MicroRNA-Related Polymorphism and Their Association with Fibromyalgia
Abstract
1. Introduction
2. Materials and Methods
2.1. Participants and Procedure
2.2. Diagnostic and Clinical Assessment
2.3. Sample Characteristics
2.4. DNA Extraction and Quality Controls, and Genotyping Array
2.5. Biostatistical Analysis
3. Results
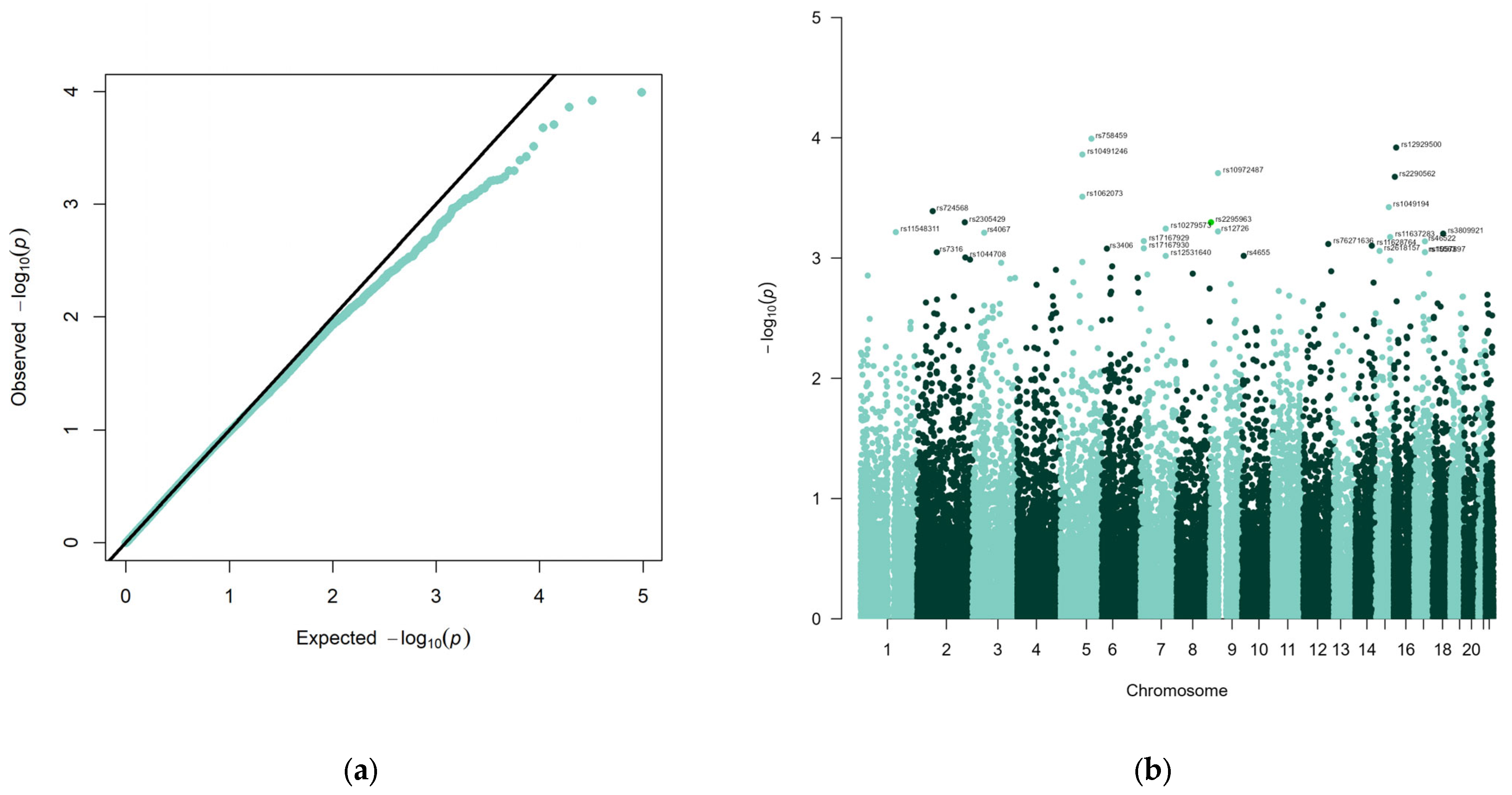
3.1. Genome-Wide DNA-Polymorphism Associations
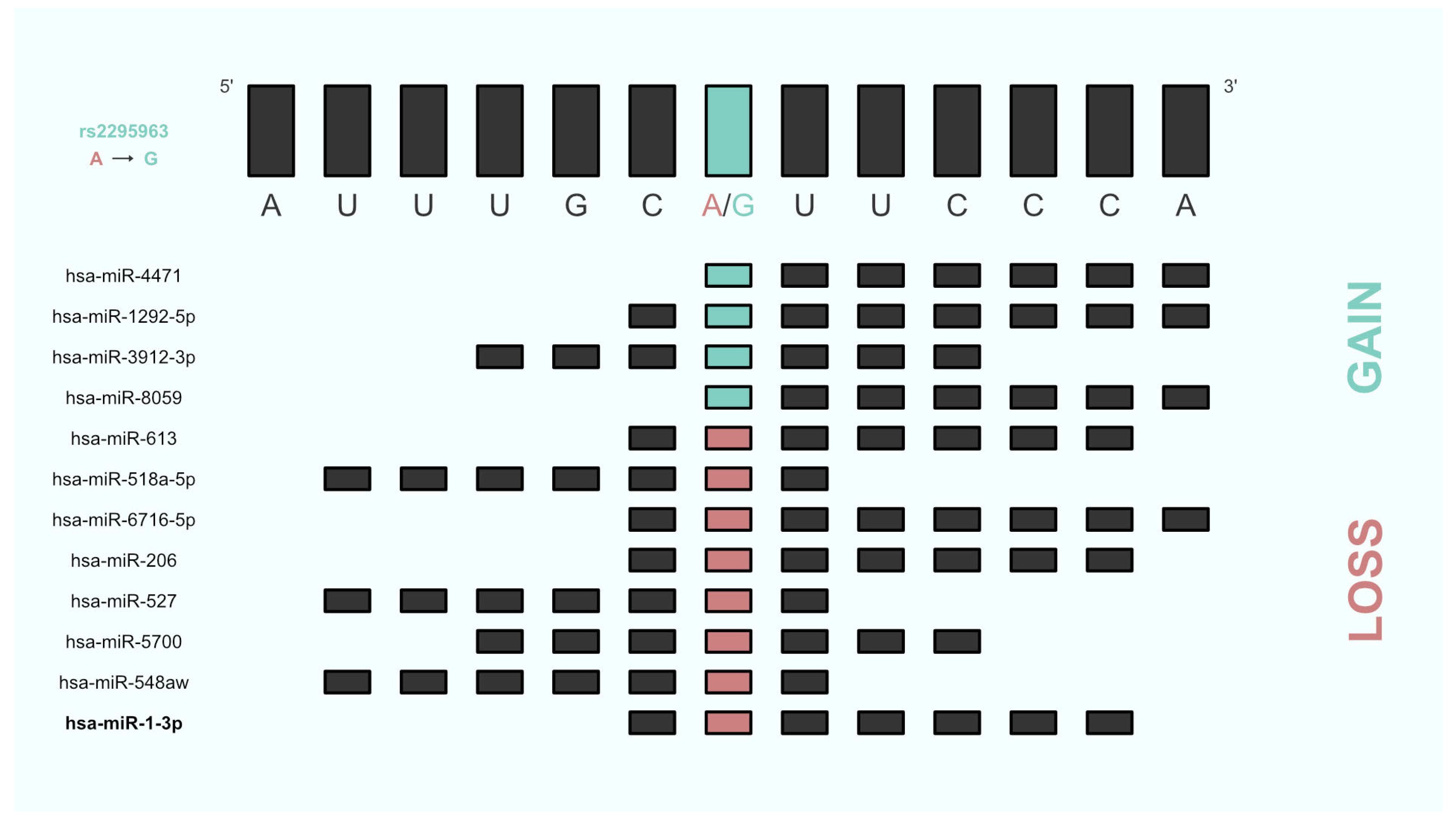
3.2. Predicted miRNA/Target Pair Gain or Loss
3.3. Overlap of Genes and Predicted miRNA Targets
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Wolfe, F.; Clauw, D.J.; Fitzcharles, M.A.; Goldenberg, D.L.; Katz, R.S.; Mease, P.; Russell, A.S.; Russell, I.J.; Winfield, J.B.; Yunus, M.B. The american college of rheumatology preliminary diagnostic criteria for fibromyalgia and measurement of symptom severity. Arthritis Care Res. 2010, 62, 600–610. [Google Scholar] [CrossRef] [PubMed]
- Diers, M. Neuroimaging the pain network—Implications for treatment. Best Pract. Res. Clin. Rheumatol. 2019, 33, 101418. [Google Scholar] [CrossRef] [PubMed]
- Hauser, W.; Zimmer, C.; Felde, E.; Kollner, V. What are the key symptoms of fibromyalgia? Schmerz 2008, 22, 176–183. [Google Scholar] [PubMed]
- Diers, M.; Koeppe, C.; Yilmaz, P.; Thieme, K.; Markela-Lerenc, J.; Schiltenwolf, M.; van Ackern, K.; Flor, H. Pain ratings and somatosensory evoked responses to repetitive intramuscular and intracutaneous stimulation in fibromyalgia syndrome. J. Clin. Neurophysiol. 2008, 25, 153–160. [Google Scholar] [CrossRef] [PubMed]
- Diers, M.; Schley, M.T.; Rance, M.; Yilmaz, P.; Lauer, L.; Rukwied, R.; Schmelz, M.; Flor, H. Differential central pain processing following repetitive intramuscular proton/prostaglandin e-2 injections in female fibromyalgia patients and healthy controls. Eur. J. Pain. 2011, 15, 716–723. [Google Scholar] [CrossRef]
- Gracely, R.H.; Petzke, F.; Wolf, J.M.; Clauw, D.J. Functional magnetic resonance imaging evidence of augmented pain processing in fibromyalgia. Arthritis Rheumatol. 2002, 46, 1333–1343. [Google Scholar] [CrossRef]
- Mosch, B.; Hagena, V.; Herpertz, S.; Ruttorf, M.; Diers, M. Neural correlates of control over pain in fibromyalgia patients. NeuroImage Clin. 2023, 37, 103355. [Google Scholar] [CrossRef]
- Ablin, J.N.; Buskila, D. Update on the genetics of the fibromyalgia syndrome. Best Pract. Res. Clin. Rheumatol. 2015, 29, 20–28. [Google Scholar] [CrossRef]
- Ablin, J.N.; Cohen, H.; Buskila, D. Mechanisms of disease: Genetics of fibromyalgia. Nat. Clin. Pract. Rheum. 2006, 2, 671–678. [Google Scholar] [CrossRef]
- Buskila, D.; Sarzi-Puttini, P.; Ablin, J.N. The genetics of fibromyalgia syndrome. Pharmacogenomics 2007, 8, 67–74. [Google Scholar] [CrossRef]
