Antibiotic-Induced Dysbiosis of the Gut Microbiota Impairs Gene Expression in Gut-Liver Axis of Mice
Abstract
1. Introduction
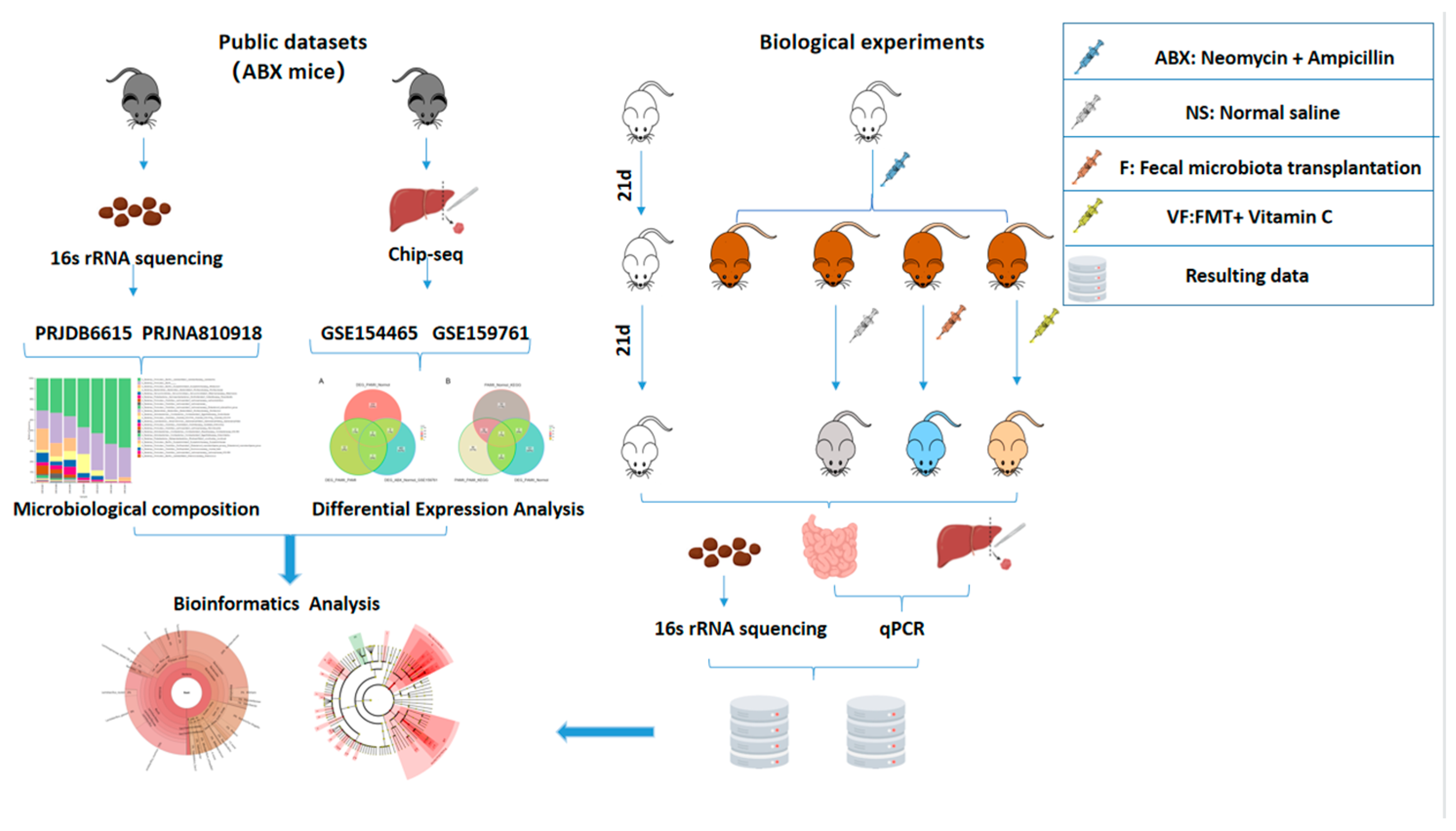
2. Materials and Methods
2.1. Data Sources
2.2. Animals and Antibiotics
2.3. Fecal Microbiota Transplantation
2.4. Conducting 16s rRNA Sequencing
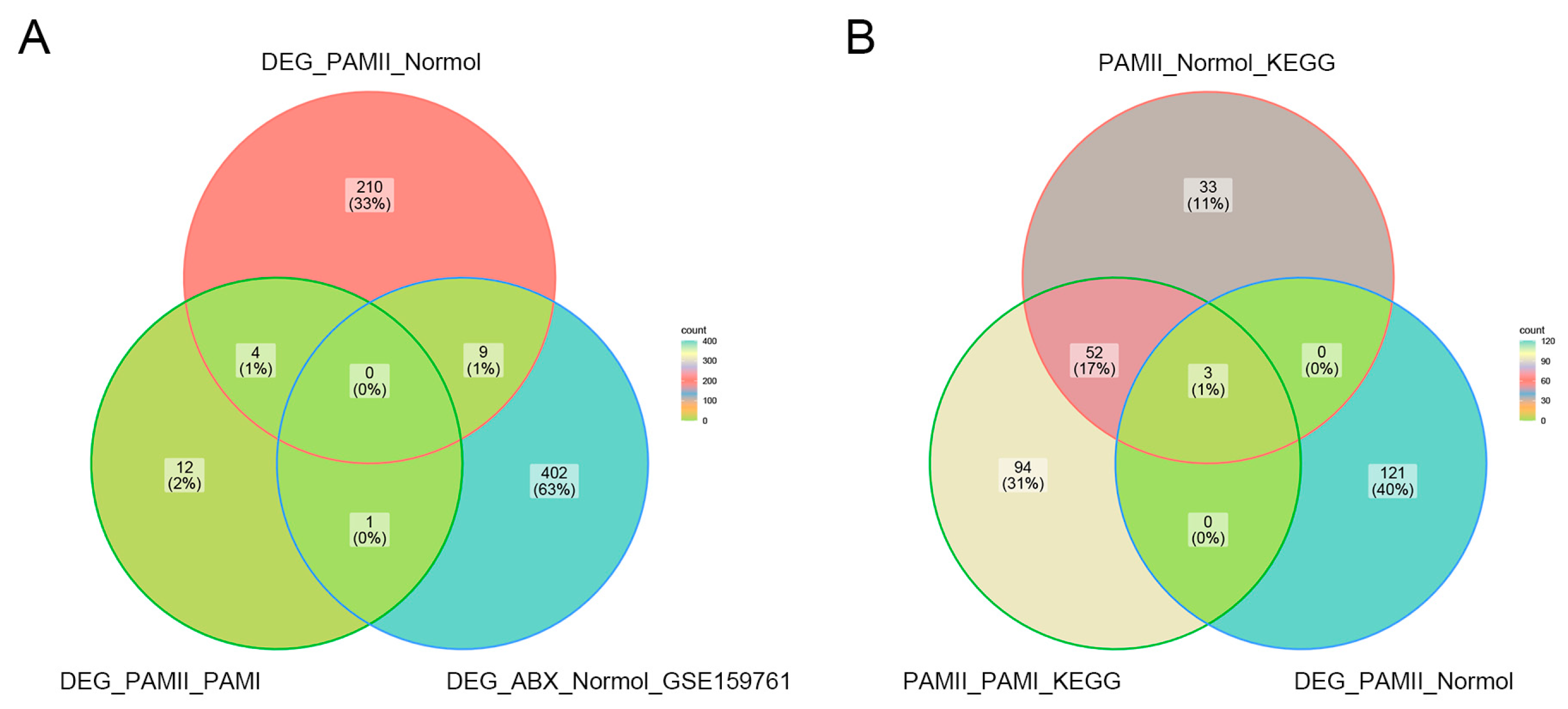
2.5. Screening for Differentially Expressed Genes
2.6. Quantitative Real-Time-PCR
2.7. Bioinformatics Analysis
2.8. Statistical Analysis
3. Results
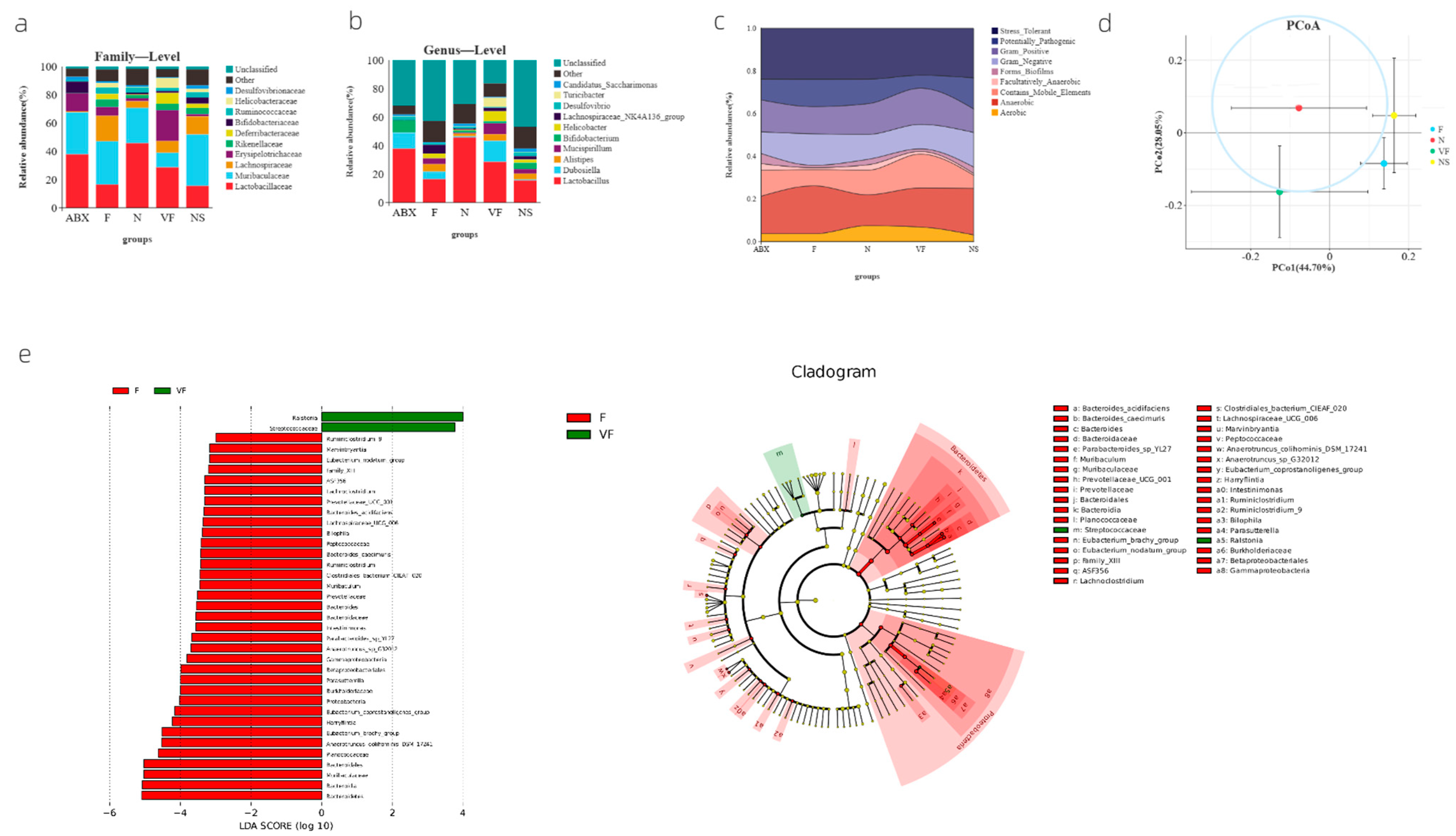
3.1. Significant Changes in Gut Microbiota Composition at the Family and Genus Level in Mice after Oral Antibiotic Dosing
3.2. Weighted UniFrac-Based PCoA Results Show That Vitamin C Contributes to the Recovery of Gut Microbiota to the Donor
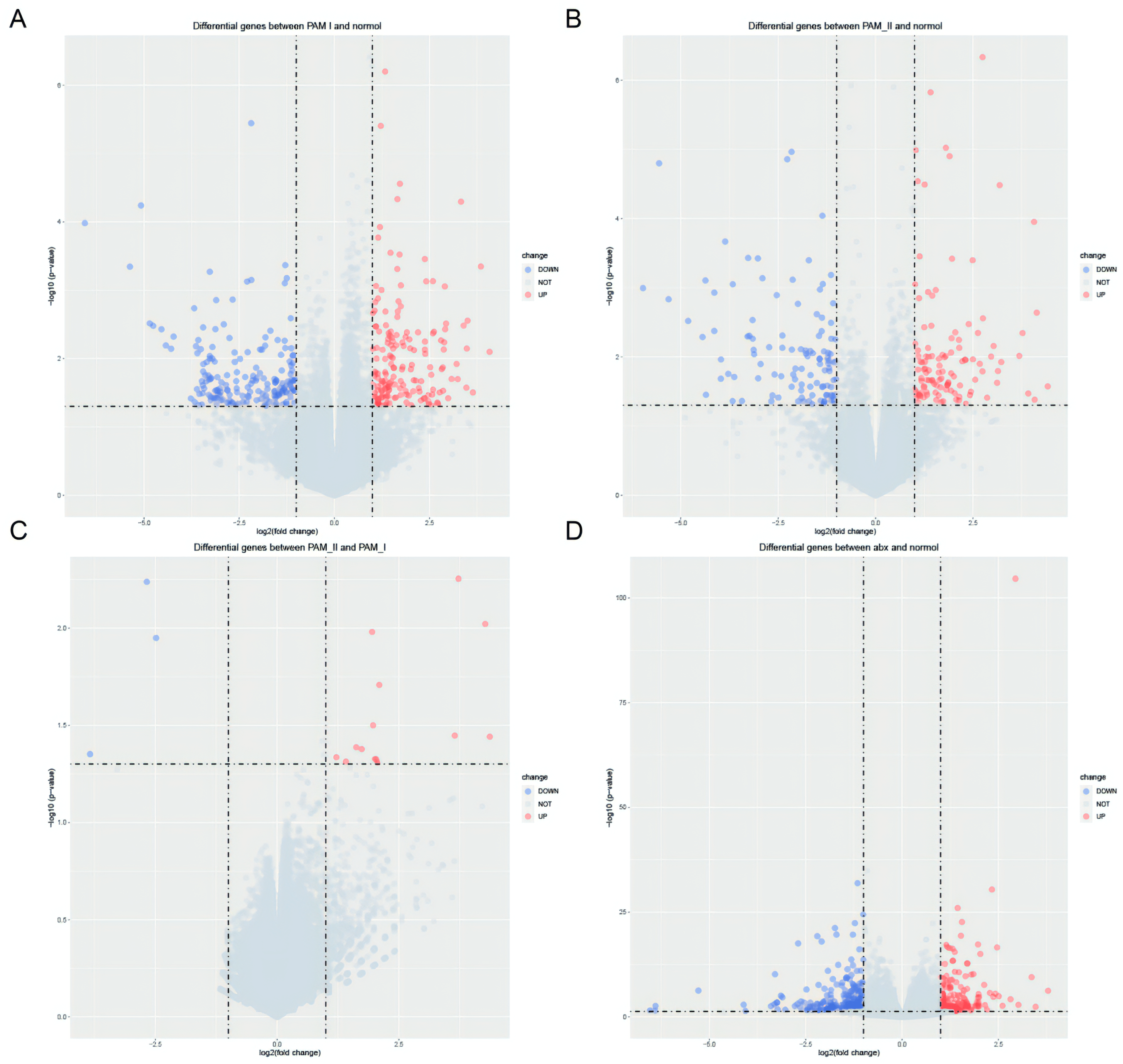
3.3. Screening for Six Differentially Expressed Genes before and after Antibiotic Treatment
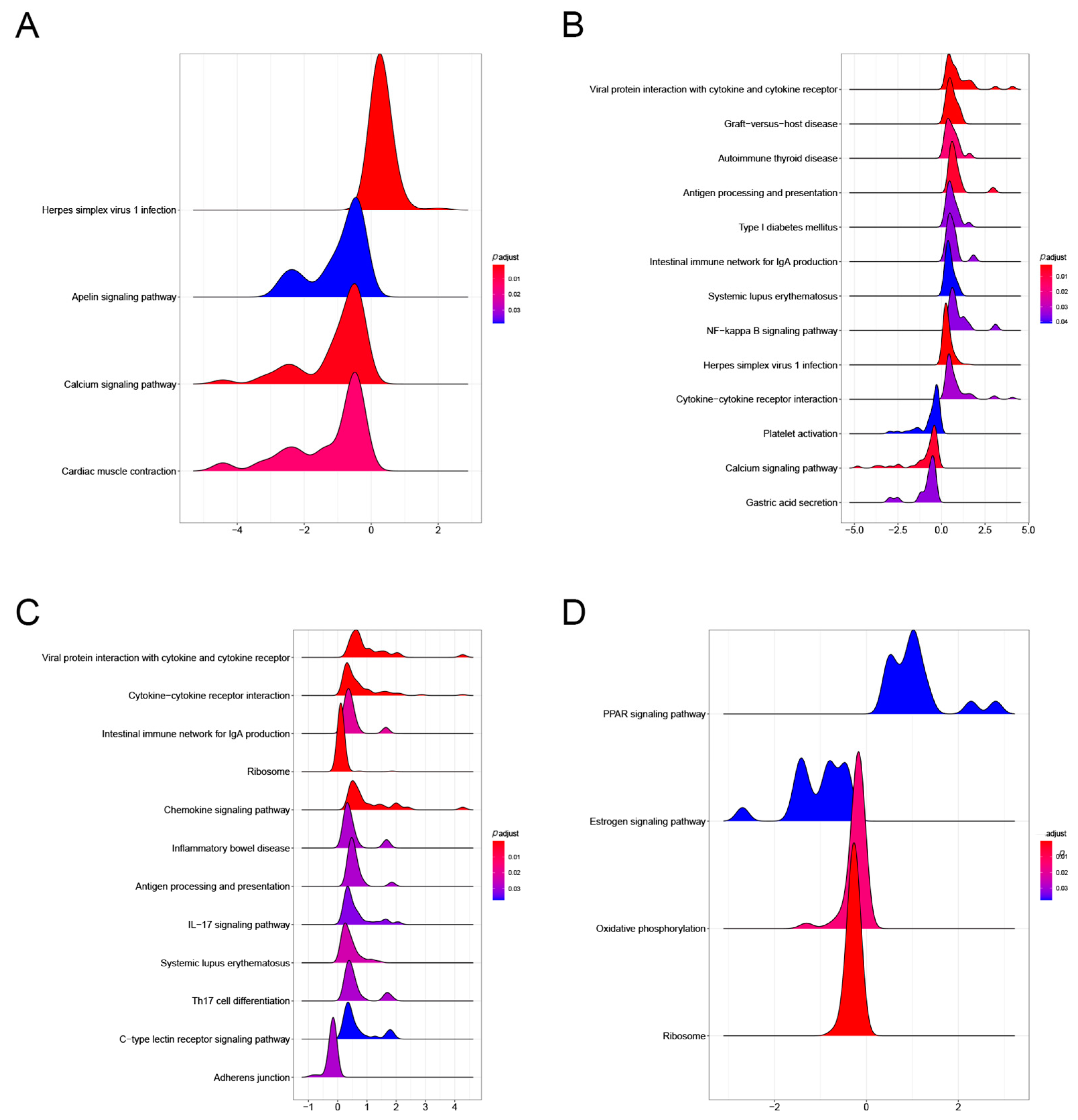
3.4. Gut Microbiota Significantly Affects Gene Expression in the Gut-Liver Axis, which FMT with Vitamin C May Help to Restore
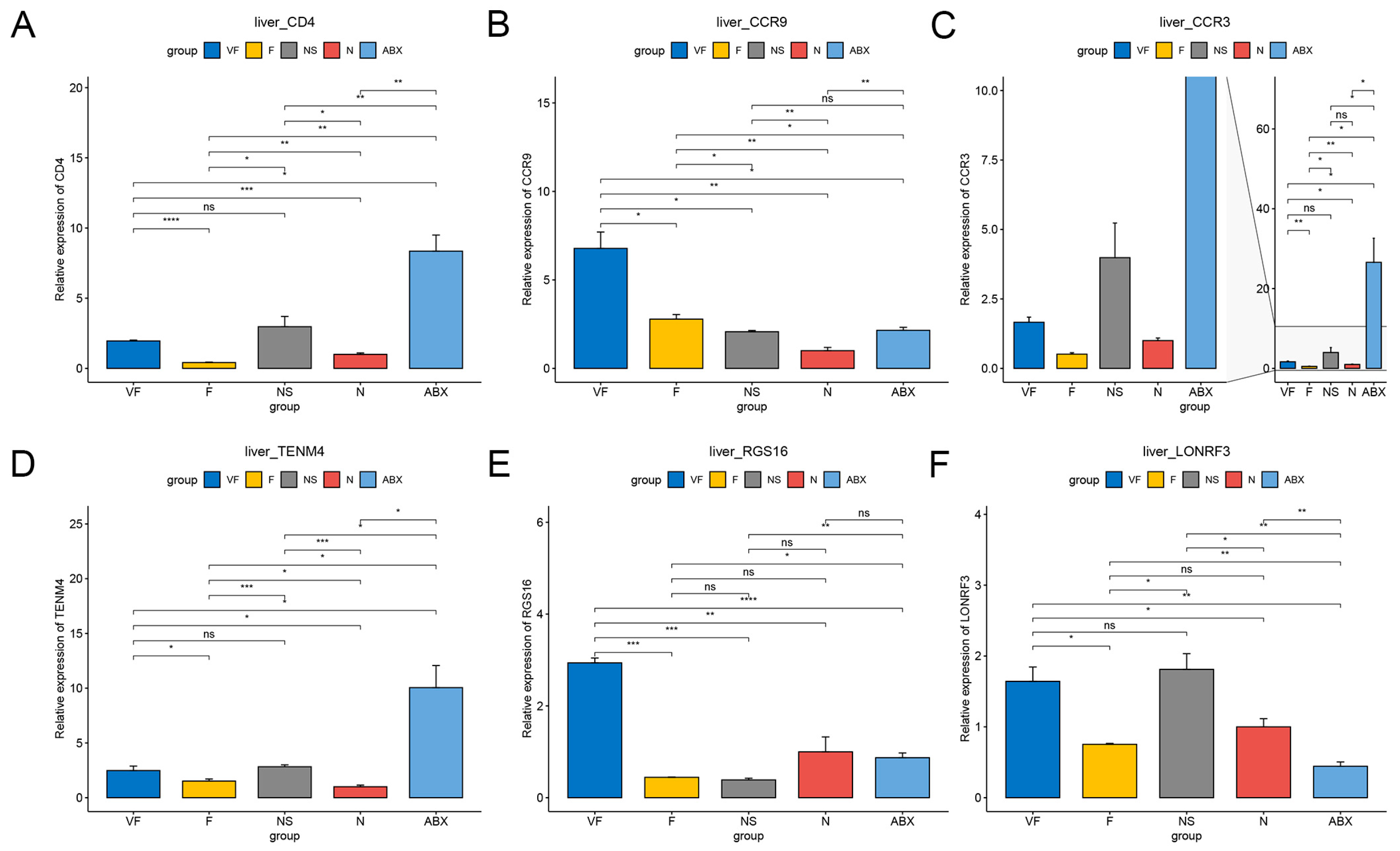
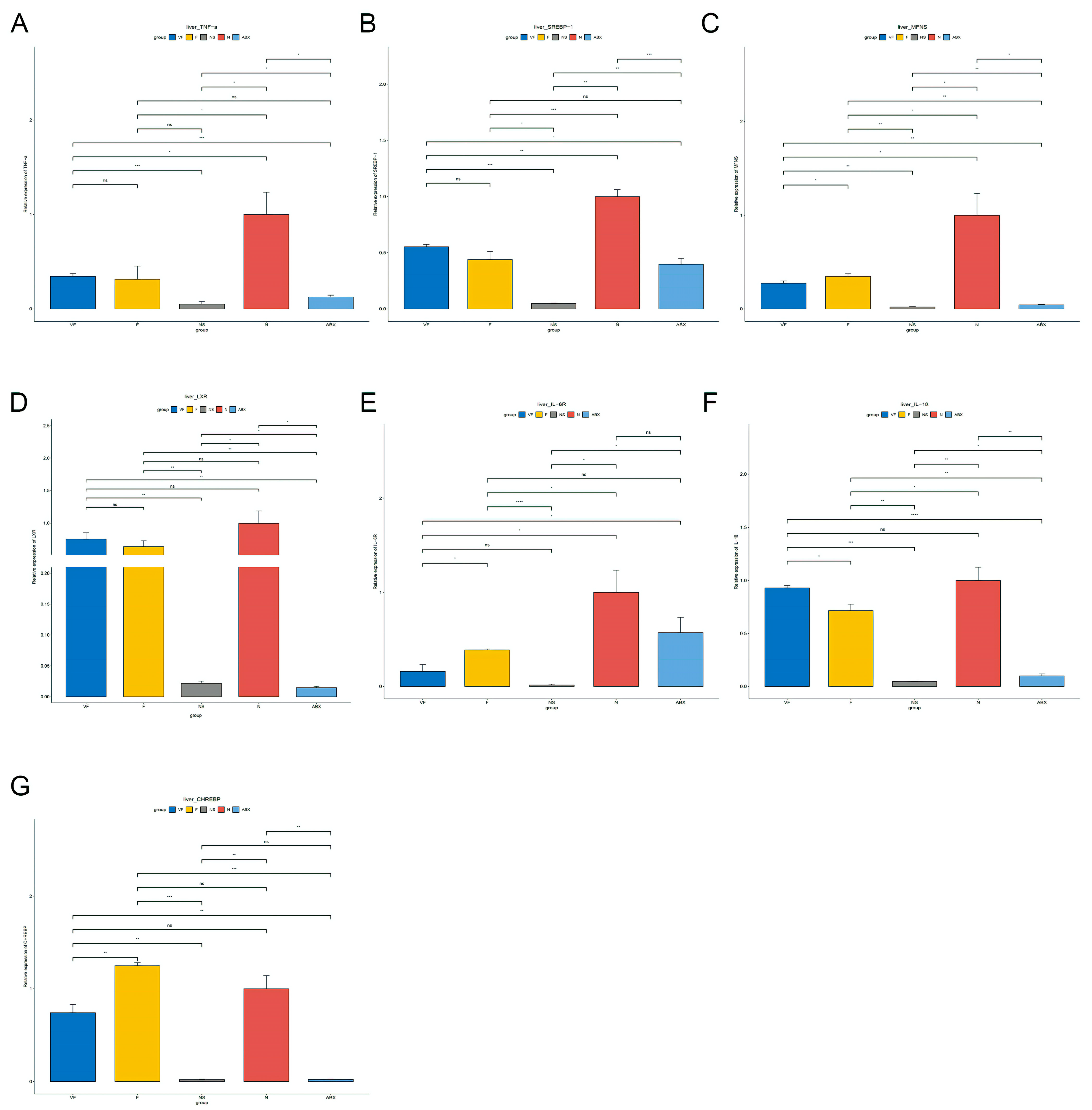
3.4.1. Gut Microbiota Significantly Affect Gene Expression in the Liver
3.4.2. Gut Microbiota Significantly Affect Gene Expression in the Liver
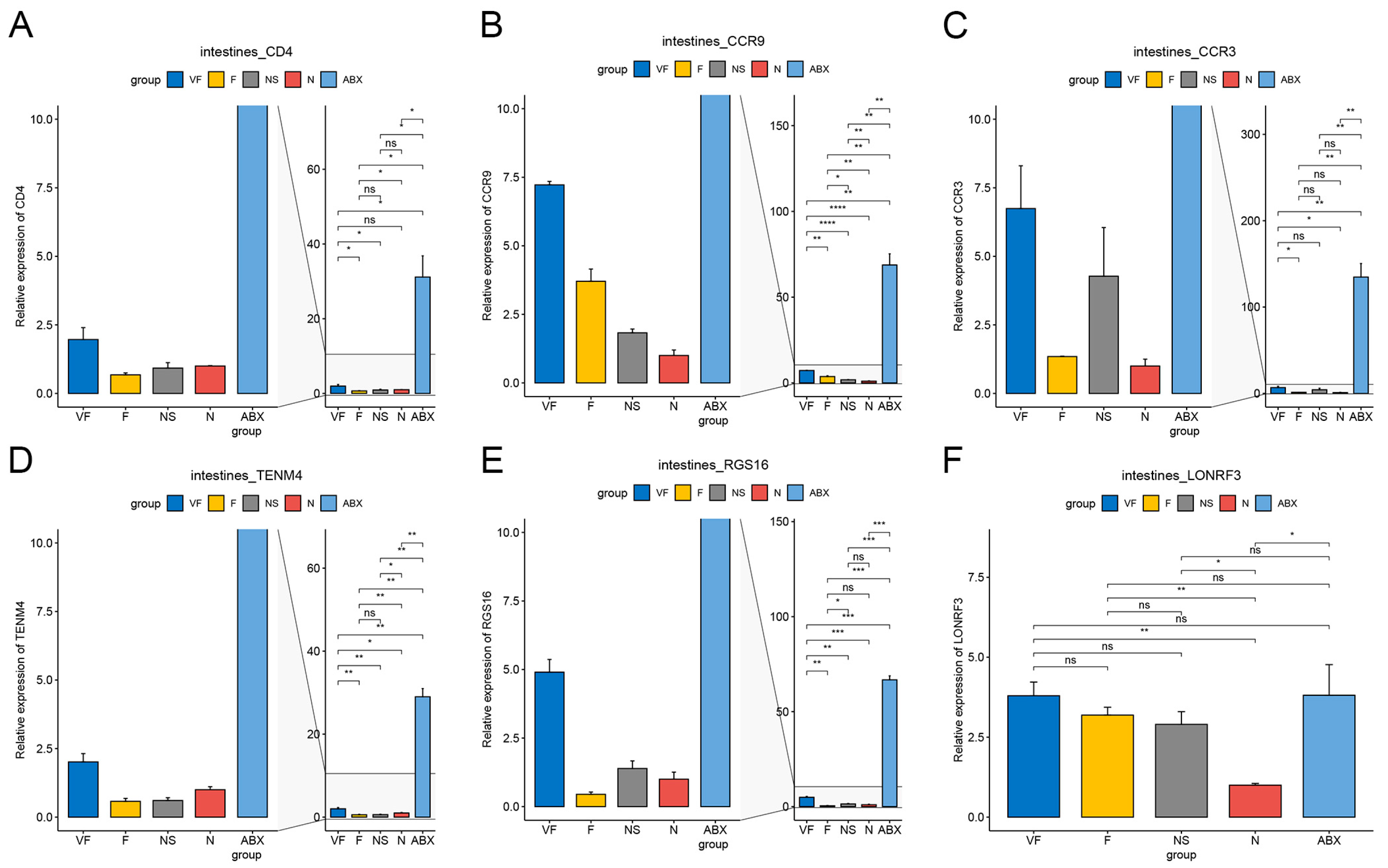
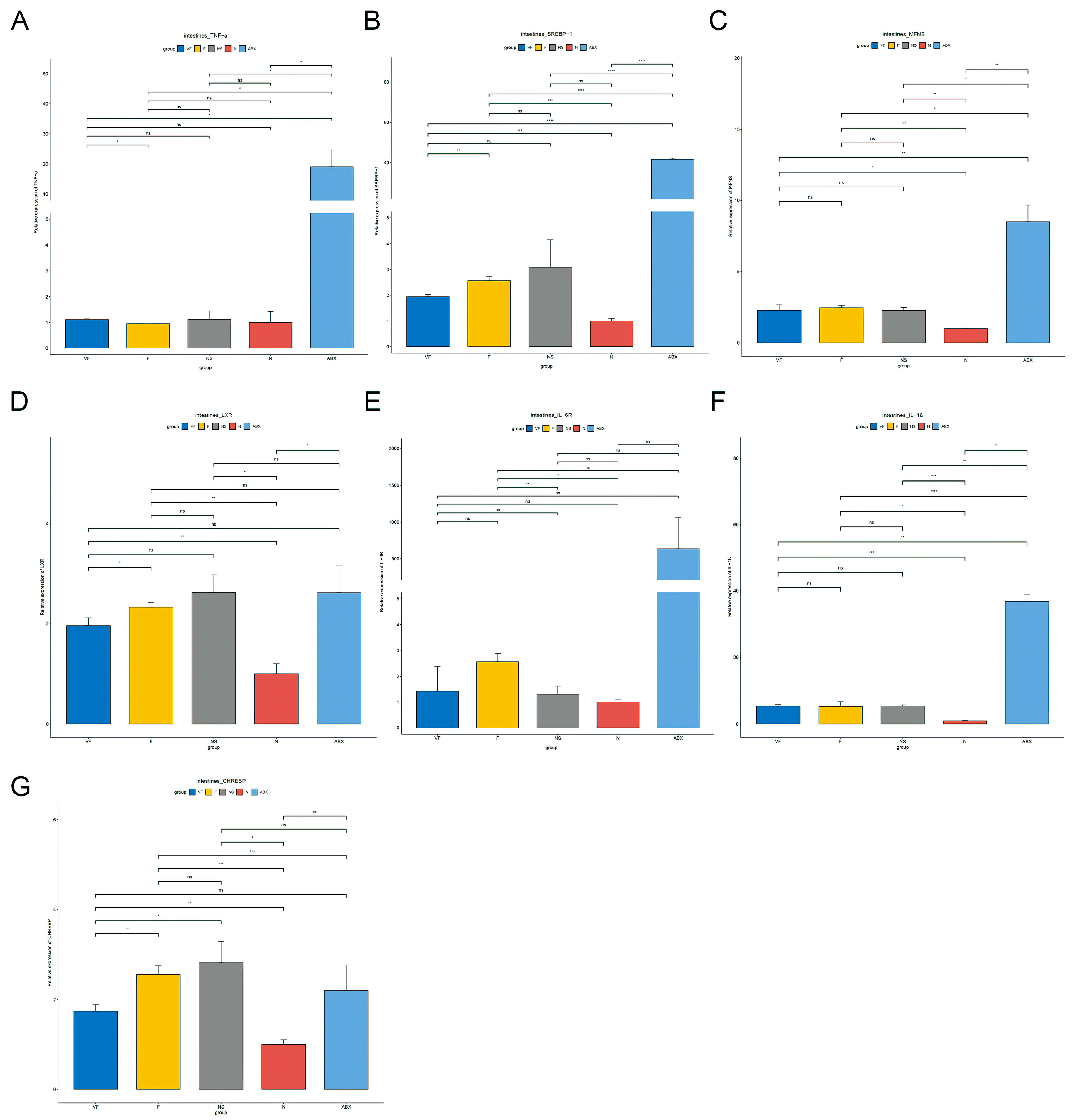
3.4.3. Gut Microbiota also have a Significant Effect on the Expression of Other Genes in the Gut-Liver Axis
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Gueimonde, M.; Sakata, S.; Kalliomäki, M.; Isolauri, E.; Benno, Y.; Salminen, S. Effect of maternal consumption of lactobacillus GG on transfer and establishment of fecal bifidobacterial microbiota in neonates. J. Pediatr. Gastroenterol. Nutr. 2006, 42, 166–170. [Google Scholar] [CrossRef]
- O’Hara, A.M.; Shanahan, F. The gut flora as a forgotten organ. EMBO Rep. 2006, 7, 688–693. [Google Scholar] [CrossRef] [PubMed]
