Abstract
Rapeseed (Brassica napus L.) is a globally important oilseed crop with various uses, including the consumption of its succulent stems as a seasonal vegetable, but its uniaxial branching habit limits the stem yield. Therefore, developing a multi-stem rapeseed variety has become increasingly crucial. In this study, a natural mutant of the wild type (ZY511, Zhongyou511) with stable inheritance of the multi-stem trait (ms) was obtained, and it showed abnormal shoot apical meristem (SAM) development and an increased main stem number compared to the WT. Histological and scanning electron microscopy analyses revealed multiple SAMs in the ms mutant, whereas only a single SAM was found in the WT. Transcriptome analyses showed significant alterations in the expression of genes involved in cytokinin (CK) biosynthesis and metabolism pathways in the ms mutant. These findings provide insight into the mechanism of multi-main-stem formation in Brassica napus L. and lay a theoretical foundation for breeding multi-main-stem rapeseed vegetable varieties.
1. Introduction
Rapeseed is one of the world’s most important oil crops, accounting for approximately 15% of the world’s vegetable oil production [1]. As the social economy develops and consumers demand more diversified products, rapeseed is becoming increasingly versatile, serving as a vegetable, a flower, a feed for bees that produce honey, fodder, and a fertilizer [1,2]. Although the crispy and succulent bolting stem of rapeseed is a seasonal vegetable commonly consumed in South China, the stem yield is lower than that of other crops, such as Chinese flowering cabbage (Brassica campestris L. ssp. Chinensis var. utilis Tsen et Lee) and Chinese kale (Brassica alboglabra) due to its single main stem. Therefore, exploring or creating new multi-main-stem rapeseed varieties could effectively enhance vegetable yield.
Branch and stem formation in plants is facilitated by the differentiation of axillary meristems (AMs) at leaf axils, which elongate and differentiate into lateral branches [3,4]. The AM initiates from the shoot apical meristem (SAM), a group of pluripotent stem cells that generate leaf, axillary branch, flower primordium, and stem tissue [5,6]. Thus, the final number of branches or stems is determined by the number of SAMs. Phytohormones, including auxin, cytokinins, and strigolactones, play an important role in activating, initiating, and regulating plant branching [7,8]. Among these, cytokinins (CKs) have been demonstrated to be positive regulators of branching and SAM development [9,10]. CKs are small molecules derived from adenine and modified with an isoprenoid or aromatic side chain at the N6 position [11]. The active forms of CKs, which are most widespread, include four isoprenoid types known as natural isoprenoid CKs or CK nucleobases: isopentenyladenine (iP), trans-zeatin (tZ), cis-zeatin (cZ), and dihydrozeatin (DHZ) [12,13]. These CK nucleobases can undergo various conversions to form different CK conjugates. Notably, they can be transformed into CK ribosides (tZR, cZR, DZR, and iPR), CK phosphates (tZRPs, cZRPs, DZRPs, and iPRPs), CK O-glucosides (tZOG, cZOG, tZROG, and cZROG), and CK N-glucosides (tZ7G, tZ9G, iP7G, and iP9G) [14,15]. Moreover, aromatic CKs, such as benzyladenine (BA), ortho-topolin (oT), meta-topolin (mT), and their methoxy-derivatives (meoT and memT), have gained significant attention in bioassays due to their remarkable biological activity [16,17].
Nishimura et al. (2004) used the Arabidopsis ahk2 ahk3 ahk4 triple CK receptor mutant and revealed that CKs function in the maintenance of SAM activity [18]. In Arabidopsis, the supershoot (sps) mutant forms multiple AMs in the leaf axils due to increased CK levels, promoting branch formation from rosette and cauline leaves [19]. Moreover, the loss of the negative feedback regulator of CK, type-A ARR homolog ABPH1, results in an increase in SAM size in maize [20]. Undoubtedly, cytokinin (CK) signaling plays a pivotal role in the regulation of WUSCHEL (WUS), a key regulator controlling SAM activity [21]. The induction of WUS transcription by CK leads to its upregulation within the SAM [22]. In Arabidopsis, the type-B Arabidopsis Response Regulators (ARRs), including ARR1, play a direct role in binding to the promoter region of WUS, thereby facilitating its transcriptional activation [23,24]. Furthermore, type-B ARRs have the ability to activate the expression of type-A ARRs, such as ARR7 and ARR15, which act as negative regulators of CK signaling [25]. Notably, WUS forms an additional positive feedback loop with CK signaling by directly suppressing the transcription of several type-A ARRs [26]. This intricate interplay between CK and WUS ensures a precise balance between the maintenance of stem cells and their differentiation during plant development.
The biosynthesis and catabolism of CK are dynamically balanced processes in plants. Cytokinin oxidase (CKX) can irreversibly cleave active CKs, decreasing cytokinin levels [27]. CKX family genes have been identified in many plant species, such as Arabidopsis thaliana [28], Nicotiana tabacum [29], Zea mays [30], Oryza sativa [31], Triticum aestivum [32], and Glycine max [33]. In rapeseed, 23 CKX genes were identified [34]. CKX has been found to play a role in SAM development and plant branching. Werner et al. (2003) found that the Arabidopsis CKX gene family has seven members (AtCKX1 to AtCKX7), and transgenic plants overexpressing CKX1 and CKX3 showed decreased SAM activity [28]. LONELY GUY (LOG) encodes a cytokinin riboside 5′-monophosphate phosphoribohydrolase that functions in the final step of bioactive CK synthesis; the analysis of a rice (Oryza sativa) mutant in which lonely guy (log) was mutated showed that it is important for the termination of shoot meristems [35]. In Arabidopsis, nine LOG genes (AtLOG1 to AtLOG9) were predicted to be homologs of rice LOG [36]. Tokunaga et al. (2011) used a multiple Atlog mutant and found that the LOG genes were important for the maintenance of the SAM in Arabidopsis [37].
Multi-main-stem (MMS) traits have been discovered in rice and wheat, which improve the growth potential and seed number and are therefore important for yield improvement [38,39]. Zhao et al. (2019) identified six candidate genes (RPT2A, HLR, CRK, LRR-RLK, AGL79, and TCTP) involved in SAM differentiation and axillary bud formation by QTL mapping and the gene-fishing technique, which were related to the formation of the MMS phenotype in rapeseed [40]. However, the mechanism underlying MMS formation and the relationship between SAM and MMS formation remain unclear. In this study, we identified a rapeseed mutant with MMS from ZY511 (WT, Zhongyou511), and after six consecutive generations with self-selection, we generated lines with stable inheritance of the multiple main branching trait. The mutant, named Multiple Stem (ms) rapeseed, develops over six main stems from the base of the plant, with small branching angles. Our results showed that the development of SAM in the ms mutant was abnormal compared to that in the WT. Furthermore, through transcriptome analysis conducted at various stages of germination, we discovered that the aberrant development of the SAM in the ms mutant initiates at 20 days after germination (DAG). Notably, within the SAM of the ms mutant, we observed significant alterations in the expression of genes associated with the CK signaling pathway. These findings establish a solid foundation for unraveling the molecular mechanisms underlying the formation of multiple main stems and provide reference for the selection and breeding of rapeseed varieties with multiple main stems.
2. Methods and Materials
2.1. Plant Materials and Growth Conditions
Wild-type and ms mutant seeds were sterilized in 75% ethanol for 15 min and then rinsed repeatedly in sterile water. The seeds were germinated on Petri dishes containing Murashige Skoog (MS) medium (see Table S1 for the recipe) in an incubator at 25 °C and were sown in culture bottles once they germinated. The bottles were placed in a light incubator with 14 h of light, 10 h of dark, a day temperature of 22 °C, a night temperature of 18 °C, and a relative humidity of 60%, with a light intensity of 10,000 lux.
2.2. Tissue Section
To prepare paraffin sections, the SAM tissues of wild-type and ms mutant plants at 20 days after germination (DAG) were cut and then placed in a stationary formaldehyde acetic acid alcohol (FAA) solution for 24 h, which was performed by Wuhan Servicebio Technology Co., Ltd. (Wuhan, China). Please refer to [41,42] for further details. The specific steps are described below.
2.2.1. Paraffin Section Preparation
The tissue was removed from the fixing solution, the target tissue was trimmed using a scalpel in a ventilated cupboard, and the trimmed tissue was labeled and put in a dehydration box.
The dehydration box was placed in a dehydrator (Diapath Donatello, Milan, Italy) for dehydration with an alcohol gradient: 75% alcohol for 4 h, 85% alcohol for 2 h, 90% alcohol for 2 h, 95% alcohol for 1 h, anhydrous ethanol for 1 h, alcohol benzene for 5~10 min, xylene for 10~20 min, and 65 °C molten paraffin for 3 h.
The wax-soaked tissue was embedded in an embedding machine (WHJJ JB-P5, Wuhan, China). First, the melted wax was placed in the embedding frame, and before the wax solidified, the tissue was removed from the dewatering box and put into the embedding frame according to the requirements of the embedding surface, and the corresponding label was affixed. This was then cooled at −20 °C on a freezing table (WHJJ JB-L5, Wuhan, China), and after the wax had solidified, the wax block was removed from the embedding frame and repaired by using a scalpel. This process ensured the production of clean and precise sections, which were suitable for subsequent experiments.