- Galvez-Sanchez, C.M.; Duschek, S.; Reyes Del Paso, G.A. Psychological impact of fibromyalgia: Current perspectives. Psychol. Res. Behav. Manag. 2019, 12, 117–127. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.A.; Keegan, D.; Gardner, G.; Sullivan, M.; Bernstein, D.; Katon, W.J. Psychosocial factors in fibromyalgia compared with rheumatoid arthritis: II. Sexual, physical, and emotional abuse and neglect. Psychosom. Med. 1997, 59, 572–577. [Google Scholar] [CrossRef] [PubMed]
- Walker, E.A.; Keegan, D.; Gardner, G.; Sullivan, M.; Katon, W.J.; Bernstein, D. Psychosocial factors in fibromyalgia compared with rheumatoid arthritis: I. Psychiatric diagnoses and functional disability. Psychosom. Med. 1997, 59, 565–571. [Google Scholar] [CrossRef] [PubMed]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small rnas with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of micrornas. Adv. Drug. Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef]
- Saunders, M.A.; Liang, H.; Li, W.H. Human polymorphism at micrornas and microrna target sites. Proc. Natl. Acad. Sci. USA 2007, 104, 3300–3305. [Google Scholar] [CrossRef]
- Brum, C.B.; Paixao-Cortes, V.R.; Carvalho, A.M.; Martins-Silva, T.; Carpena, M.X.; Ulguim, K.F.; Luquez, K.Y.S.; Salatino-Oliveira, A.; Tovo-Rodrigues, L. Genetic variants in mirnas differentially expressed during brain development and their relevance to psychiatric disorders susceptibility. World J. Biol. Psychiatry 2021, 22, 456–467. [Google Scholar] [CrossRef]
- Miller, B.H.; Wahlestedt, C. Microrna dysregulation in psychiatric disease. Brain Res. 2010, 1338, 89–99. [Google Scholar] [CrossRef]
- Cammaerts, S.; Strazisar, M.; De Rijk, P.; Del Favero, J. Genetic variants in microrna genes: Impact on microrna expression, function, and disease. Front. Genet. 2015, 6, 186. [Google Scholar] [CrossRef]
- Caputo, V.; Sinibaldi, L.; Fiorentino, A.; Parisi, C.; Catalanotto, C.; Pasini, A.; Cogoni, C.; Pizzuti, A. Brain derived neurotrophic factor (bdnf) expression is regulated by micrornas mir-26a and mir-26b allele-specific binding. PLoS ONE 2011, 6, e28656. [Google Scholar] [CrossRef]
- Gong, Y.; Wu, C.N.; Xu, J.; Feng, G.; Xing, Q.H.; Fu, W.; Li, C.; He, L.; Zhao, X.Z. Polymorphisms in microrna target sites influence susceptibility to schizophrenia by altering the binding of mirnas to their targets. Eur. Neuropsychopharmacol. 2013, 23, 1182–1189. [Google Scholar] [CrossRef]
- Rykova, E.; Ershov, N.; Damarov, I.; Merkulova, T. Snps in 3’utr mirna target sequences associated with individual drug susceptibility. Int. J. Mol. Sci. 2022, 23, 13725. [Google Scholar] [CrossRef]
- Dutta, D.; Brummett, C.M.; Moser, S.E.; Fritsche, L.G.; Tsodikov, A.; Lee, S.; Clauw, D.J.; Scott, L.J. Heritability of the fibromyalgia phenotype varies by age. Arthritis Rheumatol. 2020, 72, 815–823. [Google Scholar] [CrossRef]
