Antagonistic Bacteria Bacillus velezensis VB7 Possess Nematicidal Action and Induce an Immune Response to Suppress the Infection of Root-Knot Nematode (RKN) in Tomato
Abstract
:1. Introduction
2. Materials and Methods
2.1. Isolations and Morphological Identifications of Root-Knot Nematode from Root Galls of Tomato
2.2. Molecular Characterization of Root-Knot Nematode
2.3. Preparation of Nematode Inoculum
2.4. Testing the Nematicidal Activity of Bacterial Endophytes against RKN
2.5. Preparation and Applications of Liquid Formulations of B. velezensis VB7 and Inoculation of M. incognita
2.6. RNA Extraction and Quantitative Real-Time PCR
2.7. Protein-Protein Interaction
2.8. Statistical Analysis
3. Results
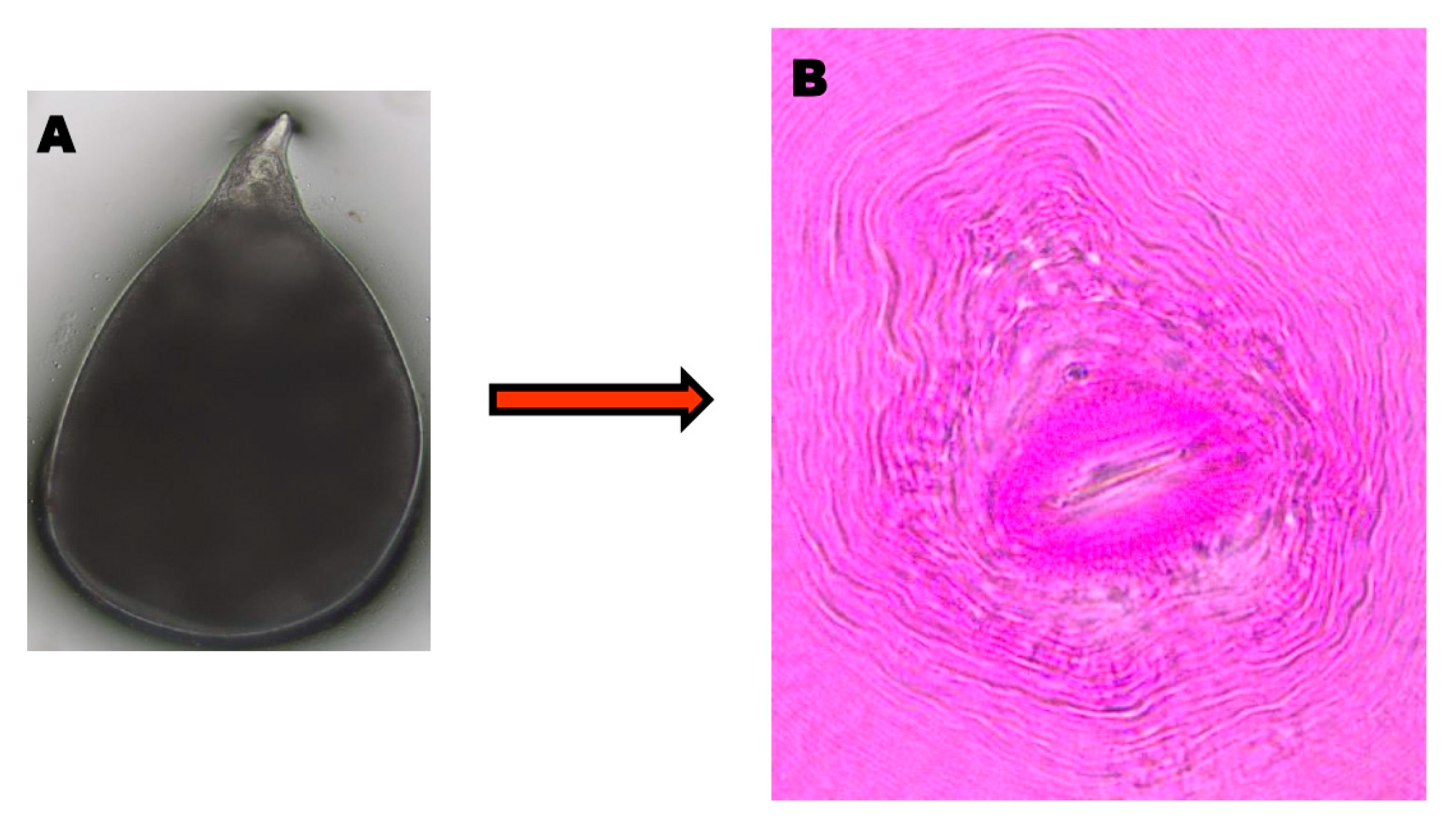
3.1. Morphological and Molecular Confirmation of RKN
3.2. Efficacy of Culture Filtrate of Bacterial Endophytes against Egg Hatching and Juveniles’ Mortality of Root Knot Nematode (RKN) M. incognita In Vitro
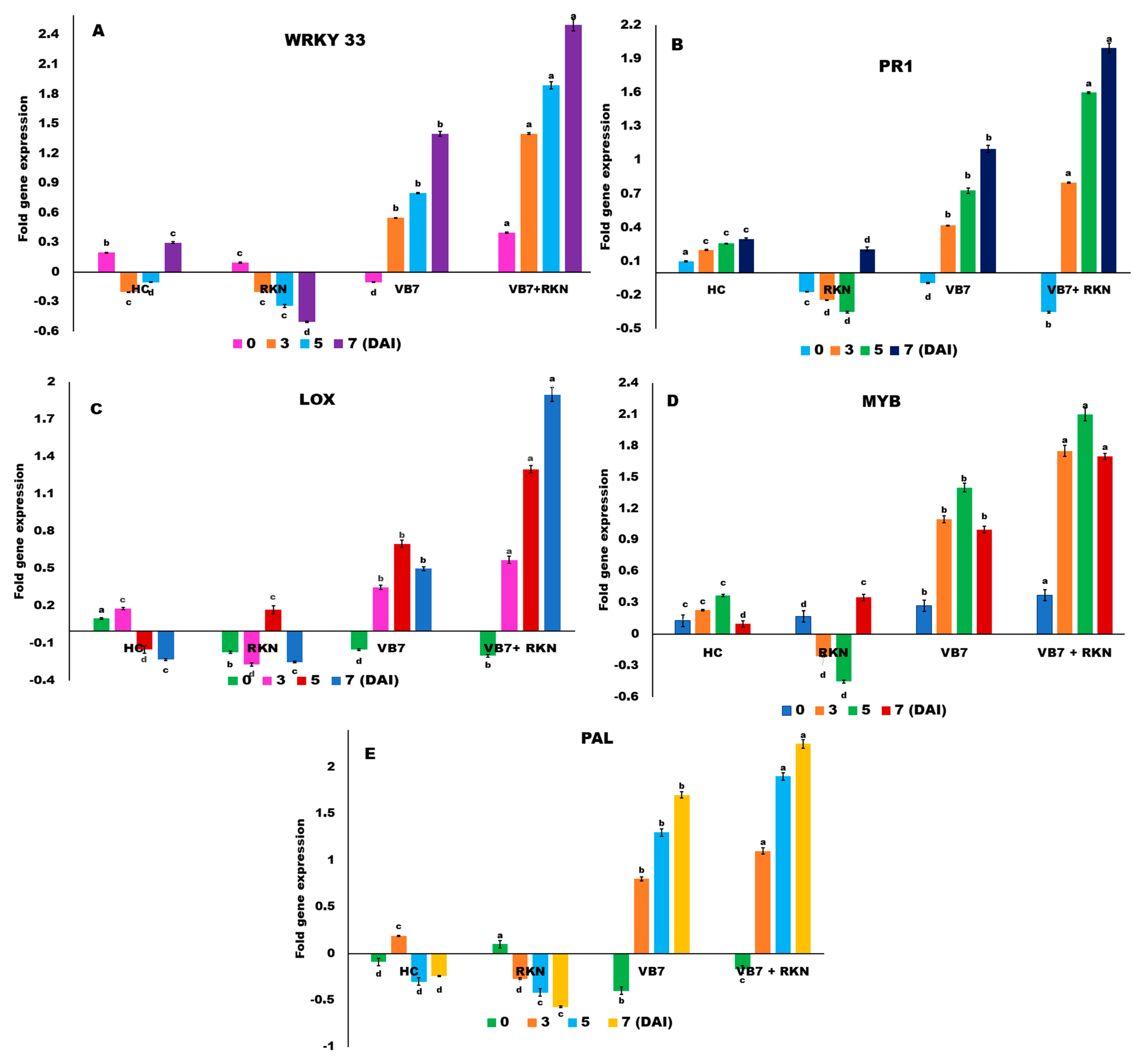
3.3. Induction of MAMP Triggered Immunity by B. velezensis for Expression of Defense Genes in Tomato against RKN
3.4. WRKY 33
3.5. PR1 Proteins
3.6. Lipoxygenase (LOX)
3.7. Myeloblastosis Related Proteins (MYB) TFs
3.8. Phenylalanine Ammonia Lyase (PAL)
3.9. Protein-Protein Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Tileubayeva, Z.; Avdeenko, A.; Avdeenko, S.; Stroiteleva, N.; Kondrashev, S. Plant-parasitic nematodes affecting vegetable crops in greenhouses. Saudi J. Biol. Sci. 2021, 28, 5428–5433. [Google Scholar] [CrossRef] [PubMed]
- Phani, V.; Khan, M.R.; Dutta, T.K. Plant-parasitic nematodes as a potential threat to protected agriculture: Current status and management options. Crop Prot. 2021, 144, 105573. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.; Askary, T. Impact of Phytonematodes on Agriculture Ecology, 3–49. Biocontrol Agents Phytonematodes; Askary, T.H., Martinelli, P.R.P., Eds.; CAB International Wallingford: Wallingford, UK, 2015; p. 480. [Google Scholar]
- Sikora, R.A.; Fernandez, E. Nematode parasites of vegetables. In Plant Parasitic Nematodes in Subtropical and Tropical Agriculture; CABI Publishing: Wallingford, UK, 2005; pp. 319–392. [Google Scholar]
- Hawk, T. The Effects of Seed-Applied Fluopyram on Root Penetration and Development of Meloidogyne Incognita on Cotton and Soybean; University of Arkansas: Fayetteville, AR, USA, 2019. [Google Scholar]
- Mazzetti, V.C.G.; Visintin, G.L.; Valério, I.P.; Camera, J.N.; Deuner, C.C.; Soares, P.L.M. Reaction of soybean cultivars to Meloidogyne javanica and Meloidogyne incognita. Rev. Ceres 2019, 66, 220–225. [Google Scholar] [CrossRef] [Green Version]
