.-C.; Argelia, L. Characterization of the response to abiotic stresses of high ascorbate Arabidopsis lines using phenomic approaches. Plant Physiol. Biochem. 2020, 151, 500–515. [Google Scholar] [CrossRef] [PubMed]
- Kuromori, T.; Fujita, M.; Takahashi, F.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Inter-tissue and inter-organ signaling in drought stress response and phenotyping of drought tolerance. Plant J. 2022, 109, 342–358. [Google Scholar] [CrossRef]
- Honsdorf, N.; March, T.J.; Berger, B.; Tester, M.; Pillen, K. High-throughput phenotyping to detect drought tolerance QTL in wild barley introgression lines. PLoS ONE 2014, 9, e97047. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Bai, G.; Stoerger, V.; Schnable, J.C. Temporal dynamics of maize plant growth, water use, and leaf water content using automated high throughput RGB and hyperspectral imaging. Comput. Electron. Agric. 2016, 127, 625–632. [Google Scholar] [CrossRef] [Green Version]
- Meng, R.; Saade, S.; Kurtek, S.; Berger, B.; Brien, C.; Pillen, K.; Tester, M.; Sun, Y. Growth curve registration for evaluating salinity tolerance in barley. Plant Methods 2017, 13, 18. [Google Scholar] [CrossRef] [Green Version]
- Hairmansis, A.; Berger, B.; Tester, M.; Roy, S.J. Image-based phenotyping for non-destructive screening of different salinity tolerance traits in rice. Rice 2014, 7, 16. [Google Scholar] [CrossRef] [Green Version]
- Humplík, J.F.; Lazár, D.; Husičková, A.; Spíchal, L. Automated phenotyping of plant shoots using imaging methods for analysis of plant stress responses—A review. Plant Methods 2015, 11, 29. [Google Scholar] [CrossRef] [Green Version]
- Briglia, N.; Nuzzo, V.; Petrozza, A.; Summerer, S.; Cellini, F.; Montanaro, G. Preliminary high-throughput phenotyping analysis in grapevines under drought. BIO Web Conf. 2019, 13, 02003. [Google Scholar] [CrossRef]
- Mutava, R.N.; Prince, S.J.K.; Syed, N.H.; Song, L.; Valliyodan, B.; Chen, W.; Nguyen, H.T. Understanding abiotic stress tolerance mechanisms in soybean: A comparative evaluation of soybean response to drought and flooding stress. Plant Physiol. Biochem. 2015, 86, 109–120. [Google Scholar] [CrossRef]
