Genetic Polymorphism and Population Genetic Structure Analysis of 21 Autosomal STR Loci for a Han-Chinese Population from Luzhou of Southwest China
Abstract
1. Introduction
2. Materials and Methods
2.1. Regents
2.2. Samples Collection and Genomic DNA Extraction
2.3. PCR Amplification and Autosomal STR Genotyping
2.4. Data Analysis
3. Results
3.1. Linkage Disequilibrium, Allele Frequencies, and Forensic Parameters
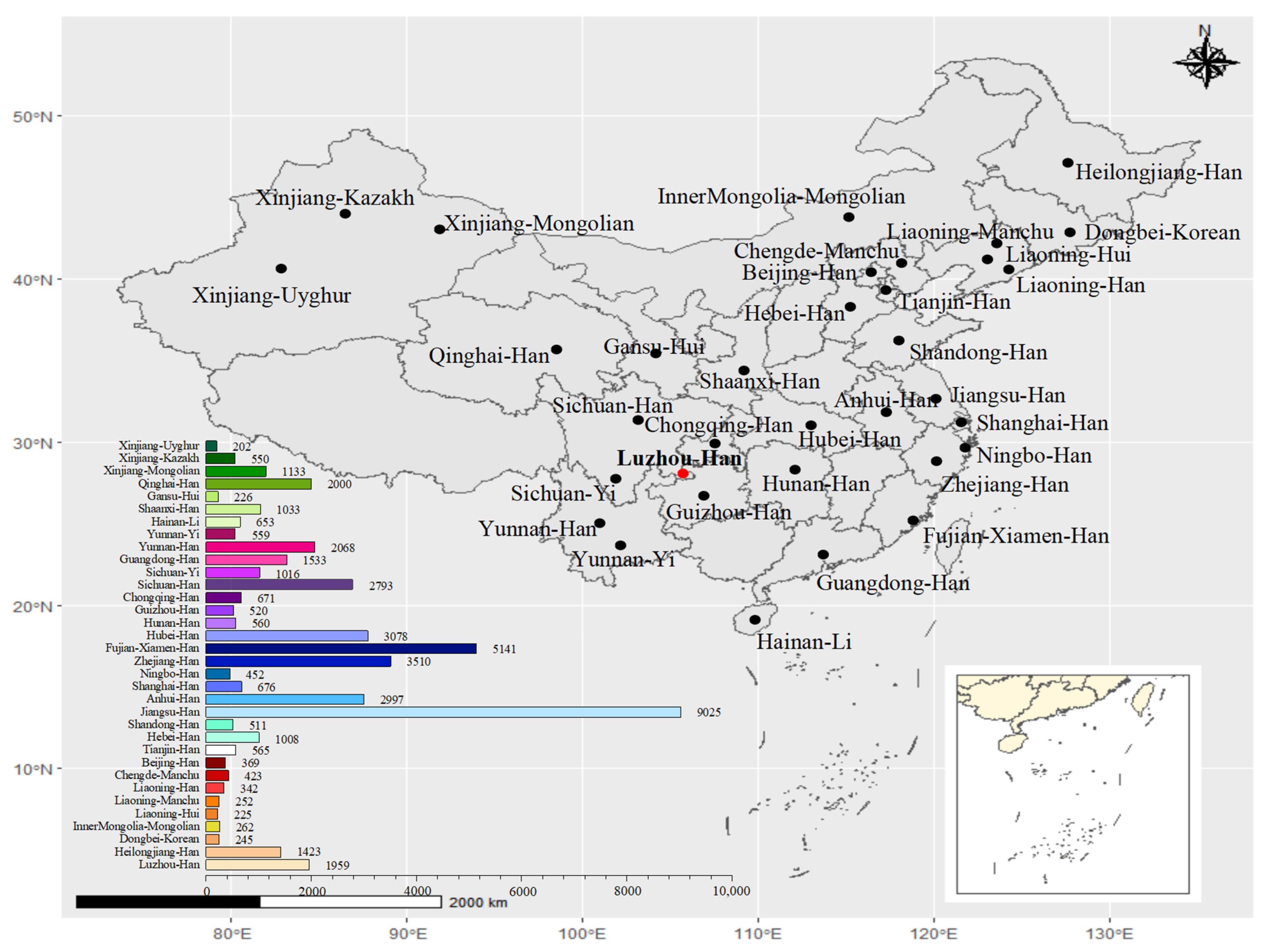
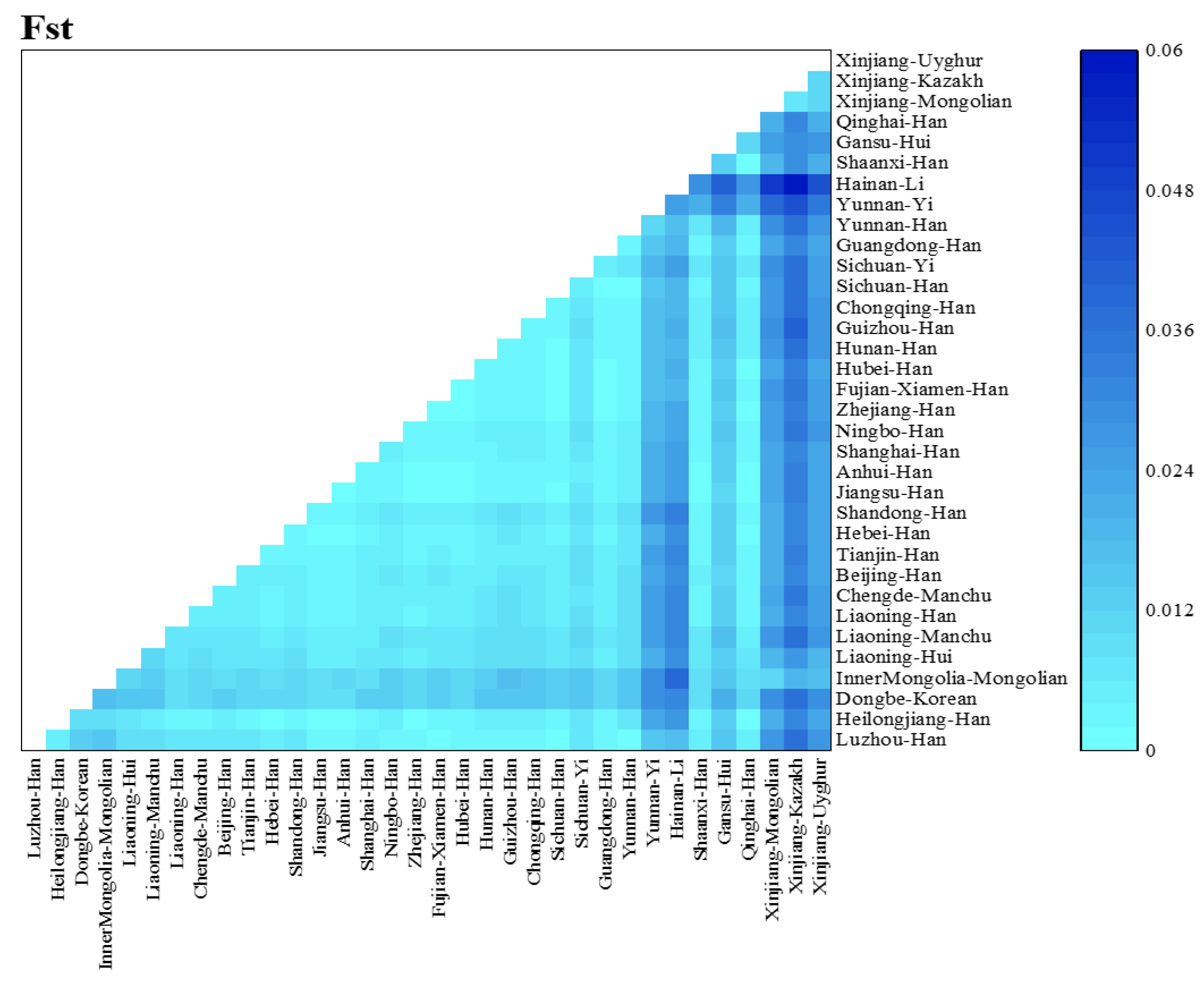
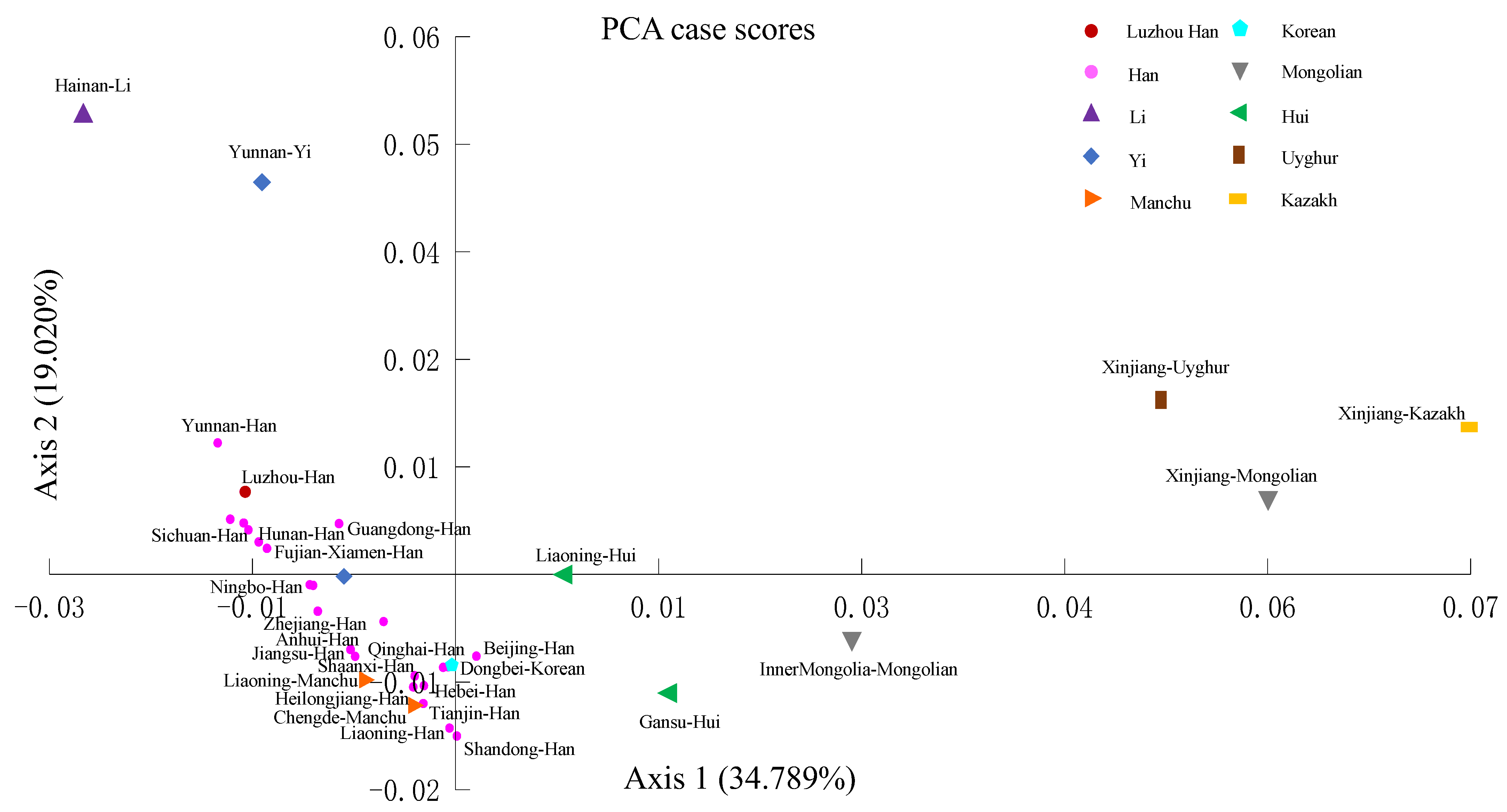
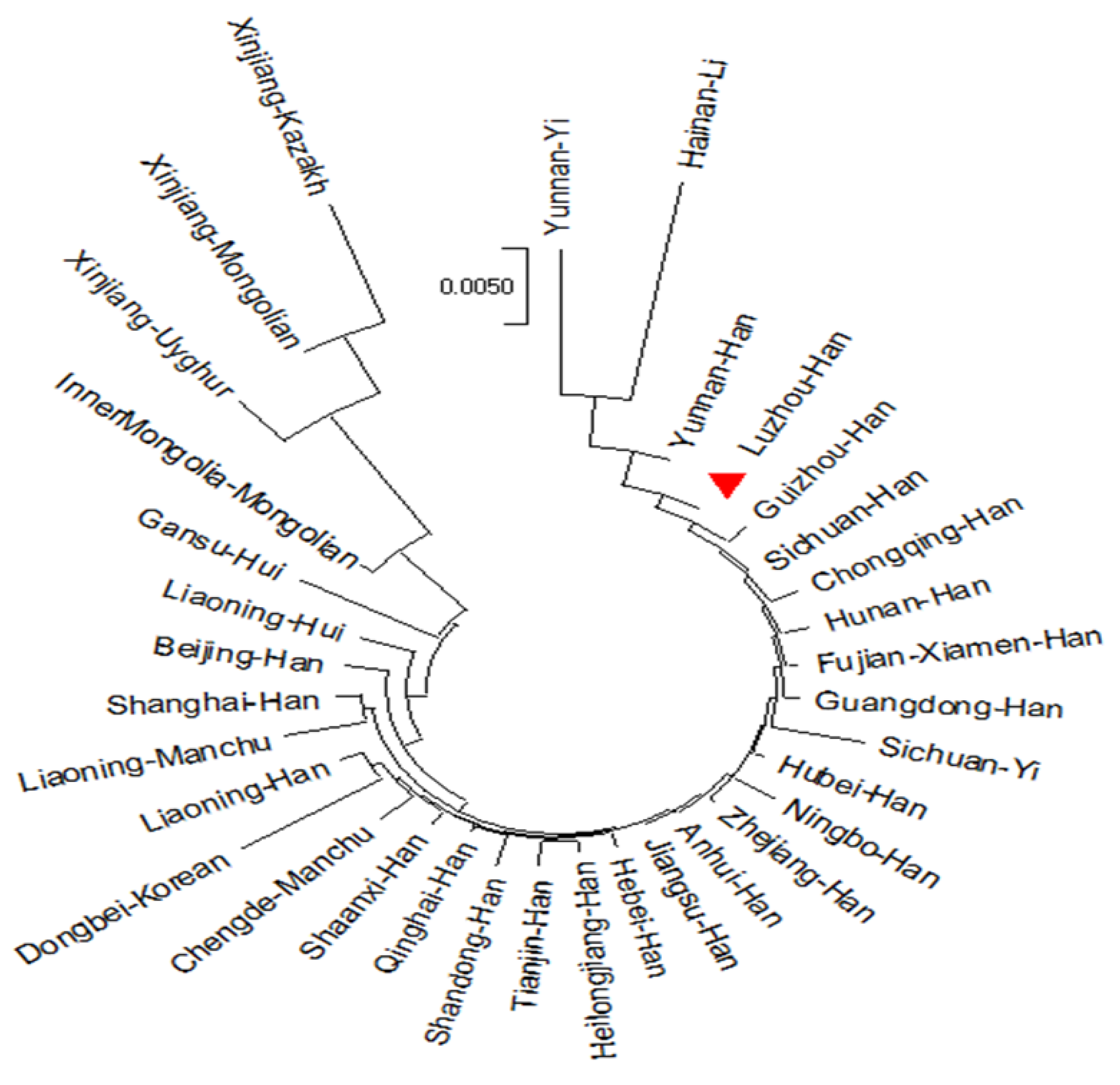
3.2. Population Structure and Population Comparisons
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- McCord, B.R.; Gauthier, Q.; Cho, S.; Roig, M.N.; Gibson-Daw, G.C.; Young, B.; Taglia, F.; Zapico, S.C.; Mariot, R.F.; Lee, S.B.; et al. Forensic DNA Analysis. Anal. Chem. 2019, 91, 673–688. [Google Scholar] [CrossRef]
- Jager, R. New Perspectives for Whole Genome Amplification in Forensic STR Analysis. Int. J. Mol. Sci. 2022, 23, 7090. [Google Scholar] [CrossRef] [PubMed]
- Wyner, N.; Barash, M.; McNevin, D. Forensic Autosomal Short Tandem Repeats and Their Potential Association With Phenotype. Front. Genet. 2020, 11, 884. [Google Scholar] [CrossRef] [PubMed]
- Al-Hazani, T.M.I.; Al-Qahtani, W.S.; Abboosh, T.S.; Safhi, F.A.; Alshaya, D.S.; Jalal, A.S.; Al-Shamrani, S.M.; Al-Ghamdi, N.A.; Alotaibi, A.M.; Alotaibi, M.A.; et al. Detecting STR profiles from degrading menstrual blood samples and their use as possible evidence in forensic investigations. Forensic Sci. Int. 2023, 343, 111562. [Google Scholar] [CrossRef]
- Shi, Y.; Niu, Y.; Zhang, P.; Luo, H.; Liu, S.; Zhang, S.; Wang, J.; Li, Y.; Liu, X.; Song, T.; et al. Characterization of genome-wide STR variation in 6487 human genomes. Nat. Commun. 2023, 14, 2092. [Google Scholar] [CrossRef]
- Bodner, M.; Parson, W. The STRidER Report on Two Years of Quality Control of Autosomal STR Population Datasets. Genes 2020, 11, 901. [Google Scholar] [CrossRef]