The trimmed wax block was placed on a −20 °C freezing table, and the modified tissue chip wax block was sliced at a thickness 4 μm using a paraffin slicer (Leica RM2016, Wetzlar, Germany). The slice of tissue was floated on 40 °C warm water in a spreading machine to flatten it (KEDEE KD-P, Jinhua, China), and the tissue was then picked up using glass slides and baked in an oven at 60 °C. After the water-baked dried wax had melted, it was taken out and stored at room temperature.
2.2.2. Paraffin Section Safranin O-Fast Green Staining
The sections were rehydrated in BioDewax and Clear Solution (Servicebio, Wuhan, China) for 40 min, 100% ethanol for 10 min, and 75% ethanol for 5 min. Finally, they were rinsed under running water.
The sections were placed in safranin O staining solution for 2 h, and then into 50%, 70%, and 80% ethanol, each for 3~8 s. The sections were then put into Fast Green staining solution (Servicebio, Wuhan, China) for 6~20 s, and dehydrated by washing three times with 100% ethanol for 5 min each. Finally, the tissue sections were mounted using neutral balsam. They were observed and photographed using an ECHO microscope (Revolve FL, San Diego, CA, USA).
2.3. Scanning Electron Microscopy
The SAM samples of 20 DAG wild-type and ms mutant plants were used for scanning electron microscopy (SEM) analysis, which was performed by Wuhan Servicebio Technology CO., Ltd. (Wuhan, China), using a Hitachi SU8100 SEM. The procedures are detailed below.
The target fresh tissues were selected in a manner that minimized mechanical damage such as pulling, contusion, and extrusion. A sharp blade was used to quickly cut and harvest the fresh tissue blocks, within 1–3 min. The area of the tissue block was to be no more than 3 mm2. The tissues were then gently washed with PBS. The target side of the tissue was labeled (the side that was to be observed). Care was taken to protect the tissue blocks, especially the target side, from mechanical damage such as extrusion with the forceps. The washed tissue blocks were immediately fixed using an electron microscopy fixative (Servicebio, Wuhan, China) for 2 h at room temperature, and then transferred into 4 °C storage for preservation and transportation.
Tissue blocks were washed with 0.1 M PB (pH 7.4) 3 times, for 15 min each time. Then, the tissue blocks were transferred into 1% OsO4 in 0.1 M PB (pH 7.4) for incubation for 1–2 h at room temperature. After that, the tissue blocks were washed in 0.1 M PB (pH 7.4) 3 times, for 15 min each time.
The samples were dehydrated as follows: 30% ethanol for 15 min; 50% ethanol for 15 min; 70% ethanol for 15 min; 80% ethanol for 15 min; 90% ethanol for 15 min; 95% ethanol for 15 min; two changes of 100% ethanol for 15 min; and isoamyl acetate for 15 min.
The samples were dried using a critical point dryer (Quorum K850, Rockville, ML, USA). The specimens were attached to metallic stubs using carbon stickers and sputter-coated (Hitachi MC1000, Tokyo, Japan) with gold for 30 s. They were observed and imaged using a scanning electron microscope (Hitachi SU8100, Tokyo, Japan).
2.4. Quantification of Cytokinins
The cytokinins in the SAMs of the wild-type and ms mutant plants at 20 DAG were quantified using HPLC-MS/MS (liquid chromatography–mass spectrometry) by Shanghai Applied Protein Technology Co., Ltd. (Shanghai, China), following a previously described method [43]. The SAMs from five individual replicate plants were pooled as one replicate, n = 3. The specific steps were as follows.
2.4.1. Sample Preparation
The samples were taken out at −80 °C. After grinding with liquid nitrogen, 100 mg samples were weighed, and 1170 µL of an acetonitrile (CAN)/water/formic acid (FA) solution was added (80:19:1, v/v). Then, 20 µL of Internal Standard (IS) was added, and the mixture was vortexed for 60 s, exposed to ultrasound at low temperature in the dark for 25 min, left to stand at −20 °C overnight, and centrifuged at 14,000 rcf at 4 °C for 20 min. Subsequently, 900 µL of the supernatant was placed in a 25 mg 96-well descaling plate for positive pressure filtration. Then, 200 µL of an ACN/water/FA solution (80:19:1, v/v) was added for elution. The supernatant was dried in liquid nitrogen. For LC-MS analysis, the samples were redissolved in 200 μL of MeOH/water (1:1, v/v), adequately vortexed, and then centrifuged (14,000 rcf, 4 °C, 15 min). The supernatants were collected for LC-MS/MS analysis.
2.4.2. HPLC-MS/MS Analysis
HPLC analysis: Analyses were performed using an UHPLC (1290 Infinity LC, Agilent Technologies, Santa Clara, CA, USA) coupled to a QTRAP (AB Sciex 5500, Sciex, Framingham, MA, USA). The mobile phase contained A: 0.05% FA in water and B: 0.05% FA in ACN. The samples were placed in the automatic sampler at 4 °C, and the column temperatures were kept constant at 45 °C. The gradients were at a flow rate of 400 µL/min, and a 4 µL aliquot of each sample was injected. Gradient B changed from 2% to 10% over 0–1 min, increased to 70% over 1–10 min, and then increased to 95% in 1 min; then, the proportion of B was reduced to 2% for 0.1 min and kept constant for 11–13 min. The QC samples used for testing and evaluating the stability and repeatability of this system, at the same time, set the standard mixture of metabolites, were also used for the correction of the chromatographic retention time.
MS/MS analysis (MRM): In ESI positive mode, the conditions were set as follows: source temperature: 550 °C; Ion Source Gas1 (Gas1): 55; Ion Source Gas2 (Gas2): 50; Curtain gas (CUR): 30; ionSapary Voltage Floating (ISVF): +4500 V. In ESI negative mode, the conditions were set as follows: source temperature: 550 °C; Ion Source Gas1 (Gas1): 55; Ion Source Gas2 (Gas2): 50; Curtain Gas (CUR): 30; ionSapary Voltage Floating (ISVF): −4500 V. The MRM mode detection ion pair was adopted.
2.5. RNA Extraction, Sequencing, and Analysis
The SAMs of wild-type and ms mutant plants at 15, 20, 25, 30, and 35 DAG were collected, and samples from five individual replicate plants were pooled as one replicate, n = 3. Samples were stored at −80 °C until use. Total RNA extraction and transcriptome sequencing were conducted as previously described. Briefly, total RNA was extracted using TRIZOL® Reagent (TRAN, Beijing, China) according to the manufacturer’s protocol and quality-checked using the QubitTM4 Fluorometer microvolume spectrophotometer (Thermo Fisher Scientific, Singapore). cDNA libraries were then prepared and sequenced on the Illumina HiSeq4000 sequencing platform by Lian Chuan Biotechnology Co., Ltd. (Hangzhou, China). Quality control for the raw RNA-seq data from the machine was performed using the fastQC v0.11.2 software. Low-quality reads and adapter sequences were deleted using Trimmomatic (0.36.5) to acquire clean, high-quality reads [44]. The obtained clean reads were mapped to the published reference genome (http://www.genoscope.cns.fr/brassicanapus/, 15 January 2022). StringTie (1.3.4) was employed to count the number of reads mapped onto each gene, and gene expression was quantified as the number of fragments per kilobase of the transcript sequence per million base pairs (FPKM). Differential expression analysis was performed using the DESeq2R package (2.11.38).
Transcripts with p values < 0.05 and |log2(fold change)| ≥ 1 were considered differentially expressed genes (DEGs). Pearson correlation analysis, principal component analysis (PCA), and Venn diagram analysis and graphing were performed using the OmicShare tools (https://www.omicshare.com/tools, accessed on 18 February 2022). GO enrichment analysis was performed using AgriGO (http://systemsbiology.cau.edu.cn/agriGOv2/index.php, accessed on 19 February 2022). KEGG analysis was conducted with reference to the Kyoto Encyclopedia of Genes and Genomes (https://www.genome.jp/kegg/, accessed on 20 February 2022) database, and KOBAS was used to detect the statistically significant enrichment of DEGs in the KEGG pathway. TBtools was employed to construct heatmaps of the transcriptome [45].
2.6. Real-Time Quantitative PCR (RT-qPCR)
To verify the reliability of the transcriptomic RNA-seq data, we randomly selected some genes for RT-qPCR validation at different developmental stages. For the gene expression analysis, first-strand cDNA was synthesized from 1 µg of total RNA for each sample using the TransScript® II All-in-One First-Strand cDNA Synthesis SuperMix for qPCR Kit (TRAN, Beijing, China). qRT-PCR was performed in a 96-well plate on a CFX96 Touch Real-Time PCR system (Bio-RAD, Hercules, CA, USA) using the TransStart® Green qPCR SuperMix (TRAN, Beijing, China). The thermal conditions were 95 °C for 30 s, followed by 39 cycles of 95 °C for 15 s, 60 °C for 30 s, and 72 °C for 15 s. Then, gene-specific primers were designed based on multiple sequence alignment. The relative expression levels of the selected DEGs were calculated using the 2−∆∆CT method [46]; specific information about the primers used in this study is shown in Table S7. Actin 7 [47] was used as an internal reference control.