- Yu, H.; Kong, B.; Hou, Y.; Xu, X.; Chen, T.; Liu, X. A critical review on applications of hyperspectral remote sensing in crop monitoring. Exp. Agric. 2022, 58, e26. [Google Scholar] [CrossRef]
- Ludovisi, R.; Tauro, F.; Salvati, R.; Khoury, S.; Mugnozza, G.S.; Harfouche, A. UAV-based thermal imaging for high-throughput field phenotyping of black poplar response to drought. Front. Plant Sci. 2017, 8, 1681. [Google Scholar] [CrossRef]
- Al-Rahbi, S.; Al-Mulla, Y.A.; Jayasuriya, H.; Choudri, B. Analysis of true-color images from unmanned aerial vehicle to assess salinity stress on date palm. J. Appl. Remote Sens. 2019, 13, 34514. [Google Scholar] [CrossRef]
- Johansen, K.; Morton, M.J.L.; Malbeteau, Y.M.; Aragon, B.; Al-Mashharawi, S.K.; Ziliani, M.G.; Angel, Y.; Fiene, G.M.; Negrão, S.S.C.; Mousa, M.A.A.; et al. Unmanned aerial vehicle-based phenotyping using morphometric and spectral analysis can quantify responses of wild tomato plants to salinity stress. Front. Plant Sci. 2019, 10, 370. [Google Scholar] [CrossRef]
- Aharon, S.; Peleg, Z.; Argaman, E.; Ben-David, R.; Lati, R.N. Image-based high-throughput phenotyping of cereals early vigor and weed-competitiveness traits. Remote Sens. 2020, 12, 3877. [Google Scholar] [CrossRef]
- Aharon, S.; Fadida-Myers, A.; Nashef, K.; Ben-David, R.; Lati, R.N.; Peleg, Z. Genetic improvement of wheat early vigor promote weed-competitiveness under Mediterranean climate. Plant Sci. 2021, 303, 110785. [Google Scholar] [CrossRef]
- Wang, D.; Cao, W.; Zhang, F.; Li, Z.; Xu, S.; Wu, X. A review of deep learning in multiscale agricultural sensing. Remote Sens. 2022, 14, 559. [Google Scholar] [CrossRef]
- Lopes, M.S.; Reynolds, M.P. Stay-green in spring wheat can be determined by spectral reflectance measurements (normalized difference vegetation index) independently from phenology. J. Exp. Bot. 2012, 63, 3789–3798. [Google Scholar] [CrossRef] [Green Version]
