Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Gene Family in Litchi chinensis
Abstract
:1. Introduction
2. Materials and Methods
2.1. Identification and Characterization of NAC Family Genes in Litchi
2.2. Phylogenetic Analysis and Classification of LcNACs
2.3. Gene Structure, Conserved Motif Analysis, and Chromosomal Location of LcNACs
2.4. Chromosomal Mapping, Gene Duplication, and Synteny Analysis
2.5. Prediction of Cis-Acting Elements within the Promoter of NAC Genes in Litchi
2.6. Expression Profiling of Litchi NAC Genes
2.7. Plant Materials and Growing Conditions of Litchi
2.8. Quantitative Real-Time PCR Analyses of NAC Expression Patterns
3. Results
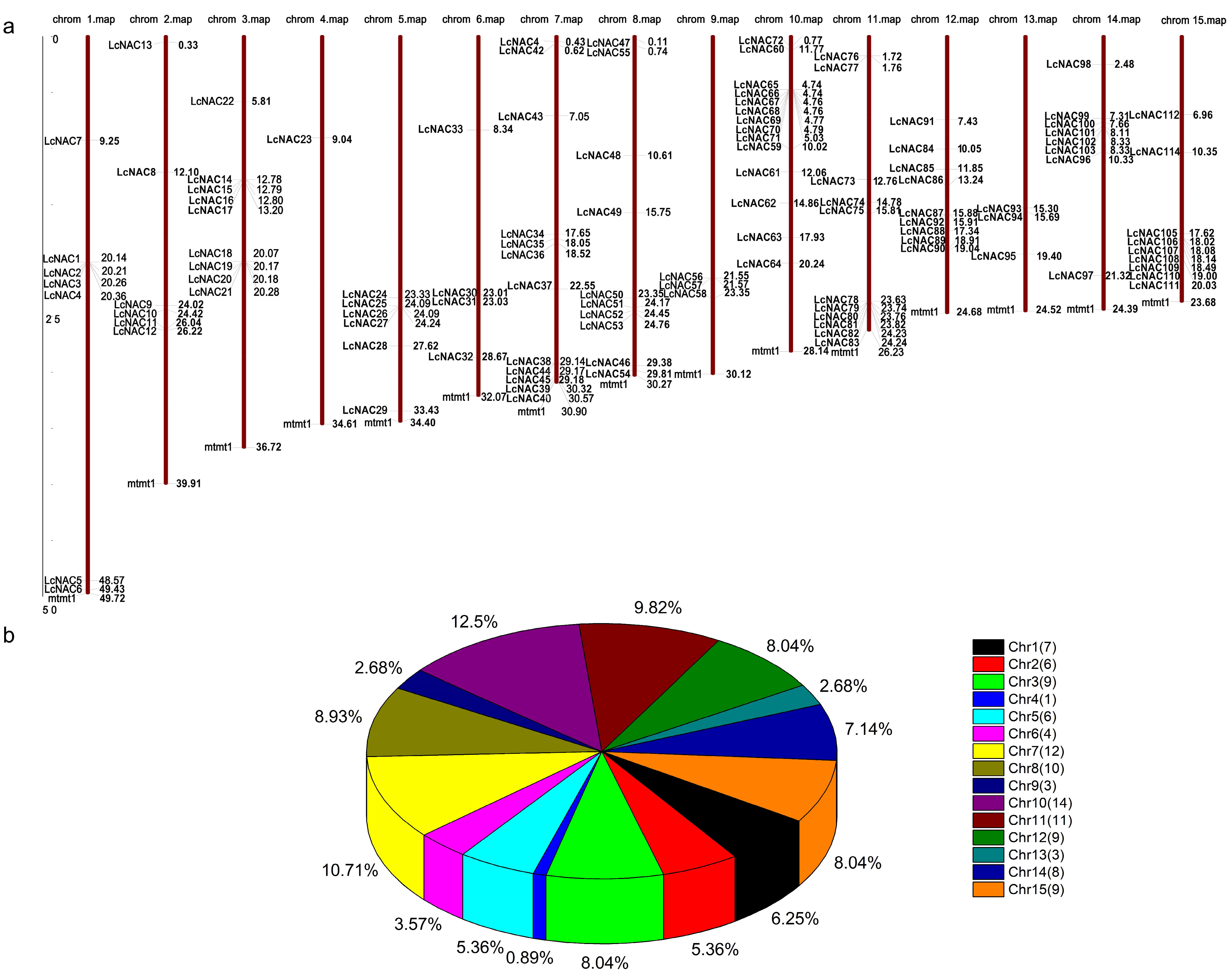
3.1. The Litchi Genome Contains 112 NAC Genes and Is Unevenly Distributed on 15 Chromosomes
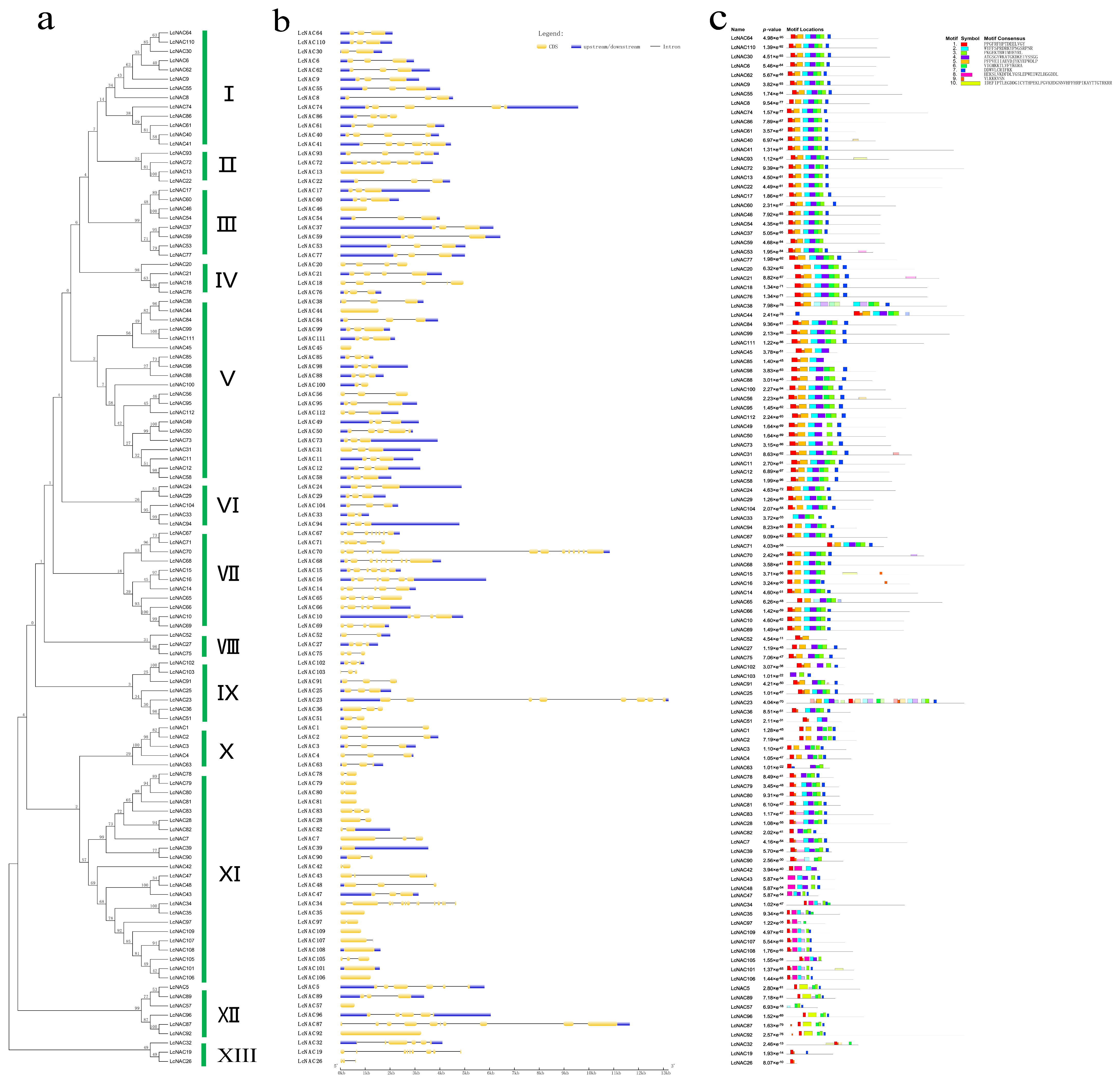
3.2. Gene Phylogenetic Analysis, Gene Structure, and Protein Motif Composition Analysis
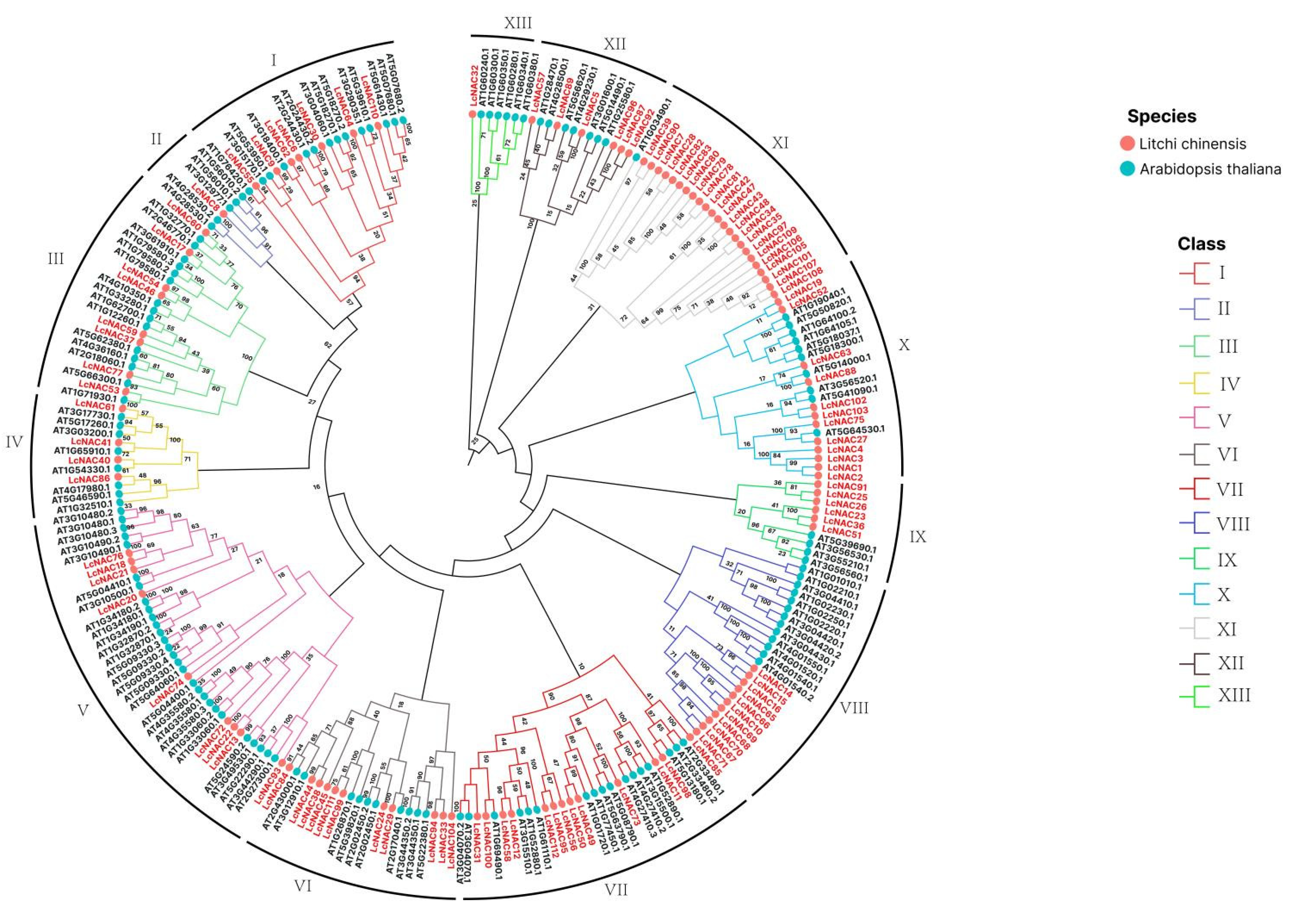
3.3. Phylogenetic Analysis of the LcNACs and A. thaliana NAC Genes
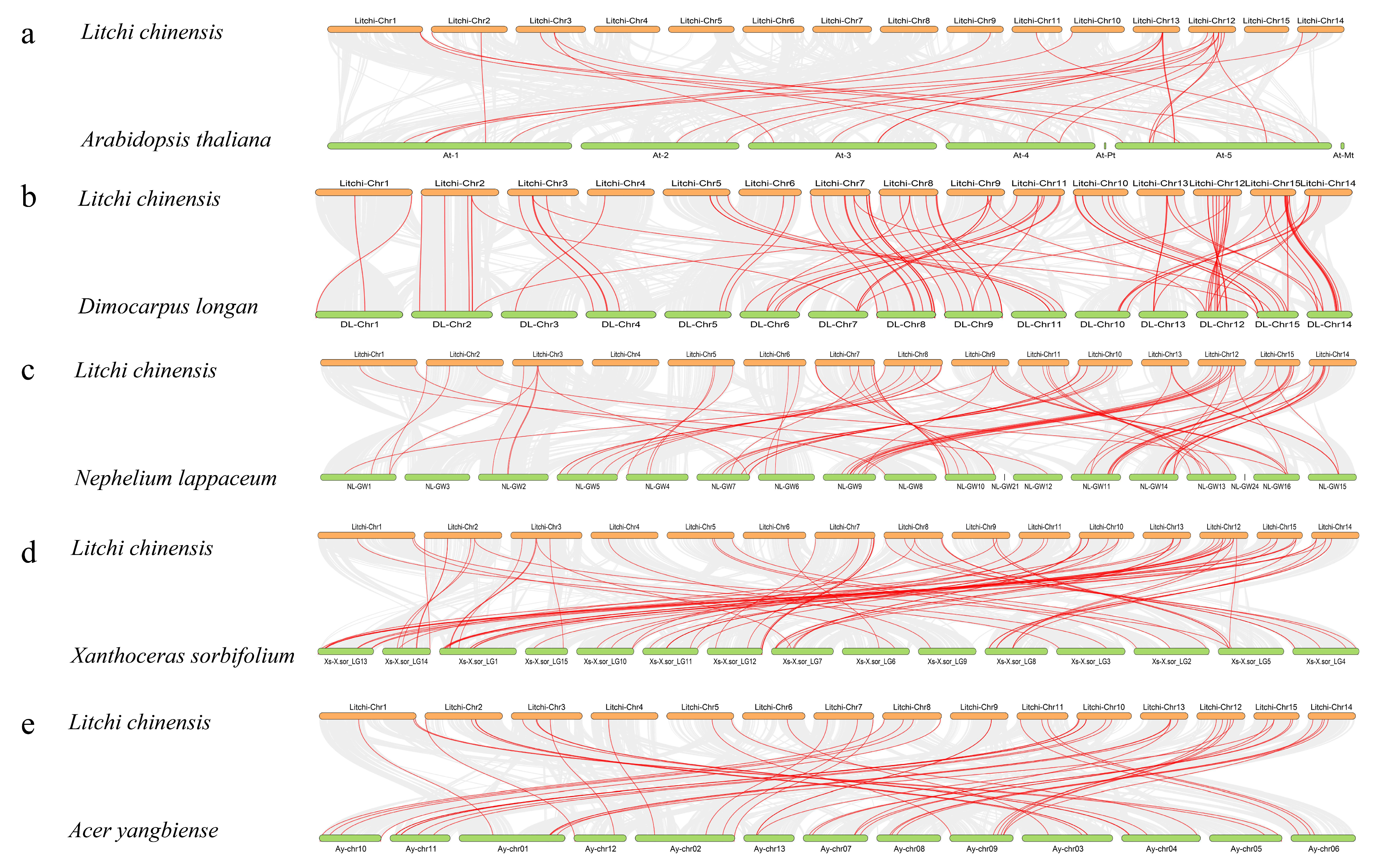
3.4. Collinearity Analysis of the LcNACs
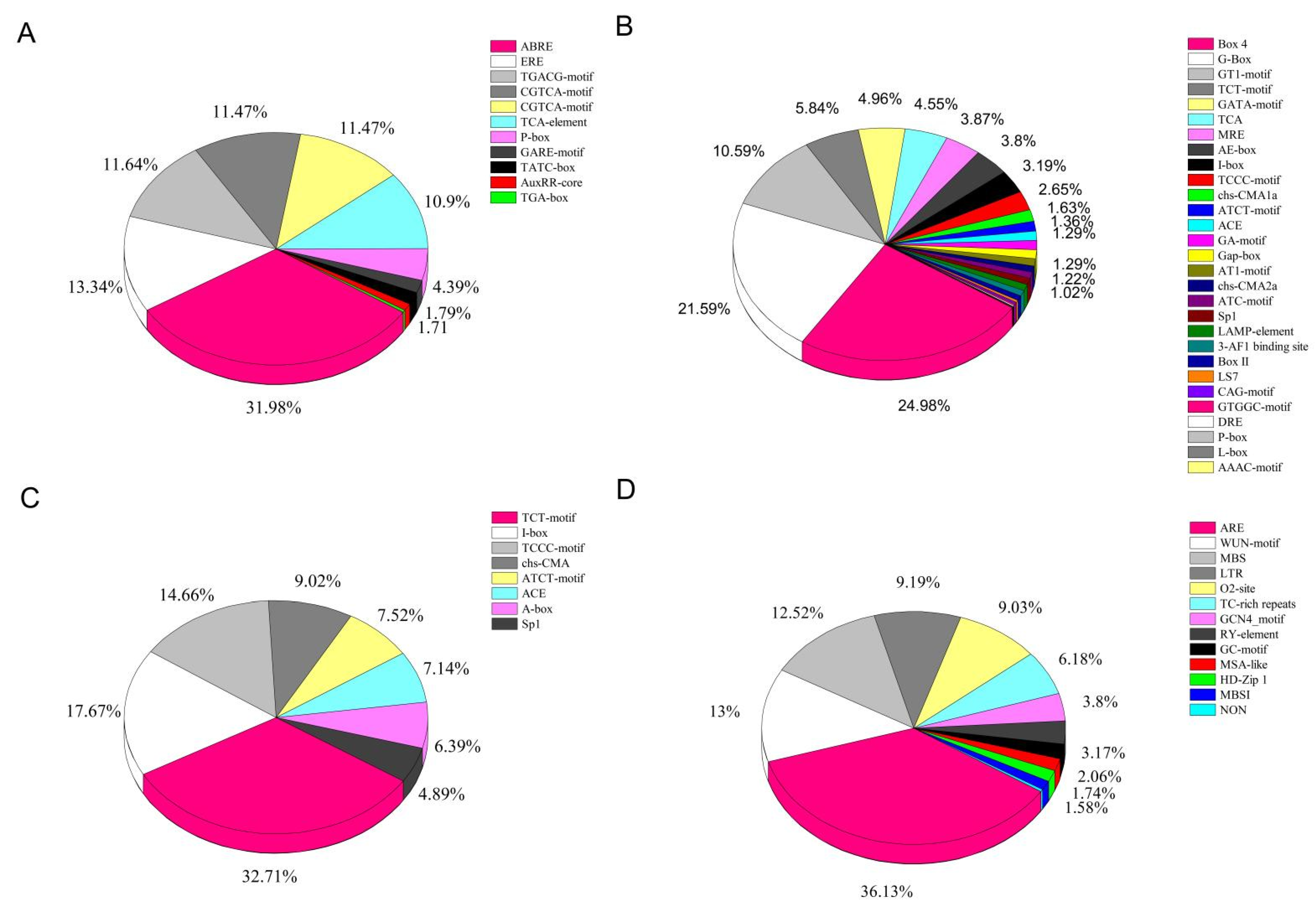
3.5. Analysis of Cis-Regulatory Elements of the LcNACs
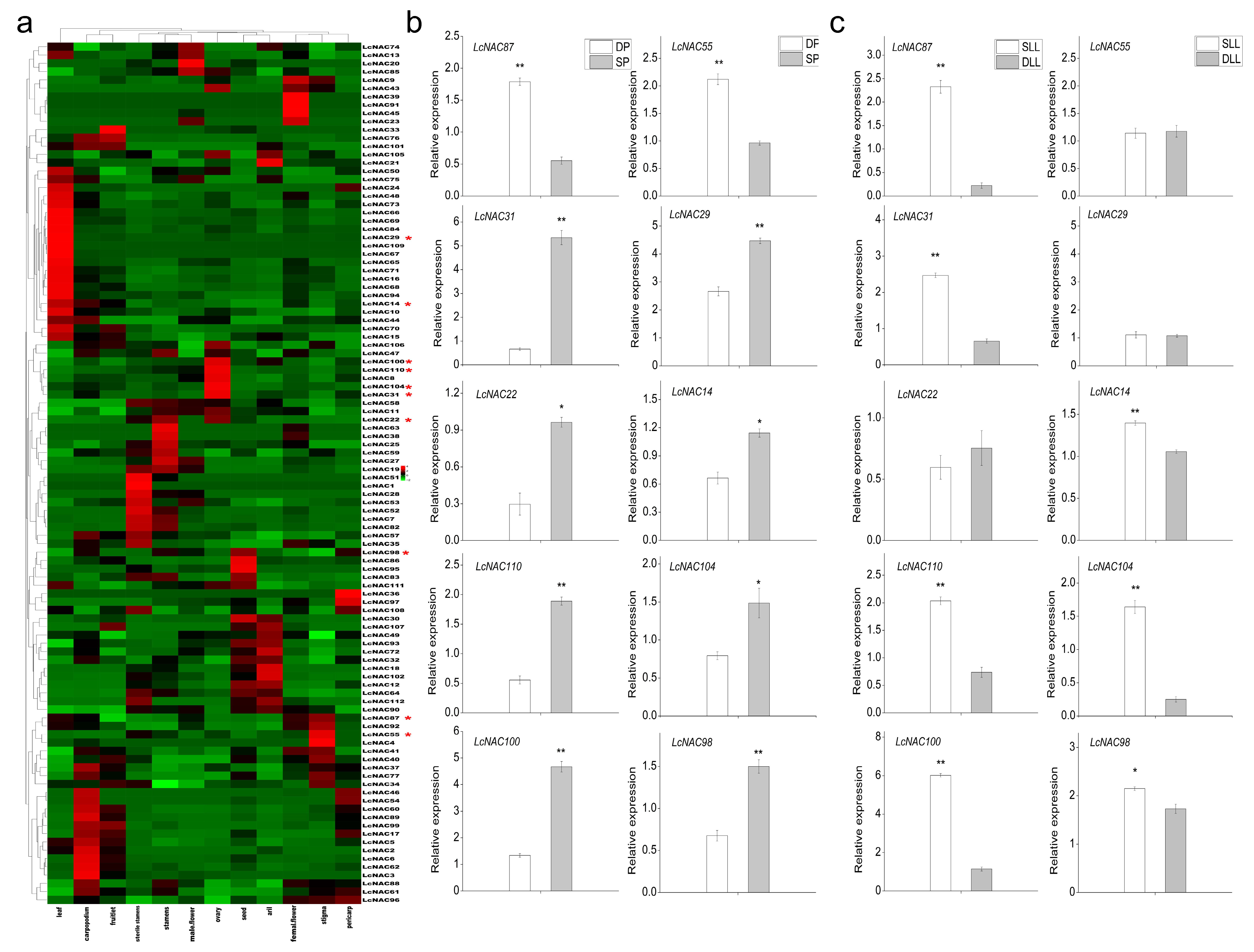
3.6. Spatiotemporal Expression Analysis of NAC Genes in Different Tissues of Litchi
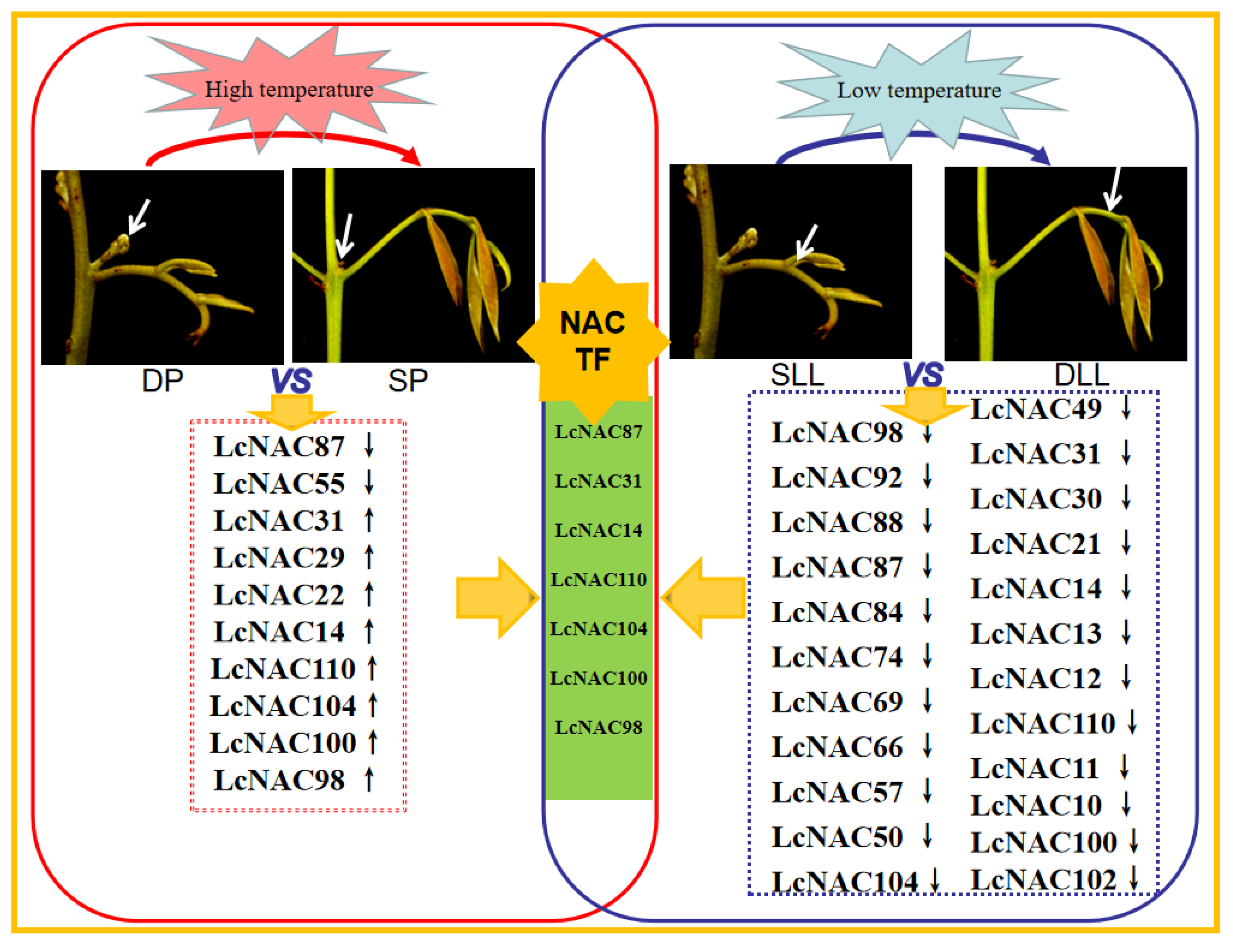
3.7. Analysis of the Role of the NAC Genes in Floral Bud Differentiation in Litchi
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Puranik, S.; Sahu, P.P.; Srivastava, P.S.; Prasad, M. NAC proteins: Regulation and role in stress tolerance. Trends Plant Sci. 2012, 17, 369–381. [Google Scholar] [CrossRef]
- Jensen, M.K.; Kjaersgaard, T.; Nielsen, M.M.; Galberg, P.; Petersen, K.; O’Shea, C.; Skriver, K. The Arabidopsis thaliana NAC transcription factor family: Structure-function relationships and determinants of ANAC019 stress signalling. Biochem. J. 2010, 426, 183–196. [Google Scholar] [CrossRef] [Green Version]
- Munir, N.; Yukun, C.; Xiaohui, C.; Nawaz, M.A.; Iftikhar, J.; Rizwan, H.M.; Xu, S.; Yuling, L.; Xuhan, X.; Zhongxiong, L. Genome-wide identification and comprehensive analyses of NAC transcription factor gene family and expression patterns during somatic embryogenesis in Dimocarpus longan Lour. Plant Physiol. Bioch. 2020, 157, 169–184. [Google Scholar] [CrossRef]