2.7. Statistical Analysis
The data were analyzed and graphed using SPSS version 18.0. An independent-sample two-tailed Student’s t-test was used to analyze the significant of differences between the wild type and the ms mutant.
3. Results
3.1. Phenotype Investigation of MS Mutant
In this study, we report the discovery of a natural mutant in the WT rapeseed background (ZY511, Zhongyou511) that exhibits a remarkable phenotype. A homozygote of the mutant was obtained by self-crossing for six generations. Unlike the WT, which develops a single main stem, the mutant produces more than two branches, typically six to seven (Figure 1A,B). Importantly, the branches in the ms mutant were formed from the base of the plant. Additionally, the branching architecture of the ms mutant is characterized by small branch angles and a compact structure (Figure 1C,D). Given its distinctive phenotype, this mutant was named ms (multi-main-stem) rapeseed. To ensure the stable inheritance of this trait, we self-crossed the mutant for several generations to obtain homozygotes.
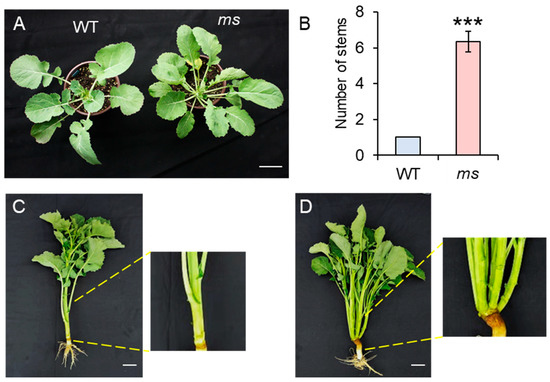
Figure 1.
(A) The phenotypes of wild-type (WT) and multi-stem (ms) mutants after germination at 60 days. Bar = 5 cm. (B) The numbers of the main stems of the WT and ms mutant. Values are the means (±SDs) (two-tailed Student’s t-tests; *** p < 0.001; n = 10). (C) The phenotypes of the WT and (D) ms mutant after germination at 120 days. Bar = 10 cm. The yellow lines indicate the enlargement of the base of the stem in the WT and ms mutant plants.
3.2. Abnormal Development of the SAM in MS Mutant
At 20 days after germination (DAG), two axillary buds had emerged in the WT (Figure 2A,B), whereas the ms mutant had multiple axillary buds at the base of the plant (Figure 2A,C). Thus, the number of main stems in the ms mutant significantly exceeded that of the WT, starting at 20 DAG. Shoot branching arises from the AMs in the axils of leaves, which are initiated by the SAMs [4]. Given that the ms mutant exhibited an increase in main stems, it was possible that SAM initiation was affected. To test this hypothesis, paraffin sectioning and scanning electron microscopy analysis of SAMs from the WT and ms mutant at 20 DAG were performed. The SAMs of WT exhibited a regular protuberance (Figure 2D,F). In contrast, the SAM in ms mutant exhibited an irregular shape and more than one SAM (Figure 2E,G). These results indicate a higher activity of the SAM in the ms mutant, which leads to the generation of multiple main stems.
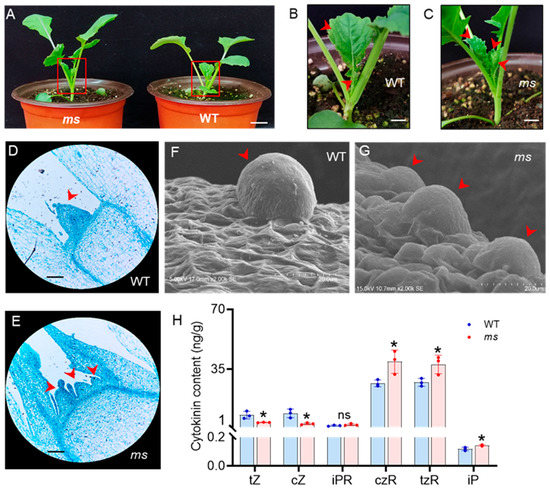
Figure 2.
The development of the shoot apical meristem in the WT and ms mutant. (A) Phenotypes of the wild-type (WT) and multi-stem (ms) mutant plants after germination at 20 days; bar = 2 cm. (B) Close-up views of the axillary buds (red arrowheads) in the WT and (C) ms mutant are shown in the red panels in (A), respectively; bar = 0.5 cm. (D) Paraffin section of the shoot apical meristems (SAMs, red arrowheads) of the WT and (E) ms mutants after germination at 20 days; ×100 magnification; bar = 100 μm; n = 10. (F) Scanned images of the SAMs (red arrowheads) in the WT (voltage: 5 kV; magnification: ×2000; and cross-section: 17 mm) and (G) ms mutant after germination at 20 days (voltage: 15 kV; magnification: ×2000; and cross-section: 10.7 mm); bar = 20 µm; n = 10. (H) The cytokinin contents in the SAMs of the WT and ms mutants after germination at 20 days. tZ: trans-zeatin; cZ: cis-zeatin; iPR: isopentenyladenine riboside; czR: cis-zeatin riboside; tzR: trans-zeatin riboside; iP: isopentenyladenine. Values are the means (±SDs). Significant differences between the WT and ms mutant are indicated (two-tailed Student’s t-tests; * p < 0.05; NS, not significant; n = 3).
3.3. Changes in Content of CKs in the SAM of the ms Mutant
CKs positively regulate the development of SAMs [9]. Specifically, we investigated the CK levels in the SAMs of both WT and ms mutant plants at 20 DAG. HPLC-MS analysis revealed that the levels of CKs were altered in the ms mutant compared to the WT. The most significant difference was observed in the levels of cis-zeatin riboside (czR) and trans-zeatin riboside (tzR) (Figure 2H). Among the active forms of CK, only isopentenyladenine (iP) was slightly increased in the ms mutant, whereas trans-zeatin (tZ) and cis-zeatin (cZ) were decreased. Compared to in the WT, the levels of the iP, CK ribosides czR and tzR were increased in the ms mutant by 21%, 47%, and 38%, respectively (Figure 2H). However, there was no significant change in the level of the nucleoside form of iP (iPR) in the ms mutant.
3.4. RNA-seq Analysis of the SAM in MS Mutant
To further elucidate the mechanism of the development of SAMs in the ms mutant, transcriptomic analysis was performed. We selected the SAMs of the WT and mutant at five developmental stages (15, 20, 25, 30, and 35 DAG) for transcriptome sequencing. First, we analyzed the reliability of the RNA-seq data. By using the FPKM values of the average of three replicates of each sample, we plotted a heatmap of the correlation between samples (Figure 3A), which showed that the coefficient of the correlation between each sample was above 0.8, suggesting high data consistency. Additionally, principal component analysis (PCA) of the samples showed that the replicates exhibited good consistency among different samples, meeting the requirements for further data analysis (Figure 3B).
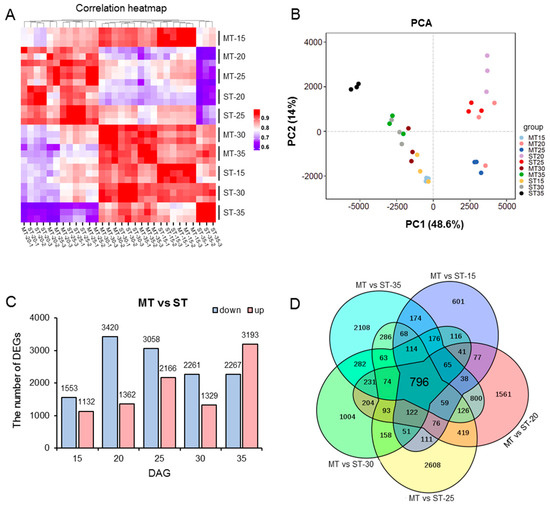
Figure 3.
Global transcript analysis of WT compared with ms mutant. The SAMs of the WT and ms mutant were collected at 15, 20, 25, 30, and 35 days after germination (DAG). (A) Heatmap of Pearson correlation between samples according to gene expression values. (B) Principal component analysis (PCA) of the 10 groups of transcriptome data. (C) The number of differentially expressed genes (DEGs) between the ms mutant (M) and WT (S) at different DAG. (D) Venn diagrams indicating the numbers of common and specific DEGs identified between the ms mutant (M) and WT (S) at different DAG. ST: time after germination of single-stem plants (WT); MT: time after germination of multi-stem (ms) mutants. Complete data can be found in Table S2.