- Sasanelli, N.; Konrat, A.; Migunova, V.; Toderas, I.; Iurcu-Straistaru, E.; Rusu, S.; Bivol, A.; Andoni, C.; Veronico, P. Review on control methods against plant parasitic nematodes applied in southern member states (C Zone) of the European Union. Agriculture 2021, 11, 602. [Google Scholar] [CrossRef]
- Abd-Elgawad, M.M. Optimizing safe approaches to manage plant-parasitic nematodes. Plants 2021, 10, 1911. [Google Scholar] [CrossRef] [PubMed]
- Poveda, J.; Abril-Urias, P.; Escobar, C. Biological control of plant-parasitic nematodes by filamentous fungi inducers of resistance: Trichoderma, mycorrhizal and endophytic fungi. Front. Microbiol. 2020, 11, 992. [Google Scholar] [CrossRef]
- Migunova, V.D.; Tomashevich, N.S.; Konrat, A.N.; Lychagina, S.V.; Dubyaga, V.M.; D’Addabbo, T.; Sasanelli, N.; Asaturova, A.M. Selection of bacterial strains for control of root-knot disease caused by Meloidogyne incognita. Microorganisms 2021, 9, 1698. [Google Scholar] [CrossRef]
- Kerry, B.R. Rhizosphere interactions and the exploitation of microbial agents for the biological control of plant-parasitic nematodes. Annu. Rev. Phytopathol. 2000, 38, 423–441. [Google Scholar] [CrossRef] [Green Version]
- Gamalero, E.; Glick, B.R. The use of plant growth-promoting bacteria to prevent nematode damage to plants. Biology 2020, 9, 381. [Google Scholar] [CrossRef]
- Tian, X.-L.; Zhao, X.-M.; Zhao, S.-Y.; Zhao, J.-L.; Mao, Z.-C. The biocontrol functions of Bacillus velezensis strain Bv-25 against Meloidogyne incognita. Front. Microbiol. 2022, 13, 843041. [Google Scholar] [CrossRef]
- Choi, T.G.; Maung, C.E.H.; Lee, D.R.; Henry, A.B.; Lee, Y.S.; Kim, K.Y. Role of bacterial antagonists of fungal pathogens, Bacillus thuringiensis KYC and Bacillus velezensis CE 100 in control of root-knot nematode, Meloidogyne incognita and subsequent growth promotion of tomato. Biocontrol Sci. Technol. 2020, 30, 685–700. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, K.Y. Antagonistic potential of Bacillus pumilus L1 against root-Knot nematode, Meloidogyne arenaria. J. Phytopathol. 2016, 164, 29–39. [Google Scholar] [CrossRef]
- Huang, Y.; Xu, C.; Ma, L.; Zhang, K.; Duan, C.; Mo, M. Characterisation of volatiles produced from Bacillus megaterium YFM3. 25 and their nematicidal activity against Meloidogyne incognita. Eur. J. Plant Pathol. 2010, 126, 417–422. [Google Scholar] [CrossRef]
- Kavitha, P.; Jonathan, E.; Nakkeeran, S. Effects of crude antibiotic of Bacillus subtilis on hatching of eggs and mortality of juveniles of Meloidogyne incognita. Nematol. Mediterr. 2012, 40, 203–206. [Google Scholar]