- Song, X.; Wang, L.; Liu, Y.; Zhang, X.; Weng, P.; Liu, L.; Zhang, R.; Wu, Z. The gut microbiota–brain axis: Role of the gut microbial metabolites of dietary food in obesity. Food Res. Int. 2022, 153, 110971. [Google Scholar] [CrossRef] [PubMed]
- Gomes, A.C.; Bueno, A.A.; de Souza, R.G.M.; Mota, J.F. Gut microbiota, probiotics and diabetes. Nutr. J. 2014, 13, 60. [Google Scholar] [CrossRef] [PubMed]
- Tao, X.; Wang, N.; Qin, W. Gut Microbiota and Hepatocellular Carcinoma. Gastrointest. Tumors 2015, 2, 33–40. [Google Scholar] [CrossRef]
- Caesar, R.; Fåk, F.; Bäckhed, F. Effects of gut microbiota on obesity and atherosclerosis via modulation of inflammation and lipid metabolism. J. Intern. Med. 2010, 268, 320–328. [Google Scholar] [CrossRef]
- Buffie, C.G.; Jarchum, I.; Equinda, M.; Lipuma, L.; Gobourne, A.; Viale, A.; Ubeda, C.; Xavier, J.; Pamer, E.G. Profound alterations of intestinal microbiota following a single dose of clindamycin results in sustained susceptibility to clostridium difficile-Induced colitis. Infect. Immun. 2012, 80, 62–73. [Google Scholar] [CrossRef]
- Noverr, M.C.; Huffnagle, G.B. The ‘microflora hypothesis’ of allergic diseases. Clin. Exp. Allergy 2005, 35, 1511–1520. [Google Scholar] [CrossRef]
- Engelbrektson, A.; Korzenik, J.R.; Pittler, A.; Sanders, M.E.; Klaenhammer, T.R.; Leyer, G.; Kitts, C.L. Probiotics to minimize the disruption of faecal microbiota in healthy subjects undergoing antibiotic therapy. J. Med Microbiol. 2009, 58, 663–670. [Google Scholar] [CrossRef]
- Wang, J.-W.; Kuo, C.-H.; Kuo, F.-C.; Wang, Y.-K.; Hsu, W.-H.; Yu, F.-J.; Hu, H.-M.; Hsu, P.-I.; Wang, J.-Y.; Wu, D.-C. Fecal microbiota transplantation: Review and update. J. Formos Med. Assoc. 2019, 118 (Suppl. 1), S23–S31. [Google Scholar] [CrossRef]
- Albesa, I.; Becerra, M.; Battán, P.C.; Páez, P.L. Oxidative stress involved in the antibacterial action of different antibiotics. Biochem. Biophys. Res. Commun. 2004, 317, 605–609. [Google Scholar] [CrossRef] [PubMed]