- Li, H.; Dong, Q.; Zhao, Q.; Shi, S.; Ran, K. Isolation, sequencing, and expression analysis of 30 AP2/ERF transcription factors in apple. PeerJ 2020, 8, e8391. [Google Scholar] [CrossRef]
- Kragelund, B.B.; Jensen, M.K.; Skriver, K. Order by disorder in plant signaling. Trends Plant Sci. 2012, 17, 625–632. [Google Scholar] [CrossRef]
- Sun, X.; Rikkerink, E.H.A.; Jones, W.T.; Uversky, V.N. Multifarious Roles of intrinsic disorder in proteins illustrate its broad impact on plant biology. Plant Cell 2013, 25, 38–55. [Google Scholar] [CrossRef] [Green Version]
- Guo, Z.; Zhang, Y.; Yao, J.; Xie, Z.; Zhang, Y.; Zhang, S.; Gu, C. The NAM/ATAF1/2/CUC2 transcription factor PpNAC.A59 enhances PpERF.A16 expression to promote ethylene biosynthesis during peach fruit ripening. Hortic. Res. 2021, 8, 209. [Google Scholar] [CrossRef]
- Olsen, A.N.; Ernst, H.A.; Leggio, L.L.; Skriver, K. NAC transcription factors: Structurally distinct, functionally diverse. Trends Plant Sci. 2005, 10, 79–87. [Google Scholar] [CrossRef]
- Forlani, S.; Mizzotti, C.; Masiero, S. The NAC side of the fruit: Tuning of fruit development and maturation. BMC Plant Biol. 2021, 21, 238. [Google Scholar] [CrossRef]
- Yuan, X.; Wang, H.; Cai, J.; Li, D.; Song, F. NAC transcription factors in plant immunity. Phytopathol. Res. 2019, 1, 3. [Google Scholar] [CrossRef]
- Trupkin, S.A.; Astigueta, F.H.; Baigorria, A.H.; Garcia, M.N.; Delfosse, V.C.; Gonzalez, S.A.; Perez, D.L.T.M.; Moschen, S.; Lia, V.V.; Fernandez, P.; et al. Identification and expression analysis of NAC transcription factors potentially involved in leaf and petal senescence in Petunia hybrida. Plant Sci. 2019, 287, 110195. [Google Scholar] [CrossRef] [PubMed]
- Yuan, X.; Wang, H.; Bi, Y.; Yan, Y.; Gao, Y.; Xiong, X.; Wang, J.; Li, D.; Song, F. ONAC066, A Stress-Responsive NAC Transcription Activator, Positively Contributes to Rice Immunity Against Magnaprothe oryzae through Modulating Expression of OsWRKY62 and Three Cytochrome P450 Genes. Front. Plant Sci. 2021, 12, 749186. [Google Scholar] [CrossRef] [PubMed]
- Fraga, O.T.; de Melo, B.P.; Quadros, I.P.S.; Reis, P.A.B.; Fontes, E.P.B. Senescence-Associated Glycine max (Gm)NAC Genes: Integration of Natural and Stress-Induced Leaf Senescence. Int. J. Mol. Sci. 2021, 22, 8287. [Google Scholar] [CrossRef] [PubMed]