- Ghahremani, Z.; Escudero, N.; Beltrán-Anadón, D.; Saus, E.; Cunquero, M.; Andilla, J.; Loza-Alvarez, P.; Gabaldón, T.; Sorribas, F.J. Bacillus firmus strain I-1582, a nematode antagonist by itself and through the plant. Front. Plant Sci. 2020, 11, 796. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Xiong, J.; Zhou, Q.; Luo, H.; Hu, S.; Xia, L.; Sun, M.; Li, L.; Yu, Z. The diverse nematicidal properties and biocontrol efficacy of Bacillus thuringiensis Cry6A against the root-knot nematode Meloidogyne hapla. J. Invertebr. Pathol. 2015, 125, 73–80. [Google Scholar] [CrossRef] [PubMed]
- Jamal, Q.; Cho, J.-Y.; Moon, J.-H.; Munir, S.; Anees, M.; Kim, K.Y. Identification for the First Time of Cyclo (d-Pro-l-Leu) Produced by Bacillus amyloliquefaciens Y1 as a Nematocide for Control of Meloidogyne incognita. Molecules 2017, 22, 1839. [Google Scholar] [CrossRef] [Green Version]
- Leonetti, P.; Zonno, M.C.; Molinari, S.; Altomare, C. Induction of SA-signaling pathway and ethylene biosynthesis in Trichoderma harzianum-treated tomato plants after infection of the root-knot nematode Meloidogyne incognita. Plant Cell Rep. 2017, 36, 621–631. [Google Scholar] [CrossRef]
- Pieterse, C.M.; Zamioudis, C.; Berendsen, R.L.; Weller, D.M.; Van Wees, S.C.; Bakker, P.A. Induced systemic resistance by beneficial microbes. Annu. Rev. Phytopathol. 2014, 52, 347–375. [Google Scholar] [CrossRef] [Green Version]
- Mou, W.; Kao, Y.-T.; Michard, E.; Simon, A.A.; Li, D.; Wudick, M.M.; Lizzio, M.A.; Feijó, J.A.; Chang, C. Ethylene-independent signaling by the ethylene precursor ACC in Arabidopsis ovular pollen tube attraction. Nat. Commun. 2020, 11, 4082. [Google Scholar] [CrossRef]
- Mbaluto, C.M.; Ahmad, E.M.; Fu, M.; Martínez-Medina, A.; van Dam, N.M. The impact of Spodoptera exigua herbivory on Meloidogyne incognita-induced root responses depends on the nematodes’ life cycle stages. AoB Plants 2020, 12, plaa029. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, L.; Birch, P. A systems biology perspective on plant–microbe interactions: Biochemical and structural targets of pathogen effectors. Plant Sci. 2011, 180, 584–603. [Google Scholar] [CrossRef] [PubMed]
- Bigeard, J.; Colcombet, J.; Hirt, H. Signaling mechanisms in pattern-triggered immunity (PTI). Mol. Plant 2015, 8, 521–539. [Google Scholar] [CrossRef] [Green Version]
- Chitwood, B. ‘Root-knot nematodes’. Part 1. A revision of the genus Meloidogyne Goeldi, 1887. Proc. Helminthol. Soc. Wash. 1949, 16, 90–114. [Google Scholar]
- Yang, Y.; Hu, X.; Liu, P.; Chen, L.; Peng, H.; Wang, Q.; Zhang, Q. A new root-knot nematode, Meloidogyne vitis sp. nov. (Nematoda: Meloidogynidae), parasitizing grape in Yunnan. PLoS ONE 2021, 16, e0245201. [Google Scholar] [CrossRef]