- Páez, P.L.; Becerra, M.C.; Albesa, I. Comparison of macromolecular oxidation by reactive oxygen species in three bacterial genera exposed to different antibiotics. Cell Biochem. Biophys. 2011, 61, 467–472. [Google Scholar] [CrossRef] [PubMed]
- Didier, A.J.; Stiene, J.; Fang, L.; Watkins, D.; Dworkin, L.D.; Creeden, J.F. Antioxidant and Anti-Tumor Effects of Dietary Vitamins A, C, and E. Antioxidants 2023, 12, 632. [Google Scholar] [CrossRef] [PubMed]
- Romano, K.A.; Martinez-Del Campo, A.; Kasahara, K.; Chittim, C.L.; Vivas, E.I.; Amador-Noguez, D.; Balskus, E.P.; Rey, F.E. Metabolic, Epigenetic, and Transgenerational Effects of Gut Bacterial Choline Consumption. Cell Host. Microbe. 2017, 22, 279–290.e7. [Google Scholar] [CrossRef]
- Wang, X.; Li, Y.; Chen, W.; Shi, H.; Eren, A.M.; Morozov, A.; He, C.; Luo, G.-Z.; Pan, T. Transcriptome-wide reprogramming of N6-methyladenosine modification by the mouse microbiome. Cell Res. 2018, 29, 167–170. [Google Scholar] [CrossRef] [PubMed]
- Kolodziejczyk, A.A.; Zheng, D.; Elinav, E. Diet–microbiota interactions and personalized nutrition. Nat. Rev. Microbiol. 2019, 17, 742–753. [Google Scholar] [CrossRef]
- Koch, B.E.V.; Yang, S.; Lamers, G.; Stougaard, J.; Spaink, H.P. Intestinal microbiome adjusts the innate immune setpoint during colonization through negative regulation of MyD88. Nat. Commun. 2018, 9, 4099, Erratum in Nat. Commun. 2019, 10, 526. [Google Scholar] [CrossRef]
- Ding, J.-H.; Jin, Z.; Yang, X.-X.; Lou, J.; Shan, W.-X.; Hu, Y.-X.; Du, Q.; Liao, Q.-S.; Xie, R.; Xu, J.-Y. Role of gut microbiota via the gut-liver-brain axis in digestive diseases. World J. Gastroenterol. 2020, 26, 6141–6162. [Google Scholar] [CrossRef]
- Arab, J.P.; Martin-Mateos, R.M.; Shah, V.H. Gut–liver axis, cirrhosis and portal hypertension: The chicken and the egg. Hepatol. Int. 2017, 12 (Suppl. 1), 24–33. [Google Scholar] [CrossRef]