3.5. Differential Transcriptional in Different Developmental Stages in the SAM of MS Mutant
The selection criteria for differentially expressed genes (DEGs) were set as follows: p value < 0.05, and |log2(fold change) | ≥ 1. The transcriptome of the SAMs of the ms mutant was compared with that of the WT at five developmental stages, and DEGs were identified. At 15 DAG, a total of 2685 DEGs were identified, including 1553 genes that were downregulated and 1132 genes that were upregulated. At 20 DAG, a total of 4782 DEGs were identified, of which 3420 were downregulated and 1362 were upregulated. At 25 DAG, a total of 5224 DEGs were identified, with 3058 downregulated genes and 2166 upregulated genes. At 30 DAG, a total of 3590 DEGs were identified, with 2261 downregulated genes and 1329 upregulated genes. At 35 DAG, 5460 DEGs were identified, of which 2269 were downregulated and 3193 were upregulated (Figure 3C, Table S2). In addition, Venn diagram analysis of DEGs across these five developmental stages revealed that a total of 796 core genes were commonly regulated in both the WT and ms mutant (Figure 3D).
3.6. GO and KEGG Analysis of All DEGs
To further elucidate the functions of DEGs between the ms mutant and WT, Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were performed. For DEGs identified at 15 DAG, we observed 50 significantly enriched GO terms, with most DEGs being enriched in the molecular function “binding”, the cellular components “cell wall” and “apoplast”, and the biological process “translation” (Figure S1, Table S3). Similarly, 48, 50, 50, and 49 significant GO terms were identified for DEGs at 20, 25, 30, and 35 DAG, respectively. Most DEGs were involved in the molecular function “DNA-binding transcription factor activity” (20 and 35 DAG), the biological process “response to chitin” (20, 30, and 35 DAG), “response to water deprivation” (25 DAG), and the cellular component “ubiquitin ligase complex” (20 DAG) and “apoplast” (25, 30, and 35 DAG) (Figures S2–S5; Table S3).
Furthermore, we identified the top 10 KEGG pathways for DEGs at 15, 20, 25, 30, and 35 DAG. The metabolism of amino acids, such as “β-alanine metabolism”, “valine, leucine, and isoleucine degradation”, and “alanine, aspartate and glutamate metabolism”, was significantly enriched, suggesting the potential involvement of these amino acids in the development of the ms mutant’s SAM. Notably, “plant hormone signal transduction” was enriched from the DEGs at 20, 30, and 35 DAG, indicating that the DEGs involved in this pathway also participate in the formation of multiple main stems in the ms mutant (Figure 4, Table S4).
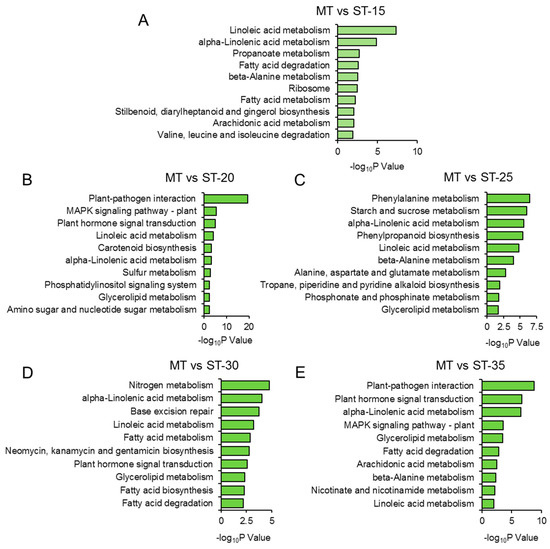
Figure 4.
KEGG enrichment analysis of DEGs between ms mutant and WT. Top 10 KEGG pathways of DEGs in ms mutant compared with WT at (A) 15, (B) 20, (C) 25, (D) 30, and (E) 35 days after germination. ST: time after germination of single-stem plants (WT); MT: time after germination of multi-stem (ms) mutants. Complete data can be found in Table S4.
3.7. The Expression of CK-Related Genes Was Affected in the SAM of MS Mutant
Given the crucial role of CKs in regulating the development of SAMs, we focused on the transcriptional levels of genes involved in CK synthesis and metabolism pathways. The LONELY GUY (LOG) are CK-activating enzymes involved in CK synthesis, and the degradation of CK is performed by cytokinin oxidase/dehydrogenase (CKX) (Figure 5A). In this study, we detected 9 LOG genes (LOC106396898 (LOG1), LOC106352601 (LOG1), LOC106446715 (LOG1), LOC106356714 (LOG4), LOC106375311 (LOG5), LOC106453249 (LOG5), LOC106384114 (LOG7), LOC106440364 (LOG8), and LOC106364020 (LOG8)) and CKX (LOC106415860 (CKX6), LOC106406332 (CKX6), LOC106384861 (CKX7), LOC106406751 (CKX7), and LOC10636555 (CKX7)) with different expression in the ms mutant at different DAGs.
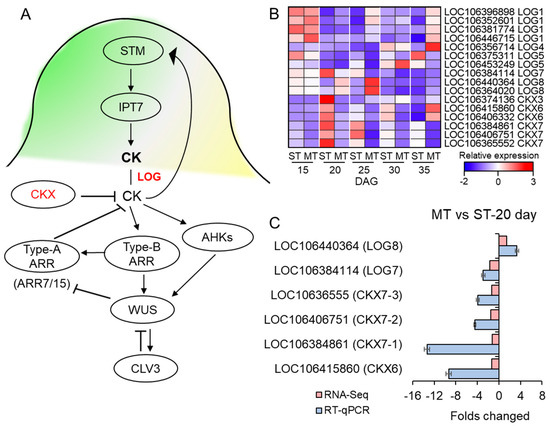
Figure 5.
The expression of genes involved in cytokinin signaling. (A) The gene networks involved in the cytokinin (CK) signaling pathway. LOG positively regulates CK synthesis, and CKX negatively regulates the synthesis of CK. CK signaling positively regulates WUS in a manner that is dependent on type-B ARRs; type-B ARRs directly activate type-A ARRs, which act as negative-feedback regulators of CK signaling, and WUS represses type-A ARRs (ARR7/15). CK also positively regulates WUS transcription via cytokinin receptors called AHKs. Adapted from [48]. (B) Heatmap of the LOG and CKX gene family in WT (S) and ms mutant (M) at 15, 20, 25, 30, and 35 days after germination (DAG). The colors correspond to the average FPKMs of three replicates, ranging from blue (low expression) to red (high expression). Complete data can be found in Table S5. (C) RT-qPCR and the RNA-seq indicated the relative expression (fold change) of the LOG and CKX genes in the ms mutant (M) relative to the WT (S) at 20 days after germination. ST: time after germination of single-stem plants (WT), MT: time after germination of multi-main-stem (ms) mutants.
During the 15 DAG, before the formation of multiple stems in the ms mutant, there was no significant change in the expression of the LOG and CKX genes in the ms mutant compared to the WT. However, during the 20 DAG when the multiple stems of the ms mutant started to form, we found that the expression of LOG8 was upregulated in the mutant but not in the WT. Conversely, the transcript levels of CKX6 and CKX7 were downregulated in the ms mutant but upregulated in the WT. The same trends were observed for these genes at 25 DAG. However, at 30 and 35 DAG, the expression of the LOG and CKX genes showed different patterns (Figure 5B, Table S5). Moreover, we observed a significant upregulation of IPT9, a key gene involved in cytokinin (CK) biosynthesis [49], at 20 DAG (Figure S6A). In terms of CK signal transduction, several members of the type-A ARABIDOPSIS REGULATOR (ARR) family, including ARR3, ARR4, ARR6, ARR8, ARR16, and ARR17, were also found to be significantly upregulated in the ms mutant at 20 DAG (Figure S6D, Table S6). These findings, combined with our previous investigations with regard to paraffin sections, CK content, and KEGG enrichment analysis, provide compelling evidence that the initiation of multi-main-stem formation in the ms mutant occurs primarily at 20 DAG.
Therefore, following 20 DAG, we selected LOG7 (LOC106384114), LOG8 (LOC106440364), CKX6 (LOC106415860), CKX7-1 (LOC106384861), CKX7-2 (LOC106406751), and CKX7-3 (LOC10636555) for RT-qPCR validation and further analysis. Both the transcriptome and RT-qPCR results showed that the expression of LOG8 was upregulated in the ms mutant compared to the WT, whereas there was a downregulation of the expression of LOG7 (Figure 5C). In contrast, the expression of CKX6, CKX7-1, CKX7-2, and CKX7-3 was downregulated in the ms mutant relative to the WT (Figure 5C). Enzymes belonging to the LOG family play a crucial role in converting iPRMP (isopentenyladenine riboside 5’-monophosphate) into its active CK form, iP [35]. The catalysis of iP is carried out by CKXs [10]. Hence, the substantial downregulation of CKX genes and the concurrent upregulation of LOG8 in the ms mutant have the potential to trigger an elevation in the levels of active cytokinin (iP) content.
4. Discussion
Multiple main stem formation is a crucial determinant of stem architecture in rapeseed and plays an essential role in improving vegetable yield. Despite its importance, the mechanism underlying the formation of multiple main stems in rapeseed remains largely unknown. In this study, we identified the ms mutant that exhibits 6–7 main stems (Figure 1). We hypothesized that the increased main branching was caused by abnormal development of the SAM. Further investigation revealed that the ms mutant had a greater number of SAMs with an irregular shape and increased axillary buds at the base than the WT at 20 DAG (Figure 2A–G). These results provide evidence that the increased number of main stems in the ms mutant is a result of an increased number of SAMs.