- Bjersing, J.L.; Lundborg, C.; Bokarewa, M.I.; Mannerkorpi, K. Profile of cerebrospinal micrornas in fibromyalgia. PLoS ONE 2013, 8, e78762. [Google Scholar] [CrossRef] [PubMed]
- Bjersing, J.L.; Bokarewa, M.I.; Mannerkorpi, K. Profile of circulating micrornas in fibromyalgia and their relation to symptom severity: An exploratory study. Rheumatol. Int. 2015, 35, 635–642. [Google Scholar] [CrossRef]
- Cerda-Olmedo, G.; Mena-Duran, A.V.; Monsalve, V.; Oltra, E. Identification of a microrna signature for the diagnosis of fibromyalgia. PLoS ONE 2015, 10, e0121903. [Google Scholar]
- Masotti, A.; Baldassarre, A.; Guzzo, M.P.; Iannuccelli, C.; Barbato, C.; Di Franco, M. Circulating microrna profiles as liquid biopsies for the characterization and diagnosis of fibromyalgia syndrome. Mol. Neurobiol. 2017, 54, 7129–7136. [Google Scholar] [CrossRef]
- Sakai, A.; Suzuki, H. Microrna and pain. Adv. Exp. Med. Biol. 2015, 888, 17–39. [Google Scholar] [PubMed]
- Wolfe, F.; Clauw, D.J.; Fitzcharles, M.A.; Goldenberg, D.L.; Hauser, W.; Katz, R.L.; Mease, P.J.; Russell, A.S.; Russell, I.J.; Walitt, B. 2016 revisions to the 2010/2011 fibromyalgia diagnostic criteria. Semin. Arthritis Rheum. 2016, 46, 319–329. [Google Scholar]
- Kerns, R.D.; Turk, D.C.; Rudy, T.E. The west haven-yale multidimensional pain inventory (whympi). Pain 1985, 23, 345–356. [Google Scholar] [CrossRef] [PubMed]
- Flor, H.; Rudy, T.E.; Birbaumer, N.; Streit, B.; Schugens, M.M. The applicability of the west haven-yale multidimensional pain inventory in german-speaking countries. Data on the reliability and validity of the mpi-d. Schmerz 1990, 4, 82–87. [Google Scholar] [CrossRef]
- Vonkorff, M.; Ormel, J.; Keefe, F.J.; Dworkin, S.F. Grading the severity of chronic pain. Pain 1992, 50, 133–149. [Google Scholar] [CrossRef] [PubMed]
- Hauser, W.; Jung, E.; Erbsloh-Moller, B.; Gesmann, M.; Kuhn-Becker, H.; Petermann, F.; Langhorst, J.; Weiss, T.; Winkelmann, A.; Wolfe, F. Validation of the fibromyalgia survey questionnaire within a cross-sectional survey. PLoS ONE 2012, 7, e37504. [Google Scholar] [CrossRef] [PubMed]
- Offenbaecher, M.; Waltz, M.; Schoeps, P. Validation of a german version of the fibromyalgia impact questionnaire (fiq-g). J. Rheumatol. 2000, 27, 1984–1988. [Google Scholar]
- Flor, H.; Behle, D.J.; Birbaumer, N. Assessment of pain-related cognitions in chronic pain patients. Behav. Res. Ther. 1993, 31, 63–73. [Google Scholar] [CrossRef] [PubMed]
- Radloff, L.S. The ces-d scale: A self-report depression scale for research in the general population. Appl. Psychol. Meas. 1977, 1, 385–401. [Google Scholar] [CrossRef]
- Hautzinger, M.; Bailer, M. Allgemeine Depressionsskala (ADS); Die Deutsche Version Des Ces—d. Beltz Test; Beltz: Weinheim, Germany, 1993. [Google Scholar]