- Holterman, M.; Van der wurff, A.; Van den Elsen, S.; Van Megen, H.; Bongers, T.; Holovachov, O.; Bakker, J.; Helder, J. Phylum-wide analysis of ssu rdna reveals deep phylogenetic relationships among nematodes and accelerated evolution toward crown clades. Mol. Biol. Evol. 2006, 23, 1792–1800. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, K.; Dudley, J.; Nei, M.; Kumar, S. Mega4: Molecular evolutionary genetics analysis (mega) software version 4.0. Mol. Biol. Evol. 2007, 24, 1596–1599. [Google Scholar] [CrossRef]
- Vinodkumar, S.; Nakkeeran, S.; Renukadevi, P.; Mohankumar, S. Diversity and antiviral potential of rhizospheric and endophytic Bacillus species and phyto-antiviral principles against tobacco streak virus in cotton. Agric. Ecosyst. Environ. 2018, 267, 42–51. [Google Scholar]
- Vanthana, M.; Nakkeeran, S.; Malathi, V.; Renukadevi, P.; Vinodkumar, S. Induction of in planta resistance by flagellin (Flg) and elongation factor-TU (EF-Tu) of Bacillus amyloliquefaciens (VB7) against groundnut bud necrosis virus in tomato. Microb. Pathog. 2019, 137, 103757. [Google Scholar] [CrossRef]
- Chomczynski, P.; Sacchi, N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal. Biochem. 1987, 162, 156–159. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]
- Szklarczyk, D.; Gable, A.L.; Nastou, K.C.; Lyon, D.; Kirsch, R.; Pyysalo, S.; Doncheva, N.T.; Legeay, M.; Fang, T.; Bork, P. The STRING database in 2021: Customizable protein–protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021, 49, D605–D612. [Google Scholar] [CrossRef]
- Weng, J.; Wang, Y.; Li, J.; Shen, Q.; Zhang, R. Enhanced root colonization and biocontrol activity of Bacillus amyloliquefaciens SQR9 by abrB gene disruption. Appl. Microbiol. Biotechnol. 2013, 97, 8823–8830. [Google Scholar] [CrossRef]
- Alfianny, R.; Aryantha, I.; Syamsudin, T. Role of indigenous rhizosphere bacteria in suppressing root-knot nematode and improve plant growth tomato. Plant Pathol. J. 2017, 16, 25–32. [Google Scholar] [CrossRef] [Green Version]
- Mhatre, P.H.; Karthik, C.; Kadirvelu, K.; Divya, K.; Venkatasalam, E.; Srinivasan, S.; Ramkumar, G.; Saranya, C.; Shanmuganathan, R. Plant growth promoting rhizobacteria (PGPR): A potential alternative tool for nematodes bio-control. Biocatal. Agric. Biotechnol. 2019, 17, 119–128. [Google Scholar] [CrossRef]
- Sidhu, H.S. Potential of plant growth-promoting rhizobacteria in the management of nematodes: A review. J. Entomol. Zool. Stud. 2018, 6, 1536–1545. [Google Scholar]
- Soliman, G.M.; Ameen, H.H.; Abdel-Aziz, S.M.; El-Sayed, G.M. In vitro evaluation of some isolated bacteria against the plant parasite nematode Meloidogyne incognita. Bull. Natl. Res. Cent. 2019, 43, 171. [Google Scholar] [CrossRef]