- Fan, K.; Shen, H.; Bibi, N.; Li, F.; Yuan, S.; Wang, M.; Wang, X. Molecular evolution and species-specific expansion of the NAP members in plants. J. Integr. Plant Biol. 2015, 57, 673–687. [Google Scholar] [CrossRef]
- Elasad, M.; Ondati, E.; Wei, H.; Wang, H.; Su, J.; Fan, S.; Pang, C.; Yu, S. Functional analysis of nine cotton genes related to leaf senescence in Gossypium hirsutum L. Physiol. Mol. Biol. Plants 2018, 24, 729–739. [Google Scholar] [CrossRef]
- Guo, Y.; Gan, S. AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J. 2006, 46, 601–612. [Google Scholar] [CrossRef]
- Raman, S.; Greb, T.; Peaucelle, A.; Blein, T.; Laufs, P.; Theres, K. Interplay of miR164, CUP-SHAPED COTYLEDON genes and LATERAL SUPPRESSOR controls axillary meristem formation in Arabidopsis thaliana. Plant J. 2008, 55, 65–76. [Google Scholar] [CrossRef]
- Fang, Y.; Xie, K.; Xiong, L. Conserved miR164-targeted NAC genes negatively regulate drought resistance in rice. J. Exp. Bot. 2014, 65, 2119–2135. [Google Scholar] [CrossRef] [Green Version]
- Geng, Y.; Jian, C.; Xu, W.; Liu, H.; Hao, C.; Hou, J.; Liu, H.; Zhang, X.; Li, T. miR164-targeted TaPSK5 encodes a phytosulfokine precursor that regulates root growth and yield traits in common wheat (Triticum aestivum L.). Plant Mol. Biol. 2020, 104, 615–628. [Google Scholar] [CrossRef]
- Li, M.; Liang, Z.; Zeng, Y.; Jing, Y.; Wu, K.; Liang, J.; He, S.; Wang, G.; Mo, Z.; Tan, F.; et al. De novo analysis of transcriptome reveals genes associated with leaf abscission in sugarcane (Saccharum officinarum L.). BMC Genom. 2016, 17, 195. [Google Scholar] [CrossRef] [Green Version]
- Corbacho, J.; Romojaro, F.; Pech, J.C.; Latche, A.; Gomez-Jimenez, M.C. Transcriptomic events involved in melon mature-fruit abscission comprise the sequential induction of cell-wall degrading genes coupled to a stimulation of endo and exocytosis. PLoS ONE 2013, 8, e58363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrero, S.; Carretero-Paulet, L.; Mendes, M.A.; Botton, A.; Eccher, G.; Masiero, S.; Colombo, L. Transcriptomic signatures in seeds of apple (Malus domestica L. Borkh) during fruitlet abscission. PLoS ONE 2015, 10, e120503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gil-Amado, J.A.; Gomez-Jimenez, M.C. Transcriptome Analysis of Mature Fruit Abscission Control in Olive. Plant Cell Physiol. 2013, 54, 244–269. [Google Scholar] [CrossRef] [Green Version]