- Miller, S.A.; Dykes, D.D.; Polesky, H.F. A simple salting out procedure for extracting DNA from human nucleated cells. Nucleic Acids Res. 1988, 16, 1215. [Google Scholar] [CrossRef]
- Purcell, S.; Neale, B.; Todd-Brown, K.; Thomas, L.; Ferreira, M.A.R.; Bender, D.; Maller, J.; Sklar, P.; de Bakker, P.I.W.; Daly, M.J.; et al. Plink: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007, 81, 559–575. [Google Scholar] [CrossRef]
- Wilkins, O.M.; Titus, A.J.; Gui, J.; Eliot, M.; Butler, R.A.; Sturgis, E.M.; Li, G.; Kelsey, K.T.; Christensen, B.C. Genome-scale identification of microrna-related snps associated with risk of head and neck squamous cell carcinoma. Carcinogenesis 2017, 38, 986–993. [Google Scholar] [CrossRef]
- Liu, C.J.; Fu, X.; Xia, M.; Zhang, Q.; Gu, Z.; Guo, A.Y. Mirnasnp-v3: A comprehensive database for snps and disease-related variations in mirnas and mirna targets. Nucleic Acids Res. 2021, 49, D1276–D1281. [Google Scholar] [CrossRef]
- Gong, J.; Liu, C.; Liu, W.; Wu, Y.; Ma, Z.; Chen, H.; Guo, A.Y. An update of mirnasnp database for better snp selection by gwas data, mirna expression and online tools. Database 2015, 2015, bav029. [Google Scholar] [CrossRef] [PubMed]
- Gong, J.; Tong, Y.; Zhang, H.M.; Wang, K.; Hu, T.; Shan, G.; Sun, J.; Guo, A.Y. Genome-wide identification of snps in microrna genes and the snp effects on microrna target binding and biogenesis. Hum. Mutat. 2012, 33, 254–263. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.H.; Wang, X.W. Mirdb: An online database for prediction of functional microrna targets. Nucleic Acids Res. 2020, 48, D127–D131. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Wang, X. Prediction of functional microrna targets by integrative modeling of microrna binding and target expression data. Genome. Biol. 2019, 20, 18. [Google Scholar] [CrossRef]
- Arnold, L.M.; Fan, J.B.; Russell, I.J.; Yunus, M.B.; Khan, M.A.; Kushner, I.; Olson, J.M.; Iyengar, S.K. The fibromyalgia family study: A genome-wide linkage scan study. Arthritis Rheumatol. 2013, 65, 1122–1128. [Google Scholar] [CrossRef]
- Docampo, E.; Escaramis, G.; Gratacos, M.; Villatoro, S.; Puig, A.; Kogevinas, M.; Collado, A.; Carbonell, J.; Rivera, J.; Vidal, J.; et al. Genome-wide analysis of single nucleotide polymorphisms and copy number variants in fibromyalgia suggest a role for the central nervous system. Pain 2014, 155, 1102–1109. [Google Scholar] [CrossRef]
- Johnston, K.J.A.; Adams, M.J.; Nicholl, B.I.; Ward, J.; Strawbridge, R.J.; Ferguson, A.; McIntosh, A.M.; Bailey, M.E.S.; Smith, D.J. Genome-wide association study of multisite chronic pain in uk biobank. PLoS Genet. 2019, 15, e1008164. [Google Scholar] [CrossRef]