- Bhambara, A.K.; Singh, A.; Sahajpal, V.; Thakur, M.; Bhandari, D.; Sharma, S.; Thakar, M.K. Evaluation of genetic polymorphisms at 21 autosomal STR loci in Ramgharia Sikh population of Punjab, India. Ann. Hum. Biol. 2022, 49, 263–268. [Google Scholar] [CrossRef] [PubMed]
- Shafique, M.; Sajid, N.; Aziz, Y.; Afzal, M.; Shahzad, M.; Shehzadi, A.; Bashir, S.; Daud, S.; Ali Shahid, A. Genetic data of 22 autosomal STR loci in Khyber-Pakhtunkhwa, Balochistan and Gilgit Baltistan population of Pakistan using PowerPlex(R) fusion system. Leg. Med. 2022, 59, 102129. [Google Scholar] [CrossRef] [PubMed]
- Preet, K.; Malhotra, S.; Shrivastava, P.; Jain, T.; Rawat, S.; Varte, L.R.; Singh, S.; Singh, I.; Sarkar, S. Genetic Diversity in Gorkhas: An Autosomal STR Study. Sci. Rep. 2016, 6, 32494. [Google Scholar] [CrossRef]
- Dung Pham, P.; Luc Hoang, T.; Tra Le, K.; Thi Le, P.; Ngoc Nguyen, N.; Linh Tran, H.; Hung Nguyen, M.; Minh Tran, D.; Hoang, H. The first data of allele frequencies for 23 autosomal STRs in the Ede ethnic group in Vietnam. Leg. Med. 2022, 57, 102072. [Google Scholar] [CrossRef]
- Srithawong, S.; Muisuk, K.; Srikummool, M.; Kampuansai, J.; Pittayaporn, P.; Ruangchai, S.; Liu, D.; Kutanan, W. Close genetic relationship between central Thai and Mon people in Thailand revealed by autosomal microsatellites. Int. J. Leg. Med. 2021, 135, 445–448. [Google Scholar] [CrossRef]
- Azeelah, A.N.; Zafarina, Z. Population data for 15 autosomal STR loci in Orang Asli subgroups of Peninsular Malaysia. Int. J. Leg. Med. 2022, 136, 547–549. [Google Scholar] [CrossRef] [PubMed]
- Hakim, H.M.; Khan, H.O.; Lalung, J.; Nelson, B.R.; Chambers, G.K.; Edinur, H.A. Autosomal STR Profiling and Databanking in Malaysia: Current Status and Future Prospects. Genes 2020, 11, 1112. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.; Oldt, R.F.; McCulloh, K.L.; Weise, J.A.; Viray, J.; Budowle, B.; Smith, D.G.; Kanthaswamy, S. Native American population data based on the Globalfiler((R)) autosomal STR loci. Forensic Sci. Int. Genet. 2016, 24, e12–e13. [Google Scholar] [CrossRef] [PubMed]