As the development of SAMs in the ms mutant was distinct from that of the WT by 20 DAG, we chose to perform transcriptome analysis at different time points, including the early germination stage (15 DAG), the day when SAM development started to produce abnormalities (20 DAG), and later time points (25, 30, and 35 DAG), to gain a comprehensive understanding of the SAM development in the ms mutant compared to the WT. At 15 DAG, there were a total of 2685 DEGs, much lower than the number observed at 20 DAG (4782), 25 DAG (5224), 30 DAG (3590), and 35 DAG (5460) (Figure 3C, Table S2). Therefore, it is reasonable to conclude that, at least by 20 DAG, the development and gene expression of SAM in the ms mutant begin to undergo significant changes compared to the WT, which ultimately lead to the formation of multiple main stems at 60 DAG and 120 DAG (Figure 1).
Furthermore, we found that the “linoleic acid metabolism” and “α-linoleic acid metabolism” pathways were enriched in the ms mutant at 20, 25, 30, and 35 DAG (Figure 4). The jasmonic acid (JA) synthesis and signaling pathways are widely recognized for their pivotal role in plant defense against herbivory [50,51]. Therefore, the perturbation of these pathways in the ms mutant suggests a potential impact on its resistance to insects. Additionally, we observed that the “plant hormone signal transduction” pathway was enriched at least three time points (20, 30, and 35 DAG), suggesting that phytohormones also play a crucial role in the development of SAMs in the ms mutant (Figure 4).
Zhu et al. (2019) made a significant discovery regarding the role of CKs as critical regulators of SAM development in the dt (dou tou) mutant of Brassica napus L., which displayed an increased number of main stems. They found that the SAM of the dt mutant exhibited elevated levels of active CKs, including tZ and iP, as well as CK ribosides such as tzR and iPR. Furthermore, the levels of DHZR (dihydrozeatin riboside) and IP7G (N6-(Δ2-isopentenyl) adenosine-7-β-D-glucoside) were also increased in the SAMs of the dt mutant [52]. However, in our study, we observed modestly elevated levels of active CK (iP) and CK ribosides (czR and tzR) in the SAMs of the ms mutant compared to the WT (Figure 2H), albeit with differences from the dt mutant.
Through transcriptome and RT-qPCR analysis, we have revealed intriguing insights into the molecular mechanisms underlying the differences shown by the ms mutant. Notably, we observed an elevation in the expression of the CK synthesis gene LOG8, whereas the CK degradation gene CKX6 and three CKX7s were downregulated in the ms mutant compared to the WT (Figure 5B,C). In addition to LOG and CKX, we also investigated the transcript levels of IPTs and genes involved in CK signal transduction in the ms mutant. Interestingly, we found minimal significant changes in gene expression at 15 DAG between the ms mutant and WT. Furthermore, the expression levels of CK receptor AHKs [53], AHPs [54], and type-B ARRs [55] exhibited relatively limited alterations in the ms mutant across all DAG (Figure S6B,C,E). However, at 20 DAG, a considerable number of genes, particularly type-A ARRs, were significantly upregulated in the ms mutant (Figure S6D). This observation aligns with our hypothesis that the alteration in the SAM of the ms mutant commenced at 20 DAG. Surprisingly, at 35 DAG, many type-A ARRs exhibited upregulation, whereas some showed slight downregulation at 30 DAG (Figure S6D). These findings appear contrary to the expected role of type-A ARR proteins as negative regulators of CK signaling and SAM function. Nevertheless, previous research by Müller et al. (2015) demonstrated that the hextuple type-A arr3,4,5,6,7,15 mutant of Arabidopsis displayed reduced rosette branching and bud activation compared to the WT [56]. Therefore, further investigation is warranted to elucidate the role of CK in SAM development and to determine whether type-A ARRs are involved in regulating the formation of multi-main stems in the ms mutant.
In addition to CK, shoot branching is also regulated by auxin and strigolactones (SLs) [57,58,59,60,61]. Previous studies have shown that CK, auxin, and SL signals can interact with each other [7,62,63]. More recently, Tang et al. (2020) identified a new high-density pod mutant of Brassica napus that displayed multiple stems and higher levels of indole-3-acetic acid (IAA) in the SAM [64]. In this study, we found that at 20 DAG, the GO enrichment analysis revealed that 45 upregulated and 12 downregulated DEGs in the ms mutant were involved in the “auxin-activated signaling pathway”, and “response to auxin” was among the pathways enriched from the 2 upregulated and 43 down-regulated DEGs in the ms mutant (Figure S2, Table S3), indicating that the auxin pathway might be involved in the formation of multiple main stems in the ms mutant.
In our investigation, we focused on CK signaling and observed a slight increase in iP content in the SAM of ms mutant, but no significant changes in other active CK types. This observation does not strongly support the hypothesis that CK regulates SAM development and shoot branching in the ms mutant. Hence, it is imperative to delve into the alterations in other phytohormones within the ms mutant in future investigations. Furthermore, determining whether auxin or other phytohormones play a role in regulating branching in the ms mutant through CK signaling necessitates further exploration.
Besides phytohormones, sugar also promotes cell division, and it plays a role in plant branching. Salam et al. (2017) reported that the branching of potato (Solanum tuberosum) was induced by the application of an exogenous sugar (sucrose, fructose, or glucose) [65]. Our KEGG analysis revealed that, at 20 and 25 DAG, when the SAM was abnormal in the ms mutant, the DEGs between the ms and WT were enriched in the “amino sugar and nucleotide sugar metabolism” and “starch and sucrose metabolism” pathways (Figure 4B,C, Table S4), suggesting that the sugar signaling may be involved in the formation of multiple main stems in the ms mutant. Thus, shoot branching is a complex quantitative trait that is under polygenic control and is regulated by phytohormones and external environmental factors; much research on CK, SLs, auxin, and sugar is needed to fully understand the increased branching in the ms mutant.
The identification of a mutation site in a newly discovered natural mutant typically requires the use of various tools and approaches. One of the most powerful tools in identifying loci or key genes associated with agronomic traits is the genome-wide association study (GWAS) [66,67]. In rapeseed, as with many other crop plants, most agronomic traits are quantitatively inherited and controlled by quantitative trait loci (QTLs) [68,69,70,71]. QTLs related to branch number have been identified in rapeseed [72,73,74,75]. Based on GWAS, numerous loci related to branch number have been identified on almost all rapeseed’s chromosomes, including A01, A03, A07, C04, C07, and C09 [76,77,78,79,80]. In addition to GWAS, Li et al. (2020) identified a major QTL related to branching (shoot branching 9, qSB.A09) on the chromosome A09 in Brassica rapa L. ssp. Chinensis by integrating QTL mapping with BSA-seq (bulked segregant analysis) [81]. Given that 43 QTLs were identified for MMS in rapeseed, and six candidate genes related to the formation of MMS were obtained from QTL mapping and using the gene-fishing technique [40], it will be worthwhile to explore the major QTLs or candidate genes associated with multiple main stems in the ms mutant using these methods in the future.
In the context of new rapeseed mutants, the high-density pod mutant (dpt247) not only exhibits multiple stems but also shows a reduced plant height and primary branch length, along with a significantly increased number of pods on the main inflorescence [64]. Similarly, the dou tou (dt) mutant displays multiple flowers from a single peduncle, and the maturation of dt siliques is considerably slower compared to that of the WT, in addition to multiple main stems being present [52]. These findings suggest that gene mutations in new mutants can lead to diverse changes beyond a single phenotype. Therefore, conducting detailed observations and analyses of the ms mutants in the future will be essential for a comprehensive understanding of their characteristics.
5. Conclusions
Our study sheds light on the mechanism underlying the formation of multiple main stems in rapeseed. Specifically, we found that the abnormal development of the SAM in the ms mutant is closely linked to the increased number of main stems. Through comprehensive transcriptome analysis conducted on both WT and ms mutant at various germination stages, we made a significant discovery that the aberrant development of the ms mutant’s SAM initiates at 20 DAG. Furthermore, our investigation unveiled noteworthy alterations in the expression profiles of genes associated with the CK signaling pathway within the SAM of the ms mutant, when compared to the WT. Intriguingly, despite conducting HPLC-MS analyses, we did not detect any substantial changes in the CK levels present in the ms mutants. However, only a subtle upregulation of iP, tzR, and czR was observed. This study not only provides insights for breeding new rapeseed varieties with multiple main stems but also provides valuable resources for future research on SAM development and shoot branching in rapeseed. However, the loci responsible for the multi-main-stem phenotype in the ms mutant remain unknown, and further GWAS and genetic fine mapping will be necessary to identify the major QTLs or candidate genes associated with multiple main stems in the ms mutant.