- Rahman, M.S.; Winsvold, B.S.; Chavez Chavez, S.O.; Borte, S.; Tsepilov, Y.A.; Sharapov, S.Z.; Pain, H.A.-I.; Aulchenko, Y.S.; Hagen, K.; Fors, E.A.; et al. Genome-wide association study identifies rnf123 locus as associated with chronic widespread musculoskeletal pain. Ann. Rheum. Dis. 2021, 80, 1227–1235. [Google Scholar] [CrossRef]
- D’Agnelli, S.; Arendt-Nielsen, L.; Gerra, M.C.; Zatorri, K.; Boggiani, L.; Baciarello, M.; Bignami, E. Fibromyalgia: Genetics and epigenetics insights may provide the basis for the development of diagnostic biomarkers. Mol. Pain 2019, 15, 1744806918819944. [Google Scholar] [CrossRef]
- Jin, Y.; Lee, C.G. Single nucleotide polymorphisms associated with microrna regulation. Biomolecules 2013, 3, 287–302. [Google Scholar] [CrossRef]
- Hummel, E.M.; Hessas, E.; Muller, S.; Beiter, T.; Fisch, M.; Eibl, A.; Wolf, O.T.; Giebel, B.; Platen, P.; Kumsta, R.; et al. Cell-free DNA release under psychosocial and physical stress conditions. Transl. Psychiatry 2018, 8, 236. [Google Scholar] [CrossRef] [PubMed]
- Moser, D.; Ekawardhani, S.; Kumsta, R.; Palmason, H.; Bock, C.; Athanassiadou, Z.; Lesch, K.P.; Meyer, J. Functional analysis of a potassium-chloride co-transporter 3 (slc12a6) promoter polymorphism leading to an additional DNA methylation site. Neuropsychopharmacology 2009, 34, 458–467. [Google Scholar] [CrossRef]
- Philippidou, D.; Schmitt, M.; Moser, D.; Margue, C.; Nazarov, P.V.; Muller, A.; Vallar, L.; Nashan, D.; Behrmann, I.; Kreis, S. Signatures of micrornas and selected microrna target genes in human melanoma. Cancer Res. 2010, 70, 4163–4173. [Google Scholar] [CrossRef]
- Jurkiewicz, M.; Moser, D.; Koller, A.; Yu, L.; Chen, E.I.; Bennett, D.A.; Canli, T. Integration of postmortem amygdala expression profiling, gwas, and functional cell culture assays: Neuroticism-associated synaptic vesicle glycoprotein 2a (sv2a) gene is regulated by mir-133a and mir-218. Transl. Psychiatry 2020, 10, 297. [Google Scholar] [CrossRef]
- Jurkiewicz, M.M.; Mueller-Alcazar, A.; Moser, D.A.; Jayatilaka, I.; Mikhailik, A.; Ferri, J.; Fogelman, N.; Canli, T. Integrated microrna and mrna gene expression in peripheral blood mononuclear cells in response to acute psychosocial stress: A repeated-measures within-subject pilot study. BMC Res. Notes 2021, 14, 222. [Google Scholar] [CrossRef] [PubMed]
- Beiter, T.; Niess, A.M.; Moser, D. Transcriptional memory in skeletal muscle. Don’t forget (to) exercise. J. Cell. Physiol. 2020, 235, 5476–5489. [Google Scholar] [CrossRef] [PubMed]
- Ehlert, T.; Simon, P.; Moser, D.A. Epigenetics in sports. Sports Med. 2013, 43, 93–110. [Google Scholar] [CrossRef]
- Hummel, E.; Elgizouli, M.; Sicorello, M.; Leitao, E.; Beygo, J.; Schroder, C.; Zeschnigk, M.; Muller, S.; Herpertz, S.; Moser, D.; et al. No evidence for intervention-associated DNA methylation changes in monocytes of patients with posttraumatic stress disorder. Sci. Rep. 2022, 12, 17347. [Google Scholar] [CrossRef]