- McCulloh, K.L.; Ng, J.; Oldt, R.F.; Weise, J.A.; Viray, J.; Budowle, B.; Glenn Smith, D.; Kanthaswamy, S. The genetic structure of native Americans in North America based on the Globalfiler(R) STRs. Leg. Med. 2016, 23, 49–54. [Google Scholar] [CrossRef] [PubMed]
- Ng, J.; Oldt, R.F.; Kanthaswamy, S. Assessing the FBI’s Native American STR database for random match probability calculations. Leg. Med. 2018, 30, 52–55. [Google Scholar] [CrossRef]
- Lu, X.; Yang, S.; Jiang, Y.; Li, K.; Zhao, F.; Yang, X.; Zhang, N.; Zhang, S.; Xia, S.; Wu, L.; et al. Genetic variation of 20 autosomal STR loci in the Han nationality in Central Yunnan, Southwest China. Leg. Med. 2021, 48, 101807. [Google Scholar] [CrossRef]
- He, G.; Wang, M.; Liu, J.; Hou, Y.; Wang, Z. Forensic features and phylogenetic analyses of Sichuan Han population via 23 autosomal STR loci included in the Huaxia Platinum System. Int. J. Leg. Med. 2018, 132, 1079–1082. [Google Scholar] [CrossRef]
- Zou, X.; Li, Y.; Li, P.; Nie, Q.; Wang, T.; Hu, Y.; Zhu, Y.; Li, J.; Tang, R. Genetic polymorphisms for 19 autosomal STR loci of Chongqing Han ethnicity and phylogenetic structure exploration among 28 Chinese populations. Int. J. Leg. Med. 2017, 131, 1539–1542. [Google Scholar] [CrossRef]
- Yang, M.; Ren, Z.; Ji, J.; Zhou, H.; Zhang, H.; Dai, J.; Wang, J.; Huang, J. Population genetic data and mutations of 22 autosomal STR loci in Guizhou Han population. Forensic Sci. Int. Genet. 2017, 29, e29–e30. [Google Scholar] [CrossRef]
- Jin, B.; Su, Q.; Luo, H.; Li, Y.; Wu, J.; Yan, J.; Hou, Y.; Liang, W.; Zhang, L. Mutational analysis of 33 autosomal short tandem repeat (STR) loci in southwest Chinese Han population based on trio parentage testing. Forensic Sci. Int. Genet. 2016, 23, 86–90. [Google Scholar] [CrossRef]