- Ramalingam, K. Exploring the disease severity by the interaction of fusarium wilt and root-knot nematode in tomato. Int. J. Fauna Biol. Stud 2019, 6, 1–5. [Google Scholar]
- AYDINLI, G.; Mennan, S. Identification of root-knot nematodes (Meloidogyne spp.) fromgreenhouses in the Middle Black Sea Region of Turkey. Turk. J. Zool. 2016, 40, 675–685. [Google Scholar] [CrossRef]
- Negretti, R.R.; Gomes, C.B.; Mattos, V.S.; Somavilla, L.; Manica-Berto, R.; Agostinetto, D.; Castagnone-Sereno, P.; Carneiro, R.M. Characterisation of a Meloidogyne species complex parasitizing rice in southern Brazil. Nematology 2017, 19, 403–412. [Google Scholar] [CrossRef] [Green Version]
- Tadigiri, S.; Das, D.; Allen, R.; Vishnu, V.; Veena, S.; Karthikeyan, S. Isolation and characterization of chemical constituents from B. amyloliquefaciens and their nematicidal activity. Mortality 2020, 8, 24. [Google Scholar]
- Mendoza, A.R.; Kiewnick, S.; Sikora, R.A. In vitro activity of Bacillus firmus against the burrowing nematode Radopholus similis, the root-knot nematode Meloidogyne incognita, and the stem nematode Ditylenchus dipsaci. Biocontrol Sci. Technol. 2008, 18, 377–389. [Google Scholar] [CrossRef]
- Vetrivelkalai, P. Evaluation of endophytic bacterial isolates against root-knot nematode, Meloidogyne incognita in tomato under glasshouse condition. Int. J. Curr. Microbiol. App. Sci. 2019, 8, 2584–2589. [Google Scholar] [CrossRef]
- Ali, M.A.; Anjam, M.S.; Nawaz, M.A.; Lam, H.-M.; Chung, G. Signal transduction in plant–nematode interactions. Int. J. Mol. Sci. 2018, 19, 1648. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ayaz, M.; Ali, Q.; Farzand, A.; Khan, A.R.; Ling, H.; Gao, X. Nematicidal volatiles from Bacillus atrophaeus GBSC56 promote growth and stimulate induced systemic resistance in tomato against Meloidogyne incognita. Int. J. Mol. Sci. 2021, 22, 5049. [Google Scholar] [CrossRef]
- Yin, N.; Zhao, J.-L.; Liu, R.; Li, Y.; Ling, J.; Yang, Y.-H.; Xie, B.-Y.; Mao, Z.-C. Biocontrol efficacy of Bacillus cereus strains Bc-cm103 against Meloidogyne incognita. Plant Dis. 2021, 105, 2061–2070. [Google Scholar] [CrossRef]
- Wu, W.; Wang, J.; Wang, Z.; Guo, L.; Zhu, S.; Zhu, Y.; Wang, Y.; He, X. Rhizosphere bacteria from Panax notoginseng against Meloidogyne hapla by rapid colonization and mediated resistance. Front. Microbiol. 2022, 13, 877082. [Google Scholar] [CrossRef]
- Grunewald, W.; Karimi, M.; Wieczorek, K.; Van de Cappelle, E.; Wischnitzki, E.; Grundler, F.; Inzé, D.; Beeckman, T.; Gheysen, G. A role for AtWRKY23 in feeding site establishment of plant-parasitic nematodes. Plant Physiol. 2008, 148, 358–368. [Google Scholar] [CrossRef] [Green Version]