- Hummel, E.M.; Piovesan, K.; Berg, F.; Herpertz, S.; Kessler, H.; Kumsta, R.; Moser, D.A. Mitochondrial DNA as a marker for treatment-response in post-traumatic stress disorder. Psychoneuroendocrinology 2023, 148, 105993. [Google Scholar] [CrossRef]
- Schwaiger, M.; Grinberg, M.; Moser, D.; Zang, J.C.; Heinrichs, M.; Hengstler, J.G.; Rahnenfuhrer, J.; Cole, S.; Kumsta, R. Altered stress-induced regulation of genes in monocytes in adults with a history of childhood adversity. Neuropsychopharmacology 2016, 41, 2530–2540. [Google Scholar] [CrossRef]
- Zang, J.C.S.; May, C.; Hellwig, B.; Moser, D.; Hengstler, J.G.; Cole, S.; Heinrichs, M.; Rahnenfuhrer, J.; Marcus, K.; Kumsta, R. Proteome analysis of monocytes implicates altered mitochondrial biology in adults reporting adverse childhood experiences. Transl. Psychiatry 2023, 13, 31. [Google Scholar] [CrossRef] [PubMed]
- Clos-Garcia, M.; Andrés-Marin, N.; Fernández-Eulate, G.; Abecia, L.; Lavín, J.L.; van Liempd, S.; Cabrera, D.; Royo, F.; Valero, A.; Errazquin, N.; et al. Gut microbiome and serum metabolome analyses identify molecular biomarkers and altered glutamate metabolism in fibromyalgia. EBioMedicine 2019, 46, 499–511. [Google Scholar]
- Leinders, M.; Doppler, K.; Klein, T.; Deckart, M.; Rittner, H.; Sommer, C.; Üçeyler, N. Increased cutaneous miR-let-7d expression correlates with small nerve fiber pathology in patients with fibromyalgia syndrome. Pain 2016, 157, 2493–2503. [Google Scholar]
- Braun, A.; Evdokimov, D.; Frank, J.; Sommer, C.; Üçeyler, N. MiR103a-3p and miR107 are related to adaptive coping in a cluster of fibromyalgia patients. PLoS ONE 2020, 15, e0239286. [Google Scholar]
- Hussein, M.; Fathy, W.; Abdelaleem, E.A.; Nasser, M.; Yehia, A.; Elanwar, R. The Impact of Micro RNA-320a Serum Level on Severity of Symptoms and Cerebral Processing of Pain in Patients with Fibromyalgia. Pain Med. 2022, 23, 2061–2072. [Google Scholar]
- Nepotchatykh, E.; Caraus, I.; Elremaly, W.; Leveau, C.; Elbakry, M.; Godbout, C.; Rostami-Afshari, B.; Petre, D.; Khatami, N.; Franco, A.; et al. Circulating microRNA expression signatures accurately discriminate myalgic encephalomyelitis from fibromyalgia and comorbid conditions. Sci. Rep. 2023, 13, 1896. [Google Scholar]
- Erbacher, C.; Vaknine, S.; Moshitzky, G.; Lobentanzer, S.; Eisenberg, L.; Evdokimov, D.; Sommer, C.; Greenberg, D.S.; Soreq, H.; Üçeyler, N. Distinct CholinomiR Blood Cell Signature as a Potential Modulator of the Cholinergic System in Women with Fibromyalgia Syndrome. Cells 2022, 11, 1276. [Google Scholar]
| FM | HC | |||||
|---|---|---|---|---|---|---|
| M | SD | Range | M | SD | Range | |