- Fu, J.; Cheng, J.; Wei, C.; Khan, M.A.; Jin, Z.; Fu, J. Assessing 23 Y-STR loci mutation rates in Chinese Han father-son pairs from southwestern China. Mol. Biol. Rep. 2020, 47, 7755–7760. [Google Scholar] [CrossRef]
- Fu, J.; Fu, S.; Yin, S.; Cheng, J.; Liu, X.; Jin, Z.; He, T.; Fu, J. Technical note: Multi-alleles at the DYS385ab locus with high frequency in a Han Chinese population from southwestern China. Int. J. Leg. Med. 2021, 135, 1737–1741. [Google Scholar] [CrossRef] [PubMed]
- Walsh, P.S.; Metzger, D.A.; Higushi, R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques 2013, 54, 134–139. [Google Scholar] [CrossRef]
- Fu, J.; Shen, S.; Cheng, J.; Lv, H.; Fu, J. A case of Usher syndrome type IIA caused by a rare USH2A homozygous frameshift variant with maternal uniparental disomy (UPD) in a Chinese family. J. Cell. Mol. Med. 2020, 24, 7743–7750. [Google Scholar] [CrossRef]
- Olaisen, B.; Bar, W.; Brinkmann, B.; Budowle, B.; Carracedo, A.; Gill, P.; Lincoln, P.; Mayr, W.R.; Rand, S. DNA recommendations 1997 of the International Society for Forensic Genetics. Vox Sang. 1998, 74, 61–63. [Google Scholar] [CrossRef] [PubMed]
- Excoffier, L.; Lischer, H.E. Arlequin suite ver 3.5: A new series of programs to perform population genetics analyses under Linux and Windows. Mol. Ecol. Resour. 2010, 10, 564–567. [Google Scholar] [CrossRef]
- Zhang, L.; Xu, C.; Chen, H.; Zhou, Y.; Li, F.; Chen, J. Genetic polymorphism of 19 STR loci in Xinjiang Barkol Kazakh population. J. Cent. South Univ. Med. Sci. 2012, 37, 934–938. [Google Scholar] [PubMed]
- Xie, J.; Shao, C.; Zhou, Y.; Zhu, W.; Xu, H.; Liu, Z.; Zhao, Z. Genetic distribution on 20 STR loci from the Han population in Shanghai, China. Forensic Sci. Int. Genet. 2014, 9, e30–e31. [Google Scholar] [CrossRef]
- Ruan, X.; Wang, W.N.; Yang, Y.R.; Xie, B.B.; Chen, J.; Liu, Y.C.; Yan, J.W. Genetic variability and phylogenetic analysis of 39 short tandem repeat loci in Beijing Han population. Hereditas 2015, 37, 683–691. [Google Scholar]