- Leclercq, S.; Matamoros, S.; Cani, P.D.; Neyrinck, A.M.; Jamar, F.; Stärkel, P.; Windey, K.; Tremaroli, V.; Bäckhed, F.; Verbeke, K.; et al. Intestinal permeability, gut-bacterial dysbiosis, and behavioral markers of alcohol-dependence severity. Proc. Natl. Acad. Sci. USA 2014, 111, E4485–E4493. [Google Scholar] [CrossRef]
- Lang, S.; Duan, Y.; Liu, J.; Torralba, M.G.; Kuelbs, C.; Ventura-Cots, M.; Abraldes, J.G.; Bosques-Padilla, F.; Verna, E.C.; Brown, R.S., Jr.; et al. Intestinal fungal dysbiosis and systemic immune response to fungi in patients with alcoholic hepatitis. Hepatology 2020, 71, 522–538. [Google Scholar] [CrossRef] [PubMed]
- Mouzaki, M.; Comelli, E.M.; Arendt, B.M.; Bonengel, J.; Fung, S.K.; Fischer, S.E.; McGilvray, I.D.; Allard, J.P. Intestinal microbiota in patients with nonalcoholic fatty liver disease. Hepatology 2013, 58, 120–127. [Google Scholar] [CrossRef]
- Bajaj, J.S.; Heuman, D.M.; Hylemon, P.B.; Sanyal, A.J.; White, M.B.; Monteith, P.; Noble, N.A.; Unser, A.B.; Daita, K.; Fisher, A.R.; et al. Altered profile of human gut microbiome is associated with cirrhosis and its complications. J. Hepatol. 2014, 60, 940–947. [Google Scholar] [CrossRef] [PubMed]
- Dapito, D.H.; Mencin, A.; Gwak, G.-Y.; Pradere, J.-P.; Jang, M.-K.; Mederacke, I.; Caviglia, J.M.; Khiabanian, H.; Adeyemi, A.; Bataller, R.; et al. Promotion of hepatocellular carcinoma by the intestinal microbiota and TLR4. Cancer Cell 2012, 21, 504–516. [Google Scholar] [CrossRef] [PubMed]
- Schnabl, B.; Brenner, D.A. Interactions between the intestinal microbiome and liver diseases. Gastroenterology 2014, 146, 1513–1524. [Google Scholar] [CrossRef]
- Segata, N.; Izard, J.; Waldron, L.; Gevers, D.; Miropolsky, L.; Garrett, W.S.; Huttenhower, C. Metagenomic biomarker discovery and explanation. Genome Biol. 2011, 12, R60. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Gonzalez Peña, A.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Ward, T.; Larson, J.; Meulemans, J.; Hillmann, B.; Lynch, J.; Sidiropoulos, D.; Spear, J.R.; Caporaso, G.; Blekhman, R.; Knight, R.; et al. BugBase predicts organism-level microbiome phenotypes. BioRxiv 2017. [Google Scholar] [CrossRef]