Supplementary Materials
The following supporting information can be downloaded at: https://www.mdpi.com/article/10.3390/genes14071396/s1, Figure S1: The GO enrichment analysis of DEGs between the ms mutant and WT at 15 days after germination (DAG). All significantly enriched GO terms (p value < 0.05) based on biological process, cellular component, and molecular function enriched from the DEGs between the ms mutant and WT at 15 DAG. Numbers of up- and downregulated DEGs of enriched GO terms are shown. Complete data can be found in Table S3. Figure S2: The GO enrichment analysis of DEGs between the ms mutant and WT at 20 days after germination (DAG). All significantly enriched GO terms (p value < 0.05) based on biological process, cellular component, and molecular function enriched from the DEGs between the ms mutant and WT at 20 DAG. Numbers of up- and downregulated DEGs of enriched GO terms are shown. Complete data can be found in Table S3. Figure S3: The GO enrichment analysis of DEGs between the ms mutant and WT at 25 days after germination (DAG). All significantly enriched GO terms (p value < 0.05) based on biological process, cellular component, and molecular function enriched from the DEGs between the ms mutant and WT at 25 DAG. Numbers of up- and downregulated DEGs of enriched GO terms are shown. Complete data can be found in Table S3. Figure S4: The GO enrichment analysis of DEGs between the ms mutant and WT at 30 days after germination (DAG). All significantly enriched GO terms (p value < 0.05) based on biological process, cellular component, and molecular function enriched from the DEGs between the ms mutant and WT at 30 DAG. Numbers of up- and downregulated DEGs of enriched GO terms are shown. Complete data can be found in Table S3. Figure S5: The GO enrichment analysis of DEGs between ms mutant and WT at 35 days after germination (DAG). All significantly enriched GO terms (p value < 0.05) based on biological process, cellular component, and molecular function enriched from the DEGs between the ms mutant and WT at 35 DAG. Numbers of up- and downregulated DEGs of enriched GO terms are shown. Complete data can be found in Table S3. Figure S6: Relative transcript levels of genes involved in the CK signaling pathway. (A) Heatmaps indicating the relative transcript levels (log2FC) of IPTs, (B) AHKs, (C) AHPs, (D) type-A ARRs, and (E) type-B ARRs in the SAMs of the ms mutant compared with the WT at 15, 20, 25, 30, and 35 DAG, respectively. * highlights genes that were differentially expressed between the ms mutant and WT (p value < 0.05; |log2(fold change) | ≥ 1). Gene names and transcript levels are listed in Table S6. Table S1: The compositions of MS medium. Table S2: The DEGs between the ms mutant and WT at different DAG; Table S3: GO analysis of DEGs between the ms mutant and WT at different DAG; Table S4: KEGG analysis of DEGs between the ms mutant and WT at different DAG; Table S5: FPKM values of CK-related genes for the WT and ms mutant at different DAG; Table S6: The expression of CK signaling-related genes in the SAMs of ms mutant compared with WT at different DAG. Table S7: The primers used for RT-qPCR.
Author Contributions
Conceptualization, Q.W., S.L. and L.L.; validation, Q.W., N.X. and C.S.; formal analysis, X.P. and J.T.; investigation, C.M. and Y.Y.; resources, Q.W. and L.L.; data curation, C.S. and N.X.; writing—original draft preparation, Q.W. and N.X.; visualization, N.X. and C.S.; supervision, M.G., R.L. and R.D.; project administration, Q.W.; funding acquisition, S.L. and L.L. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the National Natural Science Foundation of China (No. 32060681) and the Major Science and Technology Projects in Yunnan Province (No. 202205AR070001).
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data were included in the manuscript.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Mänd, M.; Williams, I.H.; Viik, E.; Karise, R. Oilseed Rape, Bees and Integrated Pest Management. In Biocontrol-Based Integrated Management of Oilseed Rape Pests; Williams, I.H., Ed.; Springer: Dordrecht, The Netherlands, 2010; pp. 357–379. [Google Scholar] [CrossRef]
- Raboanatahiry, N.; Li, H.; Yu, L.; Li, M. Rapeseed (Brassica napus): Processing, Utilization, and Genetic Improvement. Agronomy 2021, 11, 1776. [Google Scholar] [CrossRef]
- Shimizu-Sato, S.; Mori, H. Control of outgrowth and dormancy in axillary buds. Plant Physiol. 2001, 127, 1405–1413. [Google Scholar] [CrossRef] [PubMed]
- McSteen, P.; Leyser, O. Shoot branching. Annu. Rev. Plant Biol. 2005, 56, 353–374. [Google Scholar] [CrossRef] [PubMed]
- Aichinger, E.; Kornet, N.; Friedrich, T.; Laux, T. Plant stem cell niches. Annu. Rev. Plant Biol. 2012, 63, 615–636. [Google Scholar] [CrossRef] [PubMed]
- Barton, M.K. Twenty years on: The inner workings of the shoot apical meristem, a developmental dynamo. Dev. Biol. 2010, 341, 95–113. [Google Scholar] [CrossRef]
- Barbier, F.F.; Dun, E.A.; Kerr, S.C.; Chabikwa, T.G.; Beveridge, C.A. An Update on the Signals Controlling Shoot Branching. Trends Plant Sci. 2019, 24, 220–236. [Google Scholar] [CrossRef]
- Wang, B.; Smith, S.M.; Li, J. Genetic Regulation of Shoot Architecture. Annu. Rev. Plant Biol. 2018, 69, 437–468. [Google Scholar] [CrossRef]
- Shani, E.; Yanai, O.; Ori, N. The role of hormones in shoot apical meristem function. Curr. Opin. Plant Biol. 2006, 9, 484–489. [Google Scholar] [CrossRef]
- Sakakibara, H. Cytokinins: Activity, biosynthesis, and translocation. Annu. Rev. Plant Biol. 2006, 57, 431–449. [Google Scholar] [CrossRef]
- Mok, D.W.; Mok, M.C. Cytokinin Metabolism and Action. Annu. Rev. Plant Physiol. Plant Mol. Biol. 2001, 52, 89–118. [Google Scholar] [CrossRef]
- Sp Chal, L.X. Cytokinins—Recent news and views of evolutionally old molecules. Funct. Plant Biol. 2012, 39, 267–284. [Google Scholar] [CrossRef]
- Lomin, S.N.; Krivosheev, D.M.; Steklov, M.Y.; Arkhipov, D.V.; Osolodkin, D.I.; Schmulling, T.; Romanov, G.A. Plant membrane assays with cytokinin receptors underpin the unique role of free cytokinin bases as biologically active ligands. J. Exp. Bot. 2015, 66, 1851–1863. [Google Scholar] [CrossRef]
- Bajguz, A.; Piotrowska, A. Conjugates of auxin and cytokinin. Phytochemistry 2009, 70, 957–969. [Google Scholar] [CrossRef]
- Raspor, M.; Motyka, V.; Ninkovic, S.; Dobrev, P.I.; Malbeck, J.; Cosic, T.; Cingel, A.; Savic, J.; Tadic, V.; Dragicevic, I.C. Endogenous levels of cytokinins, indole-3-acetic acid and abscisic acid in in vitro grown potato: A contribution to potato hormonomics. Sci. Rep. 2020, 10, 3437. [Google Scholar] [CrossRef]
- Del Rosario Cardenas-Aquino, M.; Sarria-Guzman, Y.; Martinez-Antonio, A. Review: Isoprenoid and aromatic cytokinins in shoot branching. Plant Sci. 2022, 319, 111240. [Google Scholar] [CrossRef]
- Tarkowska, D.; Dolezal, K.; Tarkowski, P.; Astot, C.; Holub, J.; Fuksova, K.; Schmulling, T.; Sandberg, G.; Strnad, M. Identification of new aromatic cytokinins in Arabidopsis thaliana and Populus x canadensis leaves by LC-(+)ESI-MS and capillary liquid chromatography/frit-fast atom bombardment mass spectrometry. Physiol. Plant 2003, 117, 579–590. [Google Scholar] [CrossRef]
- Nishimura, C.; Ohashi, Y.; Sato, S.; Kato, T.; Tabata, S.; Ueguchi, C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell 2004, 16, 1365–1377. [Google Scholar] [CrossRef]