- Galiano, S.G. Assessment of vegetation indexes from remote sensing: Theoretical basis. Options Méditerranéennes 2012, 67, 65–75. [Google Scholar]
- Sangwan, S.; Ram, K.; Rani, P.; Munjal, R. Effect of terminal high temperature on chlorophyll content and normalized difference vegetation index in recombinant inbred lines of bread wheat. Int. J. Curr. Microbiol. Appl. Sci. 2018, 7, 1174–1183. [Google Scholar] [CrossRef]
- Beisel, N.S.; Callaham, J.B.; Sng, N.J.; Taylor, D.J.; Paul, A.L.; Ferl, R.J. Utilization of single-image normalized difference vegetation index (SI-NDVI) for early plant stress detection. Appl. Plant Sci. 2018, 6, e01186. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Castro, A.I.; Shi, Y.; Maja, J.M.; Peña, J.M. UAVs for vegetation monitoring: Overview and recent scientific contributions. Remote Sens. 2021, 13, 2139. [Google Scholar] [CrossRef]
- Jones, H.G.; Serraj, R.; Loveys, B.R.; Xiong, L.; Wheaton, A.; Price, A.H. Thermal infrared imaging of crop canopies for the remote diagnosis and quantification of plant responses to water stress in the field. Funct. Plant Biol. 2009, 36, 978–989. [Google Scholar] [CrossRef] [Green Version]
- Romero, P.; Navarro, J.M.; Ordaz, P.B. Towards a sustainable viticulture: The combination of deficit irrigation strategies and agroecological practices in Mediterranean vineyards. a review and update. Agric. Water Manag. 2022, 259, 107216. [Google Scholar] [CrossRef]
- Sweet, D.D.; Tirado, S.B.; Springer, N.M.; Hirsch, C.N.; Hirsch, C.D. Opportunities and challenges in phenotyping row crops using drone-based RGB imaging. Plant Phenome J. 2022, 5, e20044. [Google Scholar] [CrossRef]
- Tripodi, P.; Nicastro, N.; Pane, C.; Cammarano, D. Digital applications and artificial intelligence in agriculture toward next-generation plant phenotyping. Crop. Pasture Sci. 2022. [Google Scholar] [CrossRef]
- Duruflé, H.; Déjean, S. Multi-omics data integration in the context of plant abiotic stress signaling. In Plant Abiotic Stress Signaling; Methods in Molecular Biology; Couée, I., Ed.; Humana Press: New York, NY, USA, 2023; Volume 2642, pp. 295–318. [Google Scholar] [CrossRef]