- Shukla, N.; Yadav, R.; Kaur, P.; Rasmussen, S.; Goel, S.; Agarwal, M.; Jagannath, A.; Gupta, R.; Kumar, A. Transcriptome analysis of root-knot nematode (Meloidogyne incognita)-infected tomato (Solanum lycopersicum) roots reveals complex gene expression profiles and metabolic networks of both host and nematode during susceptible and resistance responses. Mol. Plant Pathol. 2018, 19, 615–633. [Google Scholar] [CrossRef] [Green Version]
- Bai, Y.; Sunarti, S.; Kissoudis, C.; Visser, R.G.; van der Linden, C. The role of tomato WRKY genes in plant responses to combined abiotic and biotic stresses. Front. Plant Sci. 2018, 9, 801. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.F.; Liu, J.K.; Yang, F.M.; Zhang, G.Y.; Wang, D.; Zhang, L.; Ou, Y.B.; Yao, Y.A. The WRKY transcription factor WRKY8 promotes resistance to pathogen infection and mediates drought and salt stress tolerance in Solanum lycopersicum. Physiol. Plant. 2020, 168, 98–117. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Li, C.; Wang, H.; Guo, Z. WRKY transcription factors: Evolution, binding, and action. Phytopathol. Res. 2019, 1, 13. [Google Scholar] [CrossRef] [Green Version]
- Ali, M.A.; Wieczorek, K.; Kreil, D.P.; Bohlmann, H. The beet cyst nematode Heterodera schachtii modulates the expression of WRKY transcription factors in syncytia to favor its development in Arabidopsis roots. PLoS ONE 2014, 9, e102360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Teixeira, M.A.; Wei, L.; Kaloshian, I. Root-knot nematodes induce pattern-triggered immunity in Arabidopsis thaliana roots. New Phytol. 2016, 211, 276–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, F.; Wu, X.; Li, T.; Jia, M.; Liao, L. Characterization of the WRKY gene family in Akebia trifoliata and their response to Colletotrichum acutatum. BMC Plant Biol. 2022, 22, 115. [Google Scholar] [CrossRef] [PubMed]
- Javed, T.; Shabbir, R.; Ali, A.; Afzal, I.; Zaheer, U.; Gao, S.-J. Transcription factors in plant stress responses: Challenges and potential for sugarcane improvement. Plants 2020, 9, 491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wani, S.H.; Anand, S.; Singh, B.; Bohra, A.; Joshi, R. WRKY transcription factors and plant defense responses: Latest discoveries and future prospects. Plant Cell Rep. 2021, 40, 1071–1085. [Google Scholar] [CrossRef]
- Hu, H.; Gao, Y.; Li, X.; Chen, S.; Yan, S.; Tian, X. Identification and nematicidal characterization of proteases secreted by endophytic bacteria Bacillus cereus BCM2. Phytopathology 2020, 110, 336–344. [Google Scholar] [CrossRef]
- Chen, Y.; Jing, X.; Wang, S.; Wang, J.; Zhang, S.; Shi, Q. Genome-wide analysis of WRKY transcription factor family in melon (Cucumis melo L.) and their response to powdery mildew. Plant Mol. Biol. Report. 2021, 39, 686–699. [Google Scholar] [CrossRef]