- Lee, Y.; Chang, J. Leafless Inflorescence Produces More Female Flowers and Fruit Yield Than Leafy Inflorescence in ‘Yu Her Pau’ Litchi. Hortscience 2019, 54, 487–491. [Google Scholar] [CrossRef] [Green Version]
- Chang, J.; Chang, Y.; Tang, L.; Chang, J. Characterization of generative development in early maturing litchi ‘Early Big’, a novel cultivar in Taiwan. Fruits 2015, 70, 289–296. [Google Scholar] [CrossRef] [Green Version]
- Xiao, Q.; Su, Z.; Chen, H.; Shen, J. Genome-wide identification and involvement of litchi SPL genes in flowering in response to cold and leaf maturity. J. Hortic. Sci. Biotechnol. 2019, 94, 428–440. [Google Scholar] [CrossRef]
- Chen, P.; Roan, S.; Lee, C.; Chen, I. Temperature model of litchi flowering-From induction to anthesis. Sci. Hortic.-Amst. 2016, 205, 106–111. [Google Scholar] [CrossRef]
- Liu, W.W.; Chen, H.B.; Lu, X.Y.; Rahman, M.J.; Zhong, S.; Zhou, B.Y. Identification of nitric oxide responsive genes in the floral buds of Litchi chinensis. Biol. Plant. 2015, 59, 115–122. [Google Scholar] [CrossRef]
- Yang, H.F.; Lu, X.Y.; Chen, H.B.; Wang, C.C.; Zhou, B.Y. Low temperature-induced leaf senescence and the expression of senescence-related genes in the panicles of Litchi chinensis. Biol. Plant. 2017, 61, 315–322. [Google Scholar] [CrossRef]
- Li, C.; Wang, Y.; Huang, X.; Li, J.; Wang, H.; Li, J. An improved fruit transcriptome and the identification of the candidate genes involved in fruit abscission induced by carbohydrate stress in litchi. Front. Plant Sci. 2015, 6, 439. [Google Scholar] [CrossRef] [Green Version]
- Huang, L.J.; Chen, H.B.; Hu, Z.Q.; Lu, X.Y.; Wang, H.Y.; Liu, H.; Zhou, B.Y. Comparative proteomics of phloem exudates reveals long-distance signals potentially involved in Litchi chinensis flowering. Biol. Plant. 2020, 64, 220–224. [Google Scholar] [CrossRef]
- Wang, F.; Zheng, Z.; Yuan, Y.; Li, J.; Zhao, M. Identification and Characterization of HAESA-Like Genes Involved in the Fruitlet Abscission in Litchi. Int. J. Mol. Sci. 2019, 20, 5945. [Google Scholar] [CrossRef] [Green Version]
- Zhao, M.; Li, C.; Ma, X.; Xia, R.; Chen, J.; Liu, X.; Ying, P.; Peng, M.; Wang, J.; Shi, C.; et al. KNOX protein KNAT1 regulates fruitlet abscission in litchi by repressing ethylene biosynthetic genes. J. Exp. Bot. 2020, 71, 4069–4082. [Google Scholar] [CrossRef] [PubMed]