- Gupta, S.; Kaur, R.; Sharma, T.; Bhardwaj, A.; Sharma, S.; Sohal, J.S.; Singh, S.V. Multi-omics approaches for understanding stressor-induced physiological changes in plants: An updated overview. Physiol. Mol. Plant Pathol. 2023, 126, 102047. [Google Scholar] [CrossRef]
- Liu, T.; Salguero, P.; Petek, M.; Martinez-Mira, C.; Balzano-Nogueira, L.; Ramšak, Ž.; McIntyre, L.; Gruden, K.; Tarazona, S.; Conesa, A. PaintOmics 4: New tools for the integrative analysis of multi-omics datasets supported by multiple pathway databases. Nucleic Acids Res. 2022, 50, W551–W559. [Google Scholar] [CrossRef]
- Mahmood, U.; Li, X.; Fan, Y.; Chang, W.; Niu, Y.; Li, J.; Qu, C.; Lu, K. Multi-omics revolution to promote plant breeding efficiency. Front. Plant Sci. 2022, 13, 1062952. [Google Scholar] [CrossRef]
- Yoosefzadeh Najafabadi, M.; Hesami, M.; Eskandari, M. Machine learning-assisted approaches in modernized plant breeding programs. Genes 2023, 14, 777. [Google Scholar] [CrossRef]
- Reimer, J.J.; Shaaban, B.; Drummen, N.; Sanjeev Ambady, S.; Genzel, F.; Poschet, G.; Wiese-Klinkenberg, A.; Usadel, B.; Wormit, A. Capsicum leaves under stress: Using multi-omics analysis to detect abiotic stress network of secondary metabolism in two species. Antioxidants 2022, 11, 671. [Google Scholar] [CrossRef]
- Gouesbet, G. Deciphering macromolecular interactions involved in abiotic stress signaling: A review of bioinformatics analysis. In Plant Abiotic Stress Signaling; Methods in Molecular Biology; Couée, I., Ed.; Humana Press: New York, NY, USA, 2023; Volume 2642, pp. 257–294. [Google Scholar] [CrossRef]
- Guerrero-Sánchez, V.M.; López-Hidalgo, C.; Rey, M.D.; Castillejo, M.Á.; Jorrín-Novo, J.V.; Escandón, M. Multiomic Data integration in the analysis of drought-responsive mechanisms in Quercus ilex seedlings. Plants 2022, 11, 3067. [Google Scholar] [CrossRef]
- Bittencourt, C.B.; Carvalho da Silva, T.L.; Rodrigues Neto, J.C.; Vieira, L.R.; Leão, A.P.; de Aquino Ribeiro, J.A.; Abdelnur, P.V.; de Sousa, C.A.F.; Souza, M.T. Insights from a multi-omics integration (MOI) study in oil palm (Elaeis guineensis Jacq.) response to abiotic stresses: Part one-salinity. Plants 2022, 11, 1755. [Google Scholar] [CrossRef]