- Xiao, C.; Zhang, W.; Wei, T.; Pan, C.; Huang, D. Population data of 21 autosomal STR loci in Chinese Han population from Hubei province in Central China. Forensic Sci. Int. Genet. 2016, 20, e13–e14. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Li, Y.; Wang, Z.; Liang, W.; Luo, H.; Liao, M.; Zhang, J.; Yan, J.; Li, Y.; Hou, Y.; et al. Genetic diversity of 21 autosomal STR loci in the Han population from Sichuan province, Southwest China. Forensic Sci. Int. Genet. 2017, 31, e33–e35. [Google Scholar] [CrossRef]
- Lu, Y.; Song, P.; Huang, J.C.; Wu, X. Genetic polymorphisms of 20 autosomal STR loci in 5141 individuals from the Han population of Xiamen, Southeast China. Forensic Sci. Int. Genet. 2017, 29, e31–e32. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Shi, K.; Tan, L.; Zhang, Q.; Fu, L.; Zhang, X.; Fu, G.; Li, S.; Cong, B. Analysis of genetic polymorphisms and mutations at 19 STR loci in Hebei Han population. Forensic Sci. Int. Genet. 2017, 31, e50–e51. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Hu, L.; Du, L.; Zheng, H.; Nie, A.; Rao, M.; Bo Pang, J.; Nie, S. Population data for 20 autosomal STR loci in the Yi ethnic minority from Yunnan Province, Southwest China. Forensic Sci. Int. Genet. 2017, 28, e43–e44. [Google Scholar] [CrossRef] [PubMed]
- Yang, L.; Zhang, X.; Zhao, L.; Sun, Y.; Li, J.; Huang, R.; Hu, L.; Nie, S. Population data of 23 autosomal STR loci in the Chinese Han population from Guangdong Province in southern China. Int. J. Leg. Med. 2018, 132, 133–135. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Liu, L.; Xie, R.; Wang, G.; Shi, Y.; Gu, T.; Hu, L.; Nie, S. Population data and mutation rates of 20 autosomal STR loci in a Chinese Han population from Yunnan Province, Southwest China. Int. J. Leg. Med. 2018, 132, 1083–1085. [Google Scholar] [CrossRef]
- Fan, H.; Wang, X.; Ren, Z.; He, G.; Long, R.; Liang, A.; Song, T.; Deng, J. Population data of 19 autosomal STR loci in the Li population from Hainan Province in southernmost China. Int. J. Leg. Med. 2019, 133, 429–431. [Google Scholar] [CrossRef]