- Topalović, O.; Bredenbruch, S.; Schleker, A.S.S.; Heuer, H. Microbes attaching to endoparasitic phytonematodes in soil trigger plant defense upon root penetration by the nematode. Front. Plant Sci. 2020, 11, 138. [Google Scholar] [CrossRef]
- Kuźniak, E.; Kopczewski, T.; Chojak-Koźniewska, J. Ascorbate-glutathione cycle and biotic stress tolerance in plants. In Ascorbic Acid in Plant Growth, Development and Stress Tolerance; Springer: Cham, Switzerland, 2017; pp. 201–231. [Google Scholar] [CrossRef]
- Vanthana, M.; Nakkeeran, S.; Malathi, V.; Renukadevi, P.; Vinodkumar, S.; Sivakumar, U.; Suganthi, A. Flagellin and elongation factor of Bacillus velezensis (VB7) reprogramme the immune response in tomato towards the management of GBNV infection. J. Virol. Methods 2022, 301, 114438. [Google Scholar] [CrossRef]
- Molinari, S.; Leonetti, P. Bio-control agents activate plant immune response and prime susceptible tomato against root-knot nematodes. PLoS ONE 2019, 14, e0213230. [Google Scholar] [CrossRef]
- Singh, R.R.; Chinnasri, B.; De Smet, L.; Haeck, A.; Demeestere, K.; Van Cutsem, P.; Van Aubel, G.; Gheysen, G.; Kyndt, T. Systemic defense activation by COS-OGA in rice against root-knot nematodes depends on stimulation of the phenylpropanoid pathway. Plant Physiol. Biochem. 2019, 142, 202–210. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghorbel, M.; Brini, F.; Sharma, A.; Landi, M. Role of jasmonic acid in plants: The molecular point of view. Plant Cell Rep. 2021, 40, 1471–1494. [Google Scholar] [CrossRef] [PubMed]
- Ambawat, S.; Sharma, P.; Yadav, N.R.; Yadav, R.C. MYB transcription factor genes as regulators for plant responses: An overview. Physiol. Mol. Biol. Plants 2013, 19, 307–321. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.-P.; Baldwin, I.T. Transport of [2-14C]jasmonic acid from leaves to roots mimics wound-induced changes in endogenous jasmonic acid pools in Nicotiana sylvestris. Planta 1997, 203, 436–441. [Google Scholar]
- Gleason, C.; Leelarasamee, N.; Meldau, D.; Feussner, I. OPDA has key role in regulating plant susceptibility to the root-knot nematode Meloidogyne hapla in Arabidopsis. Front. Plant Sci. 2016, 7, 1565. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martínez-Medina, A.; Fernandez, I.; Lok, G.B.; Pozo, M.J.; Pieterse, C.M.; Van Wees, S.C. Shifting from priming of salicylic acid-to jasmonic acid-regulated defences by Trichoderma protects tomato against the root knot nematode Meloidogyne incognita. New Phytol. 2017, 213, 1363–1377. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gattoni, K.; Xiang, N.; Lawrenace, K.; Kloepper, J. Systemic induced resistance to the root-knot nematode cause by Bacillus spp. In Proceedings of the 2018 Beltwide Cotton Conferences, San Antonio, TX, USA, 3–5 January 2018; pp. 506–510. [Google Scholar]