- Leão, A.P.; Bittencourt, C.B.; Carvalho da Silva, T.L.; Rodrigues Neto, J.C.; Braga, Í.O.; Vieira, L.R.; de Aquino Ribeiro, J.A.; Abdelnur, P.V.; de Sousa, C.A.F.; Souza Júnior, M.T. Insights from a multi-omics integration (MOI) study in oil palm (Elaeis guineensis Jacq.) response to abiotic stresses: Part two-drought. Plants 2022, 11, 2786. [Google Scholar] [CrossRef]
- Kudapa, H.; Ghatak, A.; Barmukh, R.; Chaturvedi, P.; Khan, A.; Kale, S.; Fragner, L.; Chitikineni, A.; Weckwerth, W.; Varshney, R.K. Integrated multi-omics analysis reveals drought stress response mechanism in chickpea (Cicer arietinum L.). Plant Genome 2023, e20337. [Google Scholar] [CrossRef]

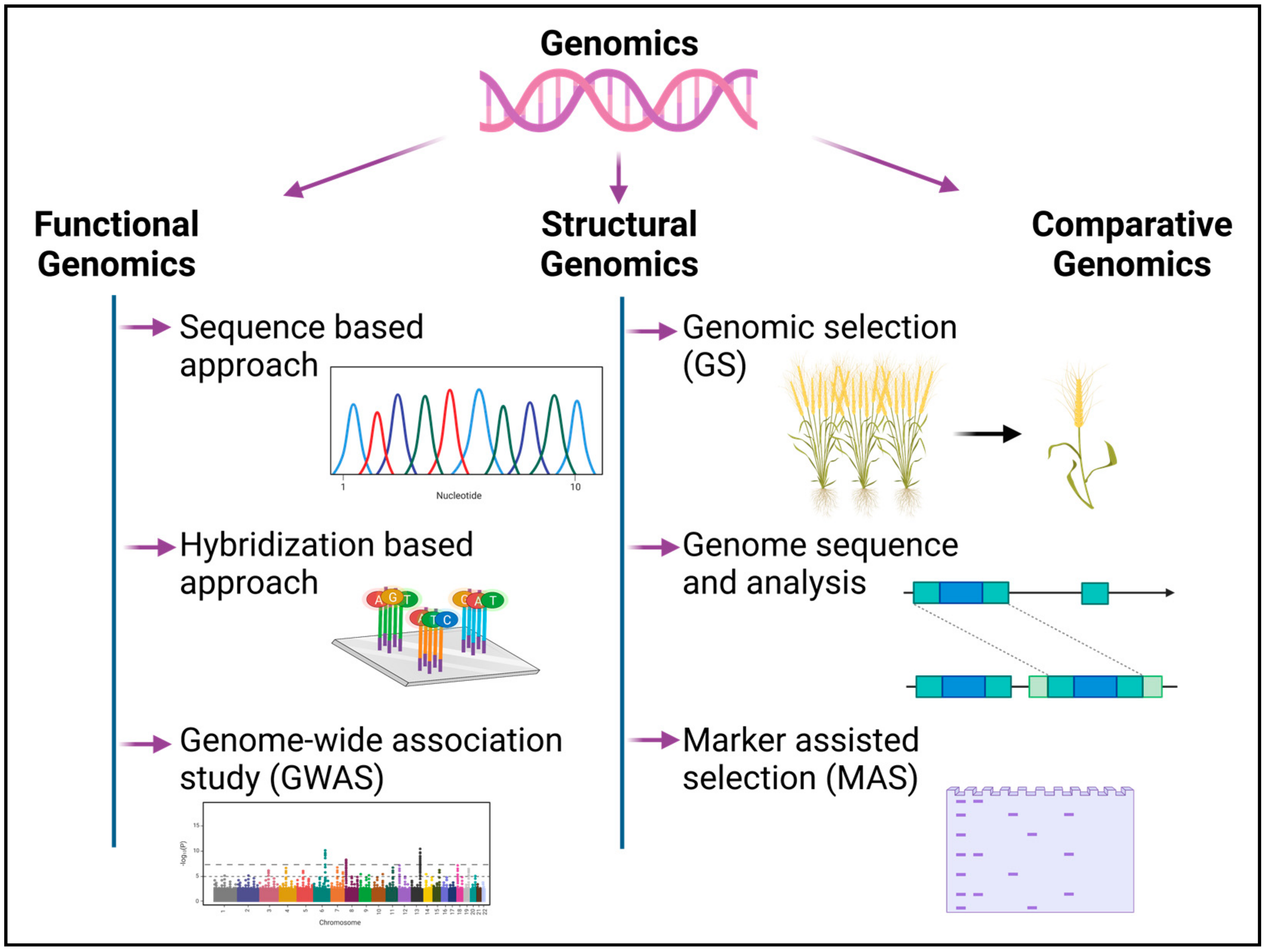
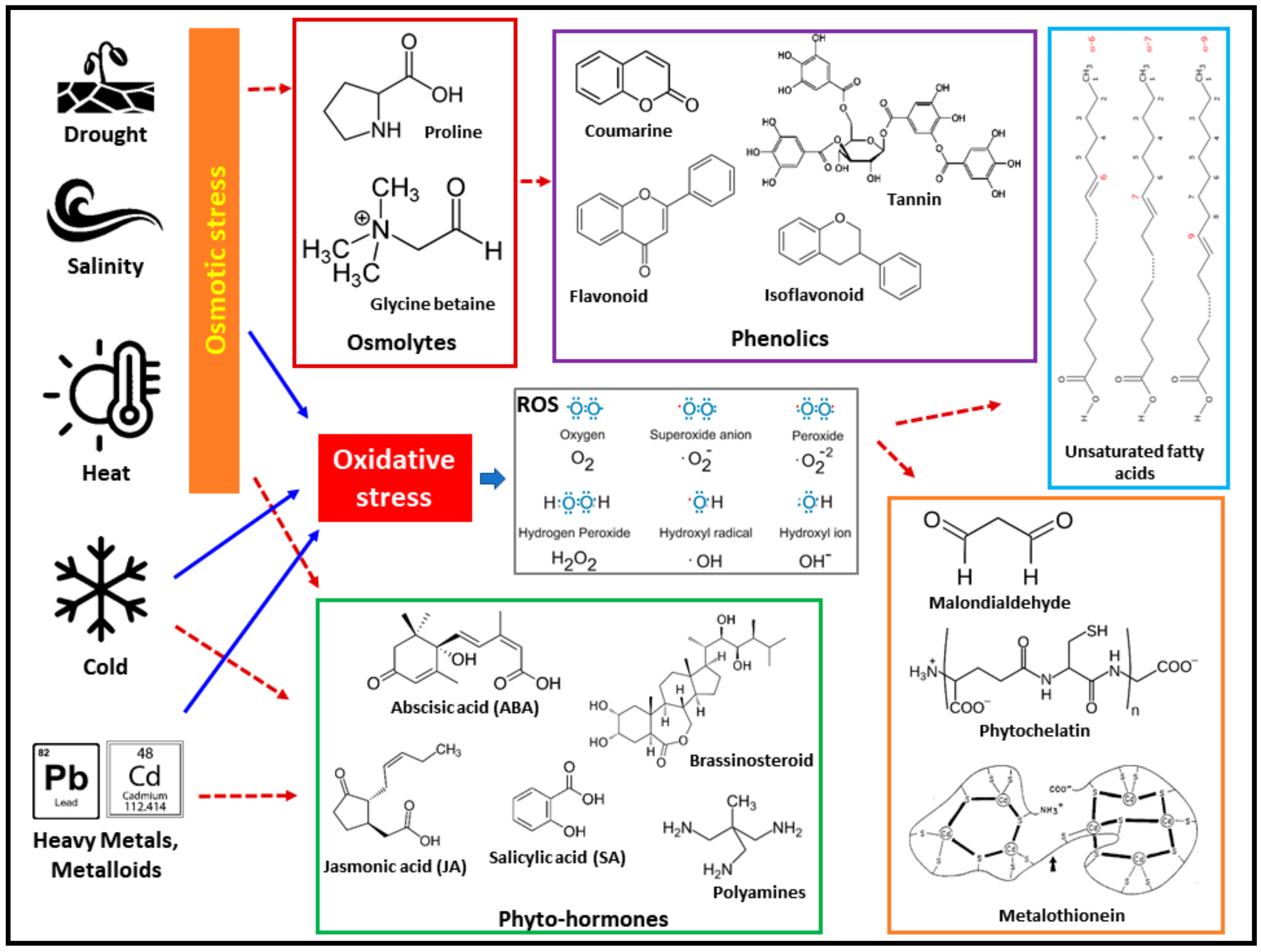


| Plants | Genes | Function | Abiotic Stresses | Note/Remarks | References |
|---|---|---|---|---|---|
| Barley | rhl1.a | Limit root hair | Drought | Loss of function mutation | [114] |
| Maize | Zm00001d010956 | Induce TFs | Salinity | [123] | |
| Tomato | SlERF.D6 | Steroidal glycoalkaloids (SGAs) biosynthesis | Drought, Salinity | [129] | |
| Alfalfa | HAMK | Signaling pathway | Heat | [135] | |
| Bread wheat | HVA1 | Signaling pathway | Drought, Heat | Hordeum vulgare aleurone 1 | [136] |
| Wheat | AtWRKY30 | TFs | Heat, Drought | Antioxidant, osmolytes biosynthesis | [137] |
| Arabidopsis | AtSZF2 | Regulate salt-responsive genes | Salinity | Loss of function mutation | [138] |
| Arabidopsis | CBF1 | TFs | Cold, Heat | C-Repeat Binding Factor1 | [139] |
| Arabidopsis | PICKLE (PKL) | CHD3-type chromatin remodeler | Cold | Trimethylation of histone H3 lysine 27 (H3K27me3) | [140] |
| Tomato | SlAREB1 | TFs | Salinity | [141] | |
| Tomato | Ethylene Response Factors (ERF) | TFs | Cold, Heat, Salinity, Drought, Submergence | [142] | |
| Tomato | SlUVR8 | UV-B photoreceptor | UV-B | [143] | |
| Wild tomato (Solanum habrochaites) | Calmodulin-like (ShCML44) | Calmodulin-like (CML) proteins Ca2+ sensors | Cold, Drought, Salinity | Enhances Antioxidants Capacity | [144] |