- Tantikanjana, T.; Yong, J.W.; Letham, D.S.; Griffith, M.; Hussain, M.; Ljung, K.; Sandberg, G.; Sundaresan, V. Control of axillary bud initiation and shoot architecture in Arabidopsis through the SUPERSHOOT gene. Genes. Dev. 2001, 15, 1577–1588. [Google Scholar] [CrossRef]
- Giulini, A.; Wang, J.; Jackson, D. Control of phyllotaxy by the cytokinin-inducible response regulator homologue ABPHYL1. Nature 2004, 430, 1031–1034. [Google Scholar] [CrossRef]
- Kieber, J.J.; Schaller, G.E. Cytokinin signaling in plant development. Development 2018, 145, dev149344. [Google Scholar] [CrossRef]
- Snipes, S.A.; Rodriguez, K.; DeVries, A.E.; Miyawaki, K.N.; Perales, M.; Xie, M.; Reddy, G.V. Cytokinin stabilizes WUSCHEL by acting on the protein domains required for nuclear enrichment and transcription. PLoS Genet. 2018, 14, e1007351. [Google Scholar] [CrossRef] [PubMed]
- Meng, W.J.; Cheng, Z.J.; Sang, Y.L.; Zhang, M.M.; Rong, X.F.; Wang, Z.W.; Tang, Y.Y.; Zhang, X.S. Type-B ARABIDOPSIS RESPONSE REGULATORs Specify the Shoot Stem Cell Niche by Dual Regulation of WUSCHEL. Plant Cell 2017, 29, 1357–1372. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Chen, H.; Huang, L.; O’Neil, R.C.; Shokhirev, M.N.; Ecker, J.R. A B-ARR-mediated cytokinin transcriptional network directs hormone cross-regulation and shoot development. Nat. Commun. 2018, 9, 1604. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Z.; Andersen, S.U.; Ljung, K.; Dolezal, K.; Miotk, A.; Schultheiss, S.J.; Lohmann, J.U. Hormonal control of the shoot stem-cell niche. Nature 2010, 465, 1089–1092. [Google Scholar] [CrossRef] [PubMed]
- Leibfried, A.; To, J.P.; Busch, W.; Stehling, S.; Kehle, A.; Demar, M.; Kieber, J.J.; Lohmann, J.U. WUSCHEL controls meristem function by direct regulation of cytokinin-inducible response regulators. Nature 2005, 438, 1172–1175. [Google Scholar] [CrossRef] [PubMed]
- Werner, T.; Köllmer, I.; Bartrina, I.; Holst, K.; Schmülling, T. New insights into the biology of cytokinin degradation. Plant Biol. 2006, 8, 371–381. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Van Onckelen, H.; Schmülling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef]
- Motyka, V.; Faiss, M.; Strand, M.; Kaminek, M.; Schmulling, T. Changes in Cytokinin Content and Cytokinin Oxidase Activity in Response to Derepression of ipt Gene Transcription in Transgenic Tobacco Calli and Plants. Plant Physiol. 1996, 112, 1035–1043. [Google Scholar] [CrossRef]
- Morris, R.O.; Bilyeu, K.D.; Laskey, J.G.; Cheikh, N.N. Isolation of a gene encoding a glycosylated cytokinin oxidase from maize. Biochem. Biophys. Res. Commun. 1999, 255, 328–333. [Google Scholar] [CrossRef]
- Ashikari, M.; Sakakibara, H.; Lin, S.; Yamamoto, T.; Takashi, T.; Nishimura, A.; Angeles, E.R.; Qian, Q.; Kitano, H.; Matsuoka, M. Cytokinin oxidase regulates rice grain production. Science 2005, 309, 741–745. [Google Scholar] [CrossRef]
- Mameaux, S.; Cockram, J.; Thiel, T.; Steuernagel, B.; Stein, N.; Taudien, S.; Jack, P.; Werner, P.; Gray, J.C.; Greenland, A.J.; et al. Molecular, phylogenetic and comparative genomic analysis of the cytokinin oxidase/dehydrogenase gene family in the Poaceae. Plant Biotechnol. J. 2012, 10, 67–82. [Google Scholar] [CrossRef]
- Le, D.T.; Nishiyama, R.; Watanabe, Y.; Vankova, R.; Tanaka, M.; Seki, M.; Ham Le, H.; Yamaguchi-Shinozaki, K.; Shinozaki, K.; Tran, L.S. Identification and expression analysis of cytokinin metabolic genes in soybean under normal and drought conditions in relation to cytokinin levels. PLoS ONE 2012, 7, e42411. [Google Scholar] [CrossRef]
- Liu, P.; Zhang, C.; Ma, J.Q.; Zhang, L.Y.; Yang, B.; Tang, X.Y.; Huang, L.; Zhou, X.T.; Lu, K.; Li, J.N. Genome-Wide Identification and Expression Profiling of Cytokinin Oxidase/Dehydrogenase (CKX) Genes Reveal Likely Roles in Pod Development and Stress Responses in Oilseed Rape (Brassica napus L.). Genes 2018, 9, 168. [Google Scholar] [CrossRef]
- Kurakawa, T.; Ueda, N.; Maekawa, M.; Kobayashi, K.; Kojima, M.; Nagato, Y.; Sakakibara, H.; Kyozuka, J. Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 2007, 445, 652–655. [Google Scholar] [CrossRef]
- Kuroha, T.; Tokunaga, H.; Kojima, M.; Ueda, N.; Ishida, T.; Nagawa, S.; Fukuda, H.; Sugimoto, K.; Sakakibara, H. Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 2009, 21, 3152–3169. [Google Scholar] [CrossRef]
- Tokunaga, H.; Kojima, M.; Kuroha, T.; Ishida, T.; Sugimoto, K.; Kiba, T.; Sakakibara, H. Arabidopsis lonely guy (LOG) multiple mutants reveal a central role of the LOG-dependent pathway in cytokinin activation. Plant J. Cell Mol. Biol. 2012, 69, 355–365. [Google Scholar] [CrossRef]
- Lu, Z.; Shao, G.; Xiong, J.; Jiao, Y.; Wang, J.; Liu, G.; Meng, X.; Liang, Y.; Xiong, G.; Wang, Y.; et al. MONOCULM 3, an ortholog of WUSCHEL in rice, is required for tiller bud formation. J. Genet. Genom. 2015, 42, 71–78. [Google Scholar] [CrossRef]
- Hu, Y.S.; Ren, T.H.; Li, Z.; Tang, Y.Z.; Ren, Z.L.; Yan, B.J. Molecular mapping and genetic analysis of a QTL controlling spike formation rate and tiller number in wheat. Gene 2017, 634, 15–21. [Google Scholar] [CrossRef]
- Zhao, W.; Chao, H.; Zhang, L.; Ta, N.; Zhao, Y.; Li, B.; Zhang, K.; Guan, Z.; Hou, D.; Chen, K.; et al. Integration of QTL Mapping and Gene Fishing Techniques to Dissect the Multi-Main Stem Trait in Rapeseed (Brassica napus L.). Front. Plant Sci. 2019, 10, 1152. [Google Scholar] [CrossRef]
- Xu, A.; Wei, C. Comprehensive comparison and applications of different sections in investigating the microstructure and histochemistry of cereal kernels. Plant Methods 2020, 16, 8. [Google Scholar] [CrossRef]
- Kong, Y.; Ebrahimpour, P.; Liu, Y.; Yang, C.; Alonso, L.C. Pancreatic Islet Embedding for Paraffin Sections. J. Vis. Exp. 2018, 136, 57931. [Google Scholar] [CrossRef]
- Tarkowski, P.; Ge, L.; Yong, J.W.H.; Tan, S.N. Analytical methods for cytokinins. TrAC Trends Anal. Chem. 2009, 28, 323–335. [Google Scholar] [CrossRef]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.J.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.H.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Yang, H.; Liu, J.; Huang, S.; Guo, T.; Deng, L.; Hua, W. Selection and evaluation of novel reference genes for quantitative reverse transcription PCR (qRT-PCR) based on genome and transcriptome data in Brassica napus L. Gene 2014, 538, 113–122. [Google Scholar] [CrossRef]
- Shi, B.; Vernoux, T. Hormonal control of cell identity and growth in the shoot apical meristem. Curr. Opin. Plant Biol. 2022, 65, 102111. [Google Scholar] [CrossRef]
- Takei, K.; Sakakibara, H.; Sugiyama, T. Identification of genes encoding adenylate isopentenyltransferase, a cytokinin biosynthesis enzyme, in Arabidopsis thaliana. J. Biol. Chem. 2001, 276, 26405–26410. [Google Scholar] [CrossRef]
- Erb, M.; Meldau, S.; Howe, G.A. Role of phytohormones in insect-specific plant reactions. Trends Plant Sci. 2012, 17, 250–259. [Google Scholar] [CrossRef]
- Koo, A.J.; Howe, G.A. The wound hormone jasmonate. Phytochemistry 2009, 70, 1571–1580. [Google Scholar] [CrossRef]
- Zhu, K.-M.; Xu, S.; Li, K.-X.; Chen, S.; Zafar, S.; Cao, W.; Wang, Z.; Ding, L.-N.; Yang, Y.-H.; Li, Y.-M.; et al. Transcriptome analysis of the irregular shape of shoot apical meristem in dt (dou tou) mutant of Brassica napus L. Mol. Breed. 2019, 39, 39. [Google Scholar] [CrossRef]