- Jiang, G.; Li, Z.; Song, Y.; Zhu, H.; Lin, S.; Huang, R.; Jiang, Y.; Duan, X. LcNAC13 Physically Interacts with LcR1MYB1 to Coregulate Anthocyanin Biosynthesis-Related Genes during Litchi Fruit Ripening. Biomolecules 2019, 9, 135. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jiang, G.; Yan, H.; Wu, F.; Zhang, D.; Zeng, W.; Qu, H.; Chen, F.; Tan, L.; Duan, X.; Jiang, Y. Litchi Fruit LcNAC1 is a Target of LcMYC2 and Regulator of Fruit Senescence Through its Interaction with LcWRKY1. Plant Cell Physiol. 2017, 58, 1075–1089. [Google Scholar] [CrossRef]
- Liu, H.; Wang, C.; Chen, H.; Zhou, B. Genome-wide transcriptome analysis reveals the molecular mechanism of high temperature-induced floral abortion in Litchi chinensis. BMC Genom. 2019, 20, 127. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.; Feng, J.; Xiang, X.; Wang, J.; Salojärvi, J.; Liu, C.; Wu, Z.; Zhang, J.; Liang, X.; Jiang, Z.; et al. Two divergent haplotypes from a highly heterozygous lychee genome suggest independent domestication events for early and late-maturing cultivars. Nat. Genet. 2022, 54, 73–83. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Anderson, J.B.; Cherukuri, P.F.; Deweese-Scott, C.; Geer, L.Y.; Gwadz, M.; He, S.; Hurwitz, D.I.; Jackson, J.D.; Ke, Z.; et al. CDD: A Conserved Domain Database for protein classification. Nucleic Acids Res. 2005, 33, D192–D196. [Google Scholar] [CrossRef]
- Marchler-Bauer, A.; Lu, S.; Anderson, J.B.; Chitsaz, F.; Derbyshire, M.K.; Deweese-Scott, C.; Fong, J.H.; Geer, L.Y.; Geer, R.C.; Gonzales, N.R.; et al. CDD: A Conserved Domain Database for the functional annotation of proteins. Nucleic Acids Res. 2011, 39, D225–D229. [Google Scholar] [CrossRef] [Green Version]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef]
- Punta, M.; Coggill, P.C.; Eberhardt, R.Y.; Mistry, J.; Tate, J.; Boursnell, C.; Pang, N.; Forslund, K.; Ceric, G.; Clements, J.; et al. The Pfam protein families database. Nucleic Acids Res. 2012, 40, D290–D301. [Google Scholar] [CrossRef] [Green Version]
- Duvaud, S.; Gabella, C.; Lisacek, F.; Stockinger, H.; Ioannidis, V.; Durinx, C. Expasy, the Swiss Bioinformatics Resource Portal, as designed by its users. Nucleic Acids Res. 2021, 49, W216–W227. [Google Scholar] [CrossRef] [PubMed]