| Age (years) | 50.20 | 9.45 | 23–68 | 47.46 | 15.35 | 19–81 |
| Pain duration in years | 16.99 | 12.82 | 0.9–52.6 | |||
| CES-D | 23.61 | 7.00 | 8–44 | 14.51 | 4.95 | 4–32 |
| FSQ | ||||||
| Symptom Severity Score | 9.61 | 1.95 | 2–12 | |||
| Widespread Pain Index | 11.49 | 4.21 | 1–19 | |||
| FIQ | ||||||
| Physical functioning | 1.46 | 0.50 | 0–2.5 | |||
| Total | 56.69 | 15.37 | 23.70–131.85 | |||
| CpG | ||||||
| Pain intensity | 72.33 | 12.63 | 40–100 | |||
| Disability score | 65.58 | 18.39 | 0–100 | |||
| Chronic pain grade | 3.35 | 0.77 | 1–4 | |||
| MPI | ||||||
| Pain severity | 4.07 | 0.98 | 1–6 | |||
| Interference | 4.25 | 1.14 | 0.5–6 | |||
| Life control | 3.16 | 1.24 | 0.33–6 | |||
| Affective distress | 3.54 | 1.30 | 0.33–6 | |||
| Social support | 3.39 | 1.68 | 0–6 | |||
| Punishing responses | 1.32 | 1.50 | 0–6 | |||
| Solicitous responses | 3.28 | 1.65 | 0–6 | |||
| Distracting responses | 2.87 | 1.42 | 0–6 | |||
| Social activities | 2.30 | 0.98 | 0.25–5.88 | |||
| General activity level | 7.53 | 2.47 | 2.08–13.18 | |||
| PRSS | ||||||
| Catastrophizing | 2.50 | 1.12 | 0–4.78 | |||
| Coping | 3.05 | 0.79 | 0.13–4.75 |
| dbSNP RS ID | CHR | BP | A1 | OR | p | Gene | SNP-Gene-Relationship | NCBI Gene ID |
|---|---|---|---|---|---|---|---|---|
| rs758459 | 5 | 134872704 | C | 0.3646 | 0.0001017 | NEUROG1 | upstream | 4762 |
| rs12929500 | 16 | 10692832 | C | 0.4219 | 0.0001205 | EMP2 | upstream | 2013 |
| rs10491246 | 5 | 94939777 | G | 2.394 | 0.0001377 | ARSK | UTR-3 | 153642 |
| rs10972487 | 9 | 35482149 | T | 0.5207 | 0.0001966 | ATP8B5P | exon | 158381 |
| rs2290562 | 16 | 4853816 | A | 0.5148 | 0.000211 | GLYR1 | UTR-3 | 84656 |
| rs1062073 | 5 | 94799799 | C | 2.282 | 0.0003089 | TTC37 | UTR-3 | 9652 |
| rs1049194 | 15 | 80478645 | C | 0.4079 | 0.0003774 | FAH | UTR-3 | 2184 |
| rs724568 | 2 | 67942480 | C | 1.773 | 0.0004073 | ETAA1 | downstream | 54465 |
| rs2305429 | 2 | 208986385 | A | 4.495 | 0.0005041 | CRYGD | UTR-3 | 1421 |
| rs2295963 | 9 | 4664852 | G | 2.126 | 0.0005043 | SPATA6L * | intron | 55064 |
| rs10279573 | 7 | 110467221 | T | 0.4421 | 0.0005692 | IMMP2L | intron | 83943 |
| rs12726 | 9 | 35404840 | A | 0.5394 | 0.0006009 | UNC13B | UTR-3 | 10497 |
| rs11548311 | 1 | 155026871 | T | 2.431 | 0.0006086 | ADAM15 | synon | 8751 |
| rs4067 | 3 | 51738256 | A | 0.4272 | 0.0006154 | TEX264 | UTR-3 | 51368 |
| rs3809921 | 18 | 46387889 | A | 0.5587 | 0.0006264 | CTIF | UTR-3 | 9811 |
| rs11637283 | 15 | 86316103 | T | 1.972 | 0.0006682 | KLHL25 | intron | 64410 |
| rs17167929 | 7 | 14187669 | C | 0.4188 | 0.0007224 | DGKB | UTR-3 | 1607 |
| rs46522 | 17 | 46988597 | C | 0.5838 | 0.0007262 | UBE2Z | intron | 65264 |
| rs76271636 | 12 | 109886494 | T | 0.264 | 0.0007639 | KCTD10 | UTR-3 | 83892 |