- Wang, D.; Zhang, T.; Xu, R.; Dong, Z.; Li, C.; Cui, W. Genetic polymorphisms of 23 STR loci of Goldeneye 25A kit in Shandong Han population. Forensic Sci. Int. Genet. 2019, 40, e256–e258. [Google Scholar] [CrossRef]
- Wang, Y.; Chen, F.; Fu, Y.; Zhang, S.; Zhang, H.; Lai, M.; Xu, E. Genetic polymorphisms of 19 autosomal STR loci in 3510 individuals from Han population of Zhejiang province, Southeast China. Forensic Sci. Int. 2020, 306, 110045. [Google Scholar] [CrossRef]
- Cheng, J.; Song, B.; Fu, J.; Zheng, X.; He, T.; Fu, J. Genetic polymorphism of 19 autosomal STR loci in the Yi ethnic minority of Liangshan Yi autonomous prefecture from Sichuan province in China. Sci. Rep. 2021, 11, 16327. [Google Scholar] [CrossRef] [PubMed]
- Shimada, M.K.; Nishida, T. A modification of the PHYLIP program: A solution for the redundant cluster problem, and an implementation of an automatic bootstrapping on trees inferred from original data. Mol. Phylogenet. Evol. 2017, 109, 409–414. [Google Scholar] [CrossRef] [PubMed]
- Moberly, J.G.; Bernards, M.T.; Waynant, K.V. Key features and updates for origin 2018. J. Cheminform. 2018, 10, 5. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular Evolutionary Genetics Analysis across Computing Platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef]
- Yao, H.; Wang, M.; Zou, X.; Li, Y.; Yang, X.; Li, A.; Yeh, H.Y.; Wang, P.; Wang, Z.; Bai, J.; et al. New insights into the fine-scale history of western-eastern admixture of the northwestern Chinese population in the Hexi Corridor via genome-wide genetic legacy. Mol. Genet. Genom. 2021, 296, 631–651. [Google Scholar] [CrossRef] [PubMed]
- Waples, R.S. Testing for Hardy-Weinberg proportions: Have we lost the plot? J. Hered. 2015, 106, 1–19. [Google Scholar] [CrossRef]
- Ye, Z.; Wang, Z.; Hou, Y. Does Bonferroni correction “rescue” the deviation from Hardy-Weinberg equilibrium? Forensic Sci. Int. Genet. 2020, 46, 102254. [Google Scholar] [CrossRef] [PubMed]