- Gasteiger, E. ExPASy: The proteomics server for in-depth protein knowledge and analysis. Nucleic Acids Res. 2003, 31, 3784–3788. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Subramanian, B.; Gao, S.; Lercher, M.J.; Hu, S.; Chen, W. Evolview v3: A webserver for visualization, annotation, and management of phylogenetic trees. Nucleic Acids Res. 2019, 47, W270–W275. [Google Scholar] [CrossRef] [PubMed]
- Hu, W.; Wei, Y.; Xia, Z.; Yan, Y.; Hou, X.; Zou, M.; Lu, C.; Wang, W.; Peng, M. Genome-Wide Identification and Expression Analysis of the NAC Transcription Factor Family in Cassava. PLoS ONE 2015, 10, e136993. [Google Scholar] [CrossRef] [PubMed]
- Voorrips, R.E. MapChart: Software for the graphical presentation of linkage maps and QTLs. J. Hered. 2002, 93, 77–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Li, J.; Paterson, A.H. MCScanX-transposed: Detecting transposed gene duplications based on multiple colinearity scans. Bioinformatics 2013, 29, 1458–1460. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Debarry, J.D.; Tan, X.; Li, J.; Wang, X.; Lee, T.H.; Jin, H.; Marler, B.; Guo, H.; et al. MCScanX: A toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 2012, 40, e49. [Google Scholar] [CrossRef] [Green Version]
- Chen, C.; Chen, H.; Zhang, Y.; Thomas, H.R.; Frank, M.H.; He, Y.; Xia, R. TBtools: An Integrative Toolkit Developed for Interactive Analyses of Big Biological Data. Mol. Plant 2020, 13, 1194–1202. [Google Scholar] [CrossRef]
- Lescot, M.; Dehais, P.; Thijs, G.; Marchal, K.; Moreau, Y.; Van de Peer, Y.; Rouze, P.; Rombauts, S. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002, 30, 325–327. [Google Scholar] [CrossRef]
- Guo, H.; Zhang, W.; Ni, C.; Cai, Z.; Chen, S.; Huang, X. Heat map visualization for electrocardiogram data analysis. BMC Cardiovasc. Disor. 2020, 20, 277. [Google Scholar] [CrossRef]
- Ning, W.; Wei, Y.; Gao, L.; Han, C.; Gou, Y.; Fu, S.; Liu, D.; Zhang, C.; Huang, X.; Wu, S.; et al. Hemi 2.0: An online service for heatmap illustration. Nucleic Acids Res. 2022, 50, W405–W411. [Google Scholar] [CrossRef]
- Singh, A.; Pandey, G.K. Primer Design Using Primer Express® for SYBR Green-Based Quantitative PCR.; Springer: New York, NY, USA, 2015; Volume 1275, pp. 153–164. ISBN 1064-3745. [Google Scholar]