| Cabbage | HSP70 | Heat Shock Protein | Heat | [145] | |
| Pearl millet | PgDREB2A | Transcription Factor | Heat, Drought, Salinity | Transgenic overexpression in tobacco | [146] |
| Rice | OsMAPK5 | Signaling pathway | Drought, Salinity, Cold | [147] | |
| Rice | OsWRKY87 | Increase DNA-binding ability | Drought, Salinity | [148] | |
| Rice | PM H+ ATPase | Proton pump | Salinity | Primary transporter | [149] |
| Rice | Cellulose synthase-like protein OsCSLD4 | Cell wall polysaccharide synthesis | Salinity | ABA-induced osmotic response | [150] |
| Rice | Sucrose non-fermenting 1-related protein kinases (OsSAPK8) | Serine/threonine (Ser/Thr) protein kinase | Cold, Drought, Salinity | [151] | |
| Soybean | MAPK | Signaling pathway | Excess light | [152] | |
| Ground nut | NAC4 | TFs | Drought | [153] | |
| Ground nut | HSP70 | Chaperon | Heat | Heat shock protein | [153] |
| Potato | LEA | Cellular protection during stress | Drought, Salinity, Heat | Late Embryogenesis-Abundant protein | [154] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Roychowdhury, R.; Das, S.P.; Gupta, A.; Parihar, P.; Chandrasekhar, K.; Sarker, U.; Kumar, A.; Ramrao, D.P.; Sudhakar, C. Multi-Omics Pipeline and Omics-Integration Approach to Decipher Plant’s Abiotic Stress Tolerance Responses. Genes 2023, 14, 1281. https://doi.org/10.3390/genes14061281
Roychowdhury R, Das SP, Gupta A, Parihar P, Chandrasekhar K, Sarker U, Kumar A, Ramrao DP, Sudhakar C. Multi-Omics Pipeline and Omics-Integration Approach to Decipher Plant’s Abiotic Stress Tolerance Responses. Genes. 2023; 14(6):1281. https://doi.org/10.3390/genes14061281
Chicago/Turabian StyleRoychowdhury, Rajib, Soumya Prakash Das, Amber Gupta, Parul Parihar, Kottakota Chandrasekhar, Umakanta Sarker, Ajay Kumar, Devade Pandurang Ramrao, and Chinta Sudhakar. 2023. "Multi-Omics Pipeline and Omics-Integration Approach to Decipher Plant’s Abiotic Stress Tolerance Responses" Genes 14, no. 6: 1281. https://doi.org/10.3390/genes14061281