| Loci | Allele Number | Genotype Number | Hexp | Hobs | MP | DP | PIC | PE | TPI | H | h | p |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| D3S1358 | 10 | 30 | 0.72390 | 0.73813 | 0.12732 | 0.72372 | 0.67410 | 0.48968 | 1.90936 | 0.26187 | 0.73813 | 0.15893 |
| D13S317 | 10 | 33 | 0.79143 | 0.78765 | 0.07384 | 0.79122 | 0.76060 | 0.57632 | 2.35457 | 0.21235 | 0.78765 | 0.68060 |
| D7S820 | 17 | 41 | 0.76764 | 0.75447 | 0.08543 | 0.76745 | 0.73458 | 0.51744 | 2.03638 | 0.24553 | 0.75447 | 0.16729 |
| D16S539 | 10 | 34 | 0.77706 | 0.76059 | 0.08369 | 0.77686 | 0.74158 | 0.52806 | 2.08849 | 0.23941 | 0.76059 | 0.07989 |
| Penta E | 29 | 171 | 0.91071 | 0.90812 | 0.01489 | 0.91048 | 0.90394 | 0.81203 | 5.44167 | 0.09188 | 0.90812 | 0.68739 |
| D2S441 | 17 | 50 | 0.78755 | 0.76161 | 0.07683 | 0.78735 | 0.75646 | 0.52984 | 2.09743 | 0.23839 | 0.76161 | 0.00501 |
| TPOX | 7 | 16 | 0.62104 | 0.62379 | 0.20662 | 0.62088 | 0.55691 | 0.32040 | 1.32904 | 0.37621 | 0.62379 | 0.80216 |
| TH01 | 17 | 41 | 0.67218 | 0.67841 | 0.15619 | 0.67201 | 0.62663 | 0.39565 | 1.55476 | 0.32159 | 0.67841 | 0.55690 |
| D2S1338 | 15 | 74 | 0.86107 | 0.85503 | 0.03448 | 0.86085 | 0.84570 | 0.70480 | 3.44894 | 0.14497 | 0.85503 | 0.43972 |
| CSF1PO | 13 | 32 | 0.72433 | 0.71261 | 0.11938 | 0.72414 | 0.67852 | 0.44803 | 1.73979 | 0.28739 | 0.71261 | 0.24061 |
| Penta D | 13 | 49 | 0.80353 | 0.80194 | 0.06140 | 0.80333 | 0.77977 | 0.60265 | 2.52448 | 0.19806 | 0.80194 | 0.85929 |
| D10S1248 | 11 | 39 | 0.75110 | 0.76110 | 0.10113 | 0.75091 | 0.71309 | 0.52895 | 2.09295 | 0.23890 | 0.76110 | 0.30603 |
| D19S433 | 19 | 79 | 0.82256 | 0.83206 | 0.05407 | 0.82235 | 0.80055 | 0.65982 | 2.97720 | 0.16794 | 0.83206 | 0.27134 |
| vWA | 12 | 35 | 0.80033 | 0.81113 | 0.07149 | 0.80013 | 0.77039 | 0.61985 | 2.64730 | 0.18887 | 0.81113 | 0.23203 |