- Lu, X.; Kim, H.; Zhong, S.; Chen, H.; Hu, Z.; Zhou, B. De novo transcriptome assembly for rudimentary leaves in Litchi chinesis Sonn. and identification of differentially expressed genes in response to reactive oxygen species. BMC Genom. 2014, 15, 805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, X.; Li, J.; Chen, H.; Hu, J.; Liu, P.; Zhou, B. RNA-seq analysis of apical meristem reveals integrative regulatory network of ROS and chilling potentially related to flowering in Litchi chinensis. Sci. Rep. 2017, 7, 15619. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Kang, H.; Su, C.; Qi, Y.; Liu, X.; Pu, J. Genome-wide identification and expression profile analysis of the NAC transcription factor family during abiotic and biotic stress in woodland strawberry. PLoS ONE 2018, 13, e197892. [Google Scholar] [CrossRef] [PubMed]
- Mohanta, T.K.; Yadav, D.; Khan, A.; Hashem, A.; Tabassum, B.; Khan, A.L.; Abd, A.E.; Al-Harrasi, A. Genomics, molecular and evolutionary perspective of NAC transcription factors. PLoS ONE 2020, 15, e231425. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gong, X.; Zhao, L.; Song, X.; Lin, Z.; Gu, B.; Yan, J.; Zhang, S.; Tao, S.; Huang, X. Genome-wide analyses and expression patterns under abiotic stress of NAC transcription factors in white pear (Pyrus bretschneideri). BMC Plant Biol. 2019, 19, 161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, N.; Zheng, Y.; Xin, H.; Fang, L.; Li, S. Comprehensive analysis of NAC domain transcription factor gene family in Vitis vinifera. Plant Cell Rep. 2013, 32, 61–75. [Google Scholar] [CrossRef] [PubMed]
- Kou, X.; Zhou, J.; Wu, C.E.; Yang, S.; Liu, Y.; Chai, L.; Xue, Z. The interplay between ABA/ethylene and NAC TFs in tomato fruit ripening: A review. Plant Mol. Biol. 2021, 106, 223–238. [Google Scholar] [CrossRef]
- Liu, G.S.; Li, H.L.; Grierson, D.; Fu, D.Q. NAC Transcription Factor Family Regulation of Fruit Ripening and Quality: A Review. Cells 2022, 11, 525. [Google Scholar] [CrossRef] [PubMed]
- Meng, X.; Liu, S.; Zhang, C.; He, J.; Ma, D.; Wang, X.; Dong, T.; Guo, F.; Cai, J.; Long, T.; et al. The unique sweet potato NAC transcription factor IbNAC3 modulates combined salt and drought stresses. Plant Physiol. 2023, 191, 747–771. [Google Scholar] [CrossRef] [PubMed]
- Thirumalaikumar, V.P.; Devkar, V.; Mehterov, N.; Ali, S.; Ozgur, R.; Turkan, I.; Mueller-Roeber, B.; Balazadeh, S. NAC transcription factor JUNGBRUNNEN1 enhances drought tolerance in tomato. Plant Biotechnol. J. 2018, 16, 354–366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, Y.; Yamaguchi, M.; Endo, H.; Rejab, N.A.; Ohtani, M. NAC-MYB-based transcriptional regulation of secondary cell wall biosynthesis in land plants. Front. Plant Sci. 2015, 6, 288. [Google Scholar] [CrossRef] [Green Version]