| rs11628764 | 14 | 92527977 | C | 0.5439 | 0.0007893 | ATXN3 | UTR-3 | 4287 |
| rs17167930 | 7 | 14187931 | C | 0.4235 | 0.0008299 | DGKB | UTR-3 | 1607 |
| rs3406 | 6 | 22194573 | A | 2.258 | 0.0008349 | CASC15 | intron | 401237 |
| rs2618157 | 15 | 39844315 | A | 2.827 | 0.0008722 | THBS1 | upstream | 7057 |
| rs1057897 | 17 | 47005509 | T | 0.5888 | 0.0008938 | UBE2Z | UTR-3 | 65264 |
| rs15563 | 17 | 47005193 | A | 0.5888 | 0.0008938 | UBE2Z | UTR-3 | 65264 |
| rs7316 | 2 | 85886013 | C | 3.151 | 0.000895 | SFTPB | UTR-3 | 6439 |
| rs4655 | 10 | 7849688 | C | 0.5598 | 0.0009588 | ATP5C1 | UTR-3 | 509 |
| rs12531640 | 7 | 110479535 | T | 0.4337 | 0.0009596 | IMMP2L | intron | 83943 |
| rs1044708 | 2 | 211298180 | T | 0.5792 | 0.0009886 | LANCL1-AS1 | intron | 102724820 |
| SNP | miRNA | Binding Start (miRmap) | Binding End (miRmap) | Binding Start (TargetScan) | Binding End (TargetScan) | Target |
|---|---|---|---|---|---|---|
| rs2295963 | hsa-miR-4471 | 4664852 | 4664858 | 4664852 | 4664858 | Gain |
| hsa-miR-1292-5p | 4664851 | 4664858 | 4664851 | 4664858 | Gain | |
| hsa-miR-3912-3p | 4664849 | 4664855 | 4664849 | 4664854 | Gain | |
| hsa-miR-8059 | 4664852 | 4664858 | 4664852 | 4664858 | Gain | |
| hsa-miR-613 | 4664851 | 4664857 | 4664851 | 4664856 | Loss | |
| hsa-miR-518a-5p | 4664847 | 4664853 | 4664847 | 4664852 | Loss | |
| hsa-miR-6716-5p | 4664851 | 4664858 | 4664851 | 4664858 | Loss | |
| hsa-miR-206 | 4664851 | 4664857 | 4664851 | 4664856 | Loss | |
| hsa-miR-527 | 4664847 | 4664853 | 4664847 | 4664852 | Loss | |
| hsa-miR-5700 | 4664849 | 4664855 | 4664849 | 4664854 | Loss | |
| hsa-miR-548aw | 4664847 | 4664853 | 4664847 | 4664852 | Loss | |
| hsa-miR-1-3p | 4664851 | 4664857 | 4664851 | 4664856 | Loss |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Berg, F.; Moser, D.A.; Hagena, V.; Streit, F.; Mosch, B.; Kumsta, R.; Herpertz, S.; Diers, M. MicroRNA-Related Polymorphism and Their Association with Fibromyalgia. Genes 2023, 14, 1312. https://doi.org/10.3390/genes14071312
Berg F, Moser DA, Hagena V, Streit F, Mosch B, Kumsta R, Herpertz S, Diers M. MicroRNA-Related Polymorphism and Their Association with Fibromyalgia. Genes. 2023; 14(7):1312. https://doi.org/10.3390/genes14071312
Chicago/Turabian StyleBerg, Fabian, Dirk A. Moser, Verena Hagena, Fabian Streit, Benjamin Mosch, Robert Kumsta, Stephan Herpertz, and Martin Diers. 2023. "MicroRNA-Related Polymorphism and Their Association with Fibromyalgia" Genes 14, no. 7: 1312. https://doi.org/10.3390/genes14071312
APA StyleBerg, F., Moser, D. A., Hagena, V., Streit, F., Mosch, B., Kumsta, R., Herpertz, S., & Diers, M. (2023). MicroRNA-Related Polymorphism and Their Association with Fibromyalgia. Genes, 14(7), 1312. https://doi.org/10.3390/genes14071312