| D21S11 | 26 | 101 | 0.81959 | 0.81674 | 0.05348 | 0.81938 | 0.79758 | 0.63048 | 2.72841 | 0.18326 | 0.81674 | 0.74339 |
| D18S51 | 19 | 94 | 0.86284 | 0.86269 | 0.03449 | 0.86262 | 0.84788 | 0.72001 | 3.64126 | 0.13731 | 0.86269 | 0.98448 |
| D6S1043 | 22 | 95 | 0.87351 | 0.88821 | 0.02993 | 0.87329 | 0.85994 | 0.77140 | 4.47260 | 0.11179 | 0.88821 | 0.05030 |
| D8S1179 | 13 | 49 | 0.84916 | 0.83512 | 0.04099 | 0.84894 | 0.83044 | 0.66576 | 3.03251 | 0.16488 | 0.83512 | 0.08255 |
| D5S818 | 12 | 34 | 0.78003 | 0.76927 | 0.08223 | 0.77983 | 0.74657 | 0.54331 | 2.16704 | 0.23073 | 0.76927 | 0.25014 |
| D12S391 | 15 | 65 | 0.84844 | 0.85860 | 0.04194 | 0.84823 | 0.83019 | 0.71189 | 3.53610 | 0.14140 | 0.85860 | 0.20985 |
| FGA | 26 | 114 | 0.86904 | 0.87034 | 0.03071 | 0.86882 | 0.85515 | 0.73533 | 3.85630 | 0.12966 | 0.87034 | 0.86458 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Song, B.; Fu, J.; Qian, J.; Yang, L.; Cheng, J.; Fu, J. Genetic Polymorphism and Population Genetic Structure Analysis of 21 Autosomal STR Loci for a Han-Chinese Population from Luzhou of Southwest China. Genes 2023, 14, 1419. https://doi.org/10.3390/genes14071419
Song B, Fu J, Qian J, Yang L, Cheng J, Fu J. Genetic Polymorphism and Population Genetic Structure Analysis of 21 Autosomal STR Loci for a Han-Chinese Population from Luzhou of Southwest China. Genes. 2023; 14(7):1419. https://doi.org/10.3390/genes14071419
Chicago/Turabian StyleSong, Binghui, Jiewen Fu, Jie Qian, Lisha Yang, Jingliang Cheng, and Junjiang Fu. 2023. "Genetic Polymorphism and Population Genetic Structure Analysis of 21 Autosomal STR Loci for a Han-Chinese Population from Luzhou of Southwest China" Genes 14, no. 7: 1419. https://doi.org/10.3390/genes14071419
APA StyleSong, B., Fu, J., Qian, J., Yang, L., Cheng, J., & Fu, J. (2023). Genetic Polymorphism and Population Genetic Structure Analysis of 21 Autosomal STR Loci for a Han-Chinese Population from Luzhou of Southwest China. Genes, 14(7), 1419. https://doi.org/10.3390/genes14071419