- Zhao, S.; Jiang, T.; Zhang, Y.; Zhang, K.; Feng, K.; Wu, P.; Li, L. Identification of the NAC Transcription Factors and Their Function in ABA and Salinity Response in Nelumbo nucifera. Int. J. Mol. Sci. 2022, 23, 12394. [Google Scholar] [CrossRef] [PubMed]
- Kim, H.J.; Nam, H.G.; Lim, P.O. Regulatory network of NAC transcription factors in leaf senescence. Curr. Opin. Plant Biol. 2016, 33, 48–56. [Google Scholar] [CrossRef]
- Wang, C.; Liu, H.; Huang, L.; Chen, H.; Lu, X.; Zhou, B. LcNAC13 Is Involved in the Reactive Oxygen Species-Dependent Senescence of the Rudimentary Leaves in Litchi chinensis. Front. Plant Sci. 2022, 13, 886131. [Google Scholar] [CrossRef]
- Yang, H.; Kim, H.; Chen, H.; Lu, Y.; Lu, X.; Wang, C.; Zhou, B. Reactive oxygen species and nitric oxide induce senescence of rudimentary leaves and the expression profiles of the related genes in Litchi chinensis. Hortic. Res. 2018, 5, 23. [Google Scholar] [CrossRef] [Green Version]
- Yi, J.W.; Wang, Y.; Ma, X.S.; Zhang, J.Q.; Zhao, M.L.; Huang, X.M.; Li, J.G.; Hu, G.B.; Wang, H.C. LcERF2 modulates cell wall metabolism by directly targeting a UDP-glucose-4-epimerase gene to regulate pedicel development and fruit abscission of litchi. Plant J. 2021, 106, 801–816. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liao, G.; Duan, Y.; Wang, C.; Zhuang, Z.; Wang, H. Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Gene Family in Litchi chinensis. Genes 2023, 14, 1416. https://doi.org/10.3390/genes14071416
Liao G, Duan Y, Wang C, Zhuang Z, Wang H. Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Gene Family in Litchi chinensis. Genes. 2023; 14(7):1416. https://doi.org/10.3390/genes14071416
Chicago/Turabian StyleLiao, Guihua, Yu Duan, Congcong Wang, Zebin Zhuang, and Haishi Wang. 2023. "Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Gene Family in Litchi chinensis" Genes 14, no. 7: 1416. https://doi.org/10.3390/genes14071416
APA StyleLiao, G., Duan, Y., Wang, C., Zhuang, Z., & Wang, H. (2023). Genome-Wide Identification, Characterization, and Expression Analysis of the NAC Gene Family in Litchi chinensis. Genes, 14(7), 1416. https://doi.org/10.3390/genes14071416