- Kuderova, A.; Gallova, L.; Kuricova, K.; Nejedla, E.; Curdova, A.; Micenkova, L.; Plihal, O.; Smajs, D.; Spichal, L.; Hejatko, J. Identification of AHK2- and AHK3-like cytokinin receptors in Brassica napus reveals two subfamilies of AHK2 orthologues. J. Exp. Bot. 2015, 66, 339–353. [Google Scholar] [CrossRef]
- Hutchison, C.E.; Li, J.; Argueso, C.; Gonzalez, M.; Lee, E.; Lewis, M.W.; Maxwell, B.B.; Perdue, T.D.; Schaller, G.E.; Alonso, J.M.; et al. The Arabidopsis histidine phosphotransfer proteins are redundant positive regulators of cytokinin signaling. Plant Cell 2006, 18, 3073–3087. [Google Scholar] [CrossRef]
- Mason, M.G.; Mathews, D.E.; Argyros, D.A.; Maxwell, B.B.; Kieber, J.J.; Alonso, J.M.; Ecker, J.R.; Schaller, G.E. Multiple type-B response regulators mediate cytokinin signal transduction in Arabidopsis. Plant Cell 2005, 17, 3007–3018. [Google Scholar] [CrossRef]
- Müller, D.; Waldie, T.; Miyawaki, K.; To, J.P.C.; Melnyk, C.W.; Kieber, J.J.; Kakimoto, T.; Leyser, O. Cytokinin is required for escape but not release from auxin mediated apical dominance. Plant J. Cell Mol. Biol. 2015, 82, 874–886. [Google Scholar] [CrossRef]
- Skoog, F.; Thimann, K.V. Further Experiments on the Inhibition of the Development of Lateral Buds by Growth Hormone. Proc. Natl. Acad. Sci. USA 1934, 20, 480–485. [Google Scholar] [CrossRef]
- Stirnberg, P.; van De Sande, K.; Leyser, H.M. MAX1 and MAX2 control shoot lateral branching in Arabidopsis. Development 2002, 129, 1131–1141. [Google Scholar] [CrossRef] [PubMed]
- Zou, J.; Chen, Z.; Zhang, S.; Zhang, W.; Jiang, G.; Zhao, X.; Zhai, W.; Pan, X.; Zhu, L. Characterizations and fine mapping of a mutant gene for high tillering and dwarf in rice (Oryza sativa L.). Planta 2005, 222, 604–612. [Google Scholar] [CrossRef]
- Beveridge, C.A. Long-distance signalling and a mutational analysis of branching in pea. Plant Growth Regul. 2000, 32, 193–203. [Google Scholar] [CrossRef]
- Gomez-Roldan, V.; Fermas, S.; Brewer, P.B.; Puech-Pages, V.; Dun, E.A.; Pillot, J.P.; Letisse, F.; Matusova, R.; Danoun, S.; Portais, J.C.; et al. Strigolactone inhibition of shoot branching. Nature 2008, 455, 189–194. [Google Scholar] [CrossRef] [PubMed]
- Hayward, A.; Stirnberg, P.; Beveridge, C.; Leyser, O. Interactions between auxin and strigolactone in shoot branching control. Plant Physiol. 2009, 151, 400–412. [Google Scholar] [CrossRef]
- Domagalska, M.A.; Leyser, O. Signal integration in the control of shoot branching. Nat. Rev. Mol. Cell Biol. 2011, 12, 211–221. [Google Scholar] [CrossRef]
- Tang, M.; Tong, C.; Liang, L.; Du, C.; Zhao, J.; Xiao, L.; Tong, J.; Dai, X.; Helal, M.; Dai, W.; et al. A recessive high-density pod mutant resource of Brassica napus. Plant Sci. 2020, 293, 110411. [Google Scholar] [CrossRef]
- Salam, B.B.; Malka, S.K.; Zhu, X.; Gong, H.; Ziv, C.; Teper-Bamnolker, P.; Ori, N.; Jiang, J.; Eshel, D. Etiolated Stem Branching Is a Result of Systemic Signaling Associated with Sucrose Level. Plant Physiol. 2017, 175, 734–745. [Google Scholar] [CrossRef]
- Xiao, Q.; Bai, X.; Zhang, C.; He, Y. Advanced high-throughput plant phenotyping techniques for genome-wide association studies: A review. J. Adv. Res. 2022, 35, 215–230. [Google Scholar] [CrossRef]
- Brachi, B.; Morris, G.P.; Borevitz, J.O. Genome-wide association studies in plants: The missing heritability is in the field. Genome Biol. 2011, 12, 232. [Google Scholar] [CrossRef]
- Khan, S.U.; Saeed, S.; Khan, M.H.U.; Fan, C.; Ahmar, S.; Arriagada, O.; Shahzad, R.; Branca, F.; Mora-Poblete, F. Advances and Challenges for QTL Analysis and GWAS in the Plant-Breeding of High-Yielding: A Focus on Rapeseed. Biomolecules 2021, 11, 1516. [Google Scholar] [CrossRef]
- Sun, F.; Liu, J.; Hua, W.; Sun, X.; Wang, X.; Wang, H. Identification of stable QTLs for seed oil content by combined linkage and association mapping in Brassica napus. Plant Sci. 2016, 252, 388–399. [Google Scholar] [CrossRef]
- Wang, T.; Wei, L.; Wang, J.; Xie, L.; Li, Y.Y.; Ran, S.; Ren, L.; Lu, K.; Li, J.; Timko, M.P.; et al. Integrating GWAS, linkage mapping and gene expression analyses reveals the genetic control of growth period traits in rapeseed (Brassica napus L.). Biotechnol. Biofuels 2020, 13, 134. [Google Scholar] [CrossRef]
- Zhao, C.; Xie, M.; Liang, L.; Yang, L.; Han, H.; Qin, X.; Zhao, J.; Hou, Y.; Dai, W.; Du, C.; et al. Genome-Wide Association Analysis Combined With Quantitative Trait Loci Mapping and Dynamic Transcriptome Unveil the Genetic Control of Seed Oil Content in Brassica napus L. Front. Plant Sci. 2022, 13, 929197. [Google Scholar] [CrossRef]
- Li, Y.; Shen, J.; Wang, T.; Chen, Q.; Zhang, X.; Fu, T.; Meng, J.; Tu, J.; Ma, C.J.C. QTL analysis of yield-related traits and their association with functional markers in Brassica napus L. Aust. J. Agric. Res. 2007, 58, 759–766. [Google Scholar] [CrossRef]
- Shi, J.; Li, R.; Qiu, D.; Jiang, C.; Long, Y.; Morgan, C.; Bancroft, I.; Zhao, J.; Meng, J. Unraveling the complex trait of crop yield with quantitative trait loci mapping in Brassica napus. Genetics 2009, 182, 851–861. [Google Scholar] [CrossRef] [PubMed]
- Ding, G.; Zhao, Z.; Liao, Y.; Hu, Y.; Shi, L.; Long, Y.; Xu, F. Quantitative trait loci for seed yield and yield-related traits, and their responses to reduced phosphorus supply in Brassica napus. Ann. Bot. 2012, 109, 747–759. [Google Scholar] [CrossRef] [PubMed]
- Chen, W.; Zhang, Y.; Liu, X.; Chen, B.; Tu, J.; Tingdong, F. Detection of QTL for six yield-related traits in oilseed rape (Brassica napus) using DH and immortalized F2 populations. Theor. Appl. Genet. 2007, 115, 849–858. [Google Scholar] [CrossRef]
- Luo, X.; Ma, C.; Yue, Y.; Hu, K.; Li, Y.; Duan, Z.; Wu, M.; Tu, J.; Shen, J.; Yi, B.; et al. Unravelling the complex trait of harvest index in rapeseed (Brassica napus L.) with association mapping. BMC Genom. 2015, 16, 379. [Google Scholar] [CrossRef]
- Li, F.; Chen, B.; Xu, K.; Gao, G.; Yan, G.; Qiao, J.; Li, J.; Li, H.; Li, L.; Xiao, X.; et al. A genome-wide association study of plant height and primary branch number in rapeseed (Brassica napus). Plant Sci. 2016, 242, 169–177. [Google Scholar] [CrossRef]
- He, Y.; Wu, D.; Wei, D.; Fu, Y.; Cui, Y.; Dong, H.; Tan, C.; Qian, W. GWAS, QTL mapping and gene expression analyses in Brassica napus reveal genetic control of branching morphogenesis. Sci. Rep. 2017, 7, 15971. [Google Scholar] [CrossRef]
- Zheng, M.; Peng, C.; Liu, H.; Tang, M.; Yang, H.; Li, X.; Liu, J.; Sun, X.; Wang, X.; Xu, J.; et al. Genome-Wide Association Study Reveals Candidate Genes for Control of Plant Height, Branch Initiation Height and Branch Number in Rapeseed (Brassica napus L.). Front. Plant Sci. 2017, 8, 1246. [Google Scholar] [CrossRef]
- Li, B.; Gao, J.; Chen, J.; Wang, Z.; Shen, W.; Yi, B.; Wen, J.; Ma, C.; Shen, J.; Fu, T.; et al. Identification and fine mapping of a major locus controlling branching in Brassica napus. Theor. Appl. Genet. 2020, 133, 771–783. [Google Scholar] [CrossRef]
- Li, P.; Su, T.; Zhang, B.; Li, P.; Xin, X.; Yue, X.; Cao, Y.; Wang, W.; Zhao, X.; Yu, Y.; et al. Identification and fine mapping of qSB.A09, a major QTL that controls shoot branching in Brassica rapa ssp. chinensis Makino. Theor. Appl. Genet. 2020, 133, 1055–1068. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).