- Lynn, M.A.; Eden, G.; Ryan, F.J.; Bensalem, J.; Wang, X.; Blake, S.J.; Choo, J.M.; Chern, Y.T.; Sribnaia, A.; James, J.; et al. The composition of the gut microbiota following early-life antibiotic exposure affects host health and longevity in later life. Cell Rep. 2021, 36, 109564. [Google Scholar] [CrossRef]
- Blake, S.J.; James, J.; Ryan, F.J.; Caparros-Martin, J.; Eden, G.L.; Tee, Y.C.; Salamon, J.R.; Benson, S.C.; Tumes, D.J.; Sribnaia, A.; et al. The immunotoxicity, but not anti-tumor efficacy, of anti-CD40 and anti-CD137 immunotherapies is dependent on the gut microbiota. Cell Rep. Med. 2021, 2, 100464. [Google Scholar] [CrossRef]
- Scott, F.I.; Horton, D.B.; Mamtani, R.; Haynes, K.; Goldberg, D.S.; Lee, D.Y.; Lewis, J.D. Administration of Antibiotics to Children Before Age 2 Years Increases Risk for Childhood Obesity. Gastroenterology 2016, 151, 120–129.e5. [Google Scholar] [CrossRef] [PubMed]
- Tsakok, T.; McKeever, T.; Yeo, L.; Flohr, C. Does early life exposure to antibiotics increase the risk of eczema? A systematic review. Br. J. Dermatol. 2013, 169, 983–991. [Google Scholar] [CrossRef] [PubMed]
- Marik, P.E. Vitamin C for the treatment of sepsis: The scientific rationale. Pharmacol. Ther. 2018, 189, 63–70. [Google Scholar] [CrossRef]
- Li, Y.; Zafar, S.; Ibrahim, R.M.S.; Chi, H.-L.; Xiao, T.; Xia, W.-J.; Li, H.-B.; Kang, Y.-M. Exercise and food supplement of vitamin C ameliorate hypertension through improvement of gut microflora in the spontaneously hypertensive rats. Life Sci. 2021, 269, 119097. [Google Scholar] [CrossRef] [PubMed]
- Mussa, A.; Idris, R.A.M.; Ahmed, N.; Ahmad, S.; Murtadha, A.H.; Din, T.A.D.A.A.T.; Yean, C.Y.; Rahman, W.F.W.A.; Lazim, N.M.; Uskoković, V.; et al. High-Dose Vitamin C for Cancer Therapy. Pharmaceuticals 2022, 15, 711. [Google Scholar] [CrossRef]