- Alves, M.S.; Dadalto, S.P.; Gonçalves, A.B.; de Souza, G.B.; Barros, V.A.; Fietto, L.G. Transcription factor functional protein-protein interactions in Plant defense responses. Proteomes 2014, 2, 85–106. [Google Scholar] [CrossRef] [Green Version]
- Chi, Y.; Yang, Y.; Zhou, Y.; Zhou, J.; Fan, B.; Yu, J.-Q.; Chen, Z. Protein–protein interactions in the regulation of WRKY transcription factors. Mol. Plant 2013, 6, 287–300. [Google Scholar] [CrossRef] [Green Version]
- Uehara, T.; Sugiyama, S.; Matsuura, H.; Arie, T.; Masuta, C. Resistant and susceptible responses in tomato to cyst nematode are differentially regulated by salicylic acid. Plant Cell Physiol. 2010, 51, 1524–1536. [Google Scholar] [CrossRef] [PubMed]
- Casey, R.; Hughes, R.K. Recombinant lipoxygenases and oxylipin metabolism in relation to food quality. Food Biotechnol. 2004, 18, 135–170. [Google Scholar] [CrossRef]
- Saravanan, R.; Nakkeeran, S.; Saranya, N.; Kavino, M.; Ragapriya, V.; Varanavasiappan, S.; Raveendran, M.; Krishnamoorthy, A.; Malathy, V.; Haripriya, S. Biohardening of Banana cv. Karpooravalli (ABB.; Pisang Awak) With Bacillus velezensis YEBBR6 Promotes Plant Growth and Reprograms the Innate Immune Response against Fusarium oxysporum f. sp. cubense. Front. Sustain. Food Syst. 2022, 6, 845512. [Google Scholar] [CrossRef]
- Mishra, M.; Slater, A. Recent advances in the genetic transformation of coffee. Biotechnol. Res. Int. 2012, 2012, 580857. [Google Scholar] [CrossRef] [Green Version]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kamalanathan, V.; Sevugapperumal, N.; Nallusamy, S. Antagonistic Bacteria Bacillus velezensis VB7 Possess Nematicidal Action and Induce an Immune Response to Suppress the Infection of Root-Knot Nematode (RKN) in Tomato. Genes 2023, 14, 1335. https://doi.org/10.3390/genes14071335
Kamalanathan V, Sevugapperumal N, Nallusamy S. Antagonistic Bacteria Bacillus velezensis VB7 Possess Nematicidal Action and Induce an Immune Response to Suppress the Infection of Root-Knot Nematode (RKN) in Tomato. Genes. 2023; 14(7):1335. https://doi.org/10.3390/genes14071335
Chicago/Turabian StyleKamalanathan, Vinothini, Nakkeeran Sevugapperumal, and Saranya Nallusamy. 2023. "Antagonistic Bacteria Bacillus velezensis VB7 Possess Nematicidal Action and Induce an Immune Response to Suppress the Infection of Root-Knot Nematode (RKN) in Tomato" Genes 14, no. 7: 1335. https://doi.org/10.3390/genes14071335