- Moya-Alvarez, V.; Koyembi, J.J.; Kayé, L.M.; Mbecko, J.; Sanke-Waîgana, H.; Djorie, S.G.; Nyasenu, Y.T.; Mad-Bondo, D.; Kongoma, J.; Nakib, S.; et al. Vitamin C levels in a Central-African mother–infant cohort: Does hypovitaminosis C increase the risk of enteric infections? Matern. Child Nutr. 2021, 17, e13215. [Google Scholar] [CrossRef]
- Pham, V.T.; Fehlbaum, S.; Seifert, N.; Richard, N.; Bruins, M.J.; Sybesma, W.; Rehman, A.; Steinert, R.E. Effects of colon-targeted vitamins on the composition and metabolic activity of the human gut microbiome—A pilot study. Gut Microbes 2021, 13, 1–20. [Google Scholar] [CrossRef]
- Sakamoto, N.; Kono, S.; Wakai, K.; Fukuda, Y.; Satomi, M.; Shimoyama, T.; Inaba, Y.; Miyake, Y.; Sasaki, S.; Okamoto, K.; et al. Dietary risk factors for inflammatory bowel disease: A multicenter case-control study in Japan. Inflamm. Bowel Dis. 2005, 11, 154–163. [Google Scholar] [CrossRef]
- Kelly, D.; King, T.; Aminov, R. Importance of microbial colonization of the gut in early life to the development of immunity. Mutat. Res. Mol. Mech. Mutagen. 2007, 622, 58–69. [Google Scholar] [CrossRef]
- Gensollen, T.; Iyer, S.S.; Kasper, D.L.; Blumberg, R.S. How colonization by microbiota in early life shapes the immune system. Science 2016, 352, 539–544. [Google Scholar] [CrossRef]
- Luciani, C.; Hager, F.T.; Cerovic, V.; Lelouard, H. Dendritic cell functions in the inductive and effector sites of intestinal immunity. Mucosal Immunol. 2022, 15, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Bene, K.; Varga, Z.; Petrov, V.O.; Boyko, N.; Rajnavolgyi, E. Gut Microbiota Species Can Provoke both Inflammatory and Tolerogenic Immune Responses in Human Dendritic Cells Mediated by Retinoic Acid Receptor Alpha Ligation. Front. Immunol. 2017, 8, 427. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Shi, Y.; Li, W.; Wang, S.; Zheng, J.; Xu, G.; Li, G.; Shen, X.; Yang, J. Gut microbiota imbalance mediates intestinal barrier damage in high-altitude exposed mice. FEBS J. 2022, 289, 4850–4868. [Google Scholar] [CrossRef] [PubMed]
- Mahnke, Y.D.; Brodie, T.M.; Sallusto, F.; Roederer, M.; Lugli, E. The who’s who of T-cell differentiation: Human memory T-cell subsets. Eur. J. Immunol. 2013, 43, 2797–2809. [Google Scholar] [CrossRef] [PubMed]
- Singer, A.; Bosselut, R. CD4⧸CD8 Coreceptors in thymocyte development, selection, and lineage commitment: Analysis of the CD4⧸CD8 lineage decision. Adv. Immunol. 2004, 83, 91–131. [Google Scholar] [CrossRef]
- Greenhill, C. Intestinal microbiota affects host physiology. Nat. Rev. Endocrinol. 2016, 13, 64. [Google Scholar] [CrossRef]
- Aktar, R.; Parkar, N.; Stentz, R.; Baumard, L.; Parker, A.; Goldson, A.; Brion, A.; Carding, S.; Blackshaw, A.; Peiris, M. Human resident gut microbe Bacteroides thetaiotaomicron regulates colonic neuronal innervation and neurogenic function. Gut Microbes 2020, 11, 1745–1757. [Google Scholar] [CrossRef]
- Xu, L.; Zhang, Q.; Dou, X.; Wang, Y.; Wang, J.; Zhou, Y.; Liu, X.; Li, J. Fecal microbiota transplantation from young donor mice improves ovarian function in aged mice. J. Genet. Genom. 2022, 49, 1042–1052. [Google Scholar] [CrossRef]
- Tian, Y.; Sun, K.-Y.; Meng, T.-Q.; Ye, Z.; Guo, S.-M.; Li, Z.-M.; Xiong, C.-L.; Yin, Y.; Li, H.-G.; Zhou, L.-Q. Gut Microbiota May Not Be Fully Restored in Recovered COVID-19 Patients After 3-Month Recovery. Front. Nutr. 2021, 8, 638825. [Google Scholar] [CrossRef]
- Zuo, T.; Zhang, F.; Lui, G.C.Y.; Yeoh, Y.K.; Li, A.Y.L.; Zhan, H.; Wan, Y.; Chung, A.C.K.; Cheung, C.P.; Chen, N.; et al. Alterations in Gut Microbiota of Patients With COVID-19 During Time of Hospitalization. Gastroenterology 2020, 159, 944–955.e948. [Google Scholar] [CrossRef]
- David, L.A.; Maurice, C.F.; Carmody, R.N.; Gootenberg, D.B.; Button, J.E.; Wolfe, B.E.; Ling, A.V.; Devlin, A.S.; Varma, Y.; Fischbach, M.A.; et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature 2014, 505, 559–563. [Google Scholar] [CrossRef]
- Zheng, X.; Wu, Y.; Bi, J.; Huang, Y.; Cheng, Y.; Li, Y.; Wu, Y.; Cao, G.; Tian, Z. The use of supercytokines, immunocytokines, engager cytokines, and other synthetic cytokines in immunotherapy. Cell. Mol. Immunol. 2022, 19, 192–209. [Google Scholar] [CrossRef]
- Munita, J.M.; Arias, C.A. Mechanisms of Antibiotic Resistance. Microbiol. Spectr. 2016, 4, 464–473. [Google Scholar] [CrossRef]
- McFarland, L. Epidemiology, Risk Factors and Treatments for Antibiotic-Associated Diarrhea. Dig. Dis. 1998, 16, 292–307. [Google Scholar] [CrossRef]
- Bassetti, S.; Tschudin-Sutter, S.; Egli, A.; Osthoff, M. Optimizing antibiotic therapies to reduce the risk of bacterial resistance. Eur. J. Intern. Med. 2022, 99, 7–12. [Google Scholar] [CrossRef]
- Miyauchi, E.; Kim, S.-W.; Suda, W.; Kawasumi, M.; Onawa, S.; Taguchi-Atarashi, N.; Morita, H.; Taylor, T.D.; Hattori, M.; Ohno, H. Gut microorganisms act together to exacerbate inflammation in spinal cords. Nature 2020, 585, 102–106. [Google Scholar] [CrossRef]




Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, P.; Zhang, Y.; Zhang, Z.; Huang, X.; Su, X.; Yang, S.; Xie, Y. Antibiotic-Induced Dysbiosis of the Gut Microbiota Impairs Gene Expression in Gut-Liver Axis of Mice. Genes 2023, 14, 1423. https://doi.org/10.3390/genes14071423
Liu P, Zhang Y, Zhang Z, Huang X, Su X, Yang S, Xie Y. Antibiotic-Induced Dysbiosis of the Gut Microbiota Impairs Gene Expression in Gut-Liver Axis of Mice. Genes. 2023; 14(7):1423. https://doi.org/10.3390/genes14071423
Chicago/Turabian StyleLiu, Pu, Yv Zhang, Zhongyuan Zhang, Xiaorong Huang, Xiaojie Su, Shilong Yang, and Yongfang Xie. 2023. "Antibiotic-Induced Dysbiosis of the Gut Microbiota Impairs Gene Expression in Gut-Liver Axis of Mice" Genes 14, no. 7: 1423. https://doi.org/10.3390/genes14071423
APA StyleLiu, P., Zhang, Y., Zhang, Z., Huang, X., Su, X., Yang, S., & Xie, Y. (2023). Antibiotic-Induced Dysbiosis of the Gut Microbiota Impairs Gene Expression in Gut-Liver Axis of Mice. Genes, 14(7), 1423. https://doi.org/10.3390/genes14071423